Abstract
Humans are feeling emotions every day, but they can still encounter difficulties understanding them. To better understand emotions, we integrated interdisciplinary knowledge about emotions from various domains such as neurosciences (e.g., neurobiology), physiology, and psychology (affective sciences, positive psychology, cognitive psychology, psychophysiology, neuropsychology, etc.). To organize the knowledge, we employ technologies such as Artificial Intelligence with Knowledge Graphs and Semantic Reasoning. Furthermore, Internet of Things (IoT) technologies can help to acquire physiological data knowledge. The goal of this paper is to aggregate the interdisciplinary knowledge and implement it within the Emotional Knowledge Graph (EmoKG). The Emotional Knowledge Graph is used within our naturopathy recommender system that suggests food to boost emotion (e.g., chocolate contains magnesium that is recommended when we feel depressed). The recommender system also answers a set of competency questions to easily retrieve emotional related-knowledge from EmoKG, such as what are the basic emotions and the more sophisticated ones, what are the neurotransmitters and hormones related to emotions, etc. To follow FAIR principles, EmoKG is mapped to existing knowledge bases found on the BioPortal biomedical ontology catalog such as SNOMEDCT, FMA, RXNORM, MedDRA, and also from emotion ontologies (when available online). We design the LOV4IoT-Emotion ontology catalog that encourages researchers from heterogeneous communities to apply FAIR principles by releasing online their (emotion) ontologies, datasets, rules, etc. The set of ontology codes shared online can be semi-automatically processed; if not available, the scientific publications describing the emotion ontologies are semi-automatically processed with Natural Language Processing (NLP) technologies. This research is also relevant for other use cases such as European projects (ACCRA for emotional robots to reduce the social isolation of aging people, StandICT for standardization, and AI4EU for Artificial Intelligence) and alliances for IoT such as AIOTI. The recommender system can be extended to address other advice such as aromatherapy and take into consideration medical devices to monitor patients’ vital signals related to emotions and mental health.
1. Introduction
What are emotions? Humans are feeling emotions every day, but they can still encounter difficulties understanding them. Emotions have been studied in various disciplines, from psychology to neuroscience. Collecting and integrating this interdisciplinary knowledge is challenging. In this paper, we integrated interdisciplinary knowledge about emotions from various domains (see Figure 1) such as neurosciences (e.g., neurobiology), physiology, and psychology (affective sciences, positive psychology, cognitive psychology, psychophysiology, neuropsychology, etc.). To organize the knowledge, we employ technologies such as Artificial Intelligence with knowledge graphs and semantic reasoning. Furthermore, Internet of Things (IoT) technologies can help to acquire physiological data knowledge. To facilitate the interoperability between sensors, software, and data, standards are also employed. In fact, the integration of heterogeneous technologies, and sensors requires an interoperable solution to describe sensors and data exchange. If the enterprises do not develop the full stack that is compatible from sensors to the final application for consumers as well as compatible with other solutions, they can fail. It demonstrates the need for interoperability between sensors, software, and the data processed. In this paper, we focused on data semantic interoperability to deduce meaningful information, which was applied to the IoT-based emotion domain.
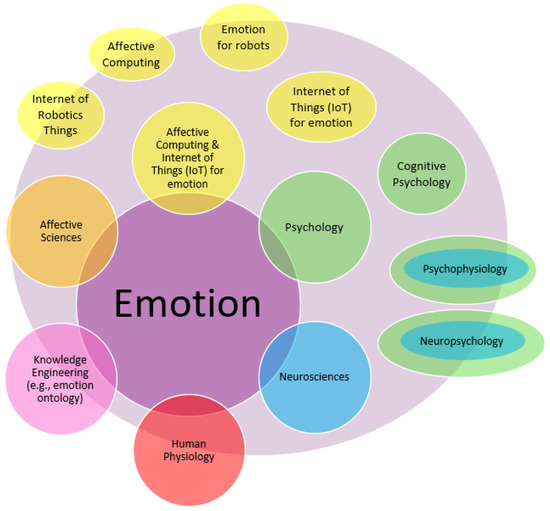
Figure 1.
Overview: Integrating emotional knowledge from heterogeneous communities: Artificial Intelligence (e.g., knowledge engineering), Affective Sciences, Internet of Things (IoT), Psychology, Neurosciences, Physiology, etc.
Ontologies in affective science are disseminated on the web (or even unshared) in different formats and different structures. Ontologies have been developed in AI to facilitate knowledge sharing and reuse. “An ontology is an explicit specification of a conceptualization” according to Gruber et al [1]. Ontology methodologies such as 101 ontology methodology [2] and NeOn [3] encourage reusing existing ontologies. Knowledge Graph (shorturl.at/duz28, accessed on 17 September 2022) [4,5] is the new term, which was popularized by Google. Finding, reusing and adding explicit mapping are time-consuming challenging tasks.
The goal of this paper is to aggregate and make interoperable the interdisciplinary knowledge and implement it within the Emotional Knowledge Graph (EmoKG). To illustrate the use of our Emotion Knowledge Graph, we build the emotion naturopathy recommender system that suggests food to boost emotion (e.g., chocolate contains magnesium that is recommended when we feel depressed). The recommender system also answers a set of competency questions such as what are the basic emotions and the more sophisticated ones, what are the neurotransmitters and hormones related to emotions, etc. The recommender system can be extended to address other advice such as aromatherapy and take into consideration medical devices to monitor patients’ vital signals related to emotions and mental health.
Recommendation Systems (RS) [6,7] are embedded in the services that we are employing every day (e.g., videos with YouTube, products with Amazon, and movies with Netflix). RS system surveys (machine learning-based RS [8]) [9,10,11] do not focus on emotions to enhance our mental health. Nouh et al. [12] introduce the research need for Well-being RS. Garcia et al. [13] survey mental health services using IoT devices. There is still a research gap to design IoT-based emotional RS for mental health. There are several kinds of recommendation systems: (1) content-based (CB), which comes from information retrieval and information filtering; (2) collaborative filtering (CF), which predicts item utility for a specific user based on the items rated by other users in the past; (3) knowledge-based, which we focus on in this paper; and (4) hybrid approaches.
There is a need to design an emotional knowledge graph to be employed within a knowledge-based recommender system to boost mental health. To reuse past expertise and follow FAIR principles [14], the EmoKG emotional knowledge graph is mapped to existing knowledge bases found on the BioPortal biomedical ontology catalog such as SNOMEDCT, FMA, RXNORM, MedDRA, and also from emotion ontologies (when available online). We design the LOV4IoT-Emotion ontology catalog that encourages researchers to apply FAIR principles [15] by releasing online their (emotion) ontologies, datasets, rules, etc.
The set of emotion ontology codes shared online can be semi-automatically processed; if the ontology code is not available yet, the scientific publications describing the emotion ontologies are semi-automatically processed with Natural Language Processing (NLP) techniques to feed the emotion reasoning engine and build the naturopathy recommender system.
We address the following research questions (RQ):
- RQ1: Can we reuse the domain expertise, and what are the shortcomings of past ontology-based emotion IoT projects?
- RQ2: What are the sensors used in the emotion domain? Are there standardized sensor emotion dictionaries? Is there emotion ontology supported by standards?
- RQ3: What are the AI technologies (e.g., rule-based inference engine) to deduce meaningful information from emotion sensor data so developers can design faster IoT-based emotion services?
- RQ4: How to prove the veracity of the reasoning engine?
The key contributions of our research are:
- C1: Ontology-based emotion projects shared though the LOV4IoT-Emotion ontology catalog tool (Table 1); it addresses RQ1 in Section 3.3.
 Table 1. Related work synthesis: Ontology-based emotional projects and reasoning mechanisms employed. Legend: Ontology Availability (OA); when the code is available, the ontologies are classified on the top. Then, the ontology-based projects are classified by year of publications. Several years can appear to highlight that the ontology was maintained from the first publications to the latest publications that we are aware of.
Table 1. Related work synthesis: Ontology-based emotional projects and reasoning mechanisms employed. Legend: Ontology Availability (OA); when the code is available, the ontologies are classified on the top. Then, the ontology-based projects are classified by year of publications. Several years can appear to highlight that the ontology was maintained from the first publications to the latest publications that we are aware of. - C2: The emotion sensor dictionary is aligned with the ETSI SmartM2M SAREF for eHealth Aging Well (SAREFEHAW) standard ontology; it addresses RQ2 in Section 4.2.
- C3: Retrieval of emotional knowledge by the reasoner, used in emotion scenarios; it addresses RQ4 in Section 4.
- C4: Provenance that keeps track of the knowledge designed by domain experts which is explicitly encoded in the ontology catalog and rule datasets to prove the veracity of the reasoning engine; it addresses RQ4 in Section 3.6.
- C5: Standardization-compliancy: Lessons learned from semantic interoperability are disseminated within the ISO/IEC 21823-3 IoT semantic interoperability [16], and the Alliance for the Internet of Things Innovation (AIOTI) Standardization WG (https://aioti.eu/aioti-wg03-reports-on-iot-standards/, accessed on 17 September 2022), which includes the Semantic Interoperability Expert Group [17,18] where the rule-based inference engine is taken as a baseline [19]. SAREF designers are also members of AIOTI Standard WG. AIOTI has other subgroups such as as AIOTI Health WG and AIOTI Urban Living WG. Furthermore, to implement the emotional knowledge graph, we employ semantic web technologies (RDF, RDFS, OWL, SPARQL), which are W3C standards.
Structure of the paper: Related work on emotion ontologies and IoT-based emotional projects are described in Section 2. Interconnecting interdisciplinary emotional knowledge is described in Section 3, which includes the set of competency questions and the ontology catalog for emotions. The emotional recommender system is described in Section 4, which includes the sensor dictionary for emotion, its reasoner and emotion scenarios. Evaluation is included in Section 5. The paper concludes and envisions future work in Section 6.
2. Related Work: Ontology and IoT-Based Emotion Aware Recommender Systems
Existing surveys on emotion ontology-based projects are investigated in Section 2.1. We selected a set of emotion ontology-based projects since the ontology code is shared online in Section 2.2. Well-being recommender systems are introduced in Section 2.3. Shortcomings of the literature study are summarized in Section 2.4. More than 20 ontology-based emotion projects are sumamrized in Table 1, including those not sharing online the ontology code. Additional information about ontology-based emotion projects can be found in Appendix A.
2.1. Existing Surveys on Ontology-Based Emotion Aware Projects
Twenty ontology-based affective state projects which include affective state influences (Abaalkhail et al. [31]) are reviewed. In our paper, we synthesize the work in Table 1, and we focus on the availability on ontologies, reasoning employed within the project, and sensors employed.
2.2. Selected Ontology-Based Emotion Aware Projects Due to Ontology Availability
We collected the ontology-based emotion projects that share online the ontology code and emphasize them in this section.
Emotion Ontology (EMO) (Hastings et al. [22]) describes types of emotion for the affective science domain. EMO is aligned with Basic Formal Ontology (BFO) and the Ontology of Mental Disease (OMD). Mental health and disease ontologies (MFOEM) (Hastings et al. [48]) provides a taxonomy of emotions. Affective science, psychology, and the psychiatric domains are interconnected with the Mental Functioning Ontology (MF) (Larsen, Hastings et al. [23]), which describes concepts such as consciousness, perception, thinking, and believing. Within their state of the art [23]), only few ontologies are mentioned, such as Gene Ontology (GO) and Hastings’s ontology [48]. For this reason, we deeply investigated ontology-based emotion projects (see Table 1).
Patient-facing software tool, based on Visualized Emotion Ontology (VEO) and Emotion and Mood Ontology (Lin et al. [24,25]) describes and references 25 emotions and their visualizations to enhance patient interaction in clinical environments. The Visualized Emotion Ontology is based on the revised Ortony, Clore, and Collins’ models of emotions (OCC model). OntoKeeper is used to evaluate ontology quality.
EEG neuroheadset-based online learning used the Emotion and Cognition ontology to understand emotions (Gil et al. [26]).
Bodily expression-based emotion detection uses the Emotion Ontology (EmOCA) for Context Awareness (Berthelon et al. [28,29]). The ontology references six emotions and focuses on describing philia/phobia and their impact on emotion. Reasoning is achived with the CORESE inference engine, a SPARQL request, and the regression algorithm. This ontology-based motion detection is experimented with six men and four women between 20 and 36 years old.
Emotion annotation of corpora using Web of Data (Linked data) is achieved within the Eurosentiment EU FP7 project, which develops the Onyx ontology (Sanchez-Rada et al. [30]). The Onyx ontology is aligned with the Provenance Ontology, Lexicon Model for Ontologies (LEMON), EmotionML, NLP Interchange Format (NIF), OpenAnnotation, and WordNet-Affect. Differences between Onyx and Human Emotion Ontology (HEO) from Grassi et al. [42] are analyzed. Automatic reasoning, based on SPIN rules, enables: (1) composition of emotions such as Anticipation and Joy result in Optimism, and (2) applying categories based on the dimensions of an emotion.
Emotion-aware applications is based on an ontology describing emotions (Lopez et al. [27]), their detection, and expression systems. The ontology reuses DOLCE (http://www.loa.istc.cnr.it/dolce/overview.html, accessed on 17 September 2022) and FrameNet (https://framenet.icsi.berkeley.edu/fndrupal/, accessed on 17 September 2022) to model descriptions and situations.
Conclusion: Finding and reusing the right emotion ontology for our needs is challenging. There is a need for more tools to help choose the ontology fitting our needs and encouraging ontology best practices with FAIR pinciples.
2.3. Well-Being Recommendation Systems
Well-being recommendation systems are summarized in Table “Well-being and IoT-based emotion applications (positive and negative) related work synthesis” within our past publication [49].
Healthy food smart Recommender System of Hybrid Learning (SRHL) [12] personalizes well-being and prevents diseases. The hybrid RS (content-based and collaborative filtering) employs unsupervised machine learning algorithms and considers time, activity, location, monetary costs, ingredients, health, nutritional value, availability, and the effects of combining the ingredients.
A personalized well-being and health-care support system, called MiningMinds [50], is a rule-based system used in 40 contextual scenarios varying in location, activity, weather, and emotion. MiningMinds is evaluated with forty users (thirty males, ten females in the middle-aged group: 25–49 years, ten different nationalities) and ten domain experts.
The I-Wellness personalized therapy Recommender System [51] is a hybrid Case-Based Reasoning (CBR), integrated within wellness websites, which helps users search for personalized therapy treatment based on their health condition. The I-Wellness online system arranges flexible appointments for patients with a wellness center. I-Wellness comprises six modules: (1) user wellness information, (2) wellness recommendation, (3) package selection, (4) appointment scheduling, (5) point allocation, and (6) wellness monitoring. The prototype is not accessible online to be tested.
A healthy and active lifestyle personalized context-aware RS Android smartphone application, called Motivate [52], recommends twenty types of activities according to location, agenda, weather, profile (e.g., can cycle), and time. The Motivate application is evaluated from 15 November to 25 December 2010 (5 weeks) with six Android phone participants (five male, one female, range: 24–63 years) that are colleagues or friends and worked five days a week. Five participants have a healthy weight body mass index (BMI), and one is slightly overweight.
Conclusion: Those recommender systems use contextual information but do not exploit the emotions of the users. Recommender systems for enhancing people’s emotion and mental health are still lacking.
2.4. Limitations of the Ontology and IoT-Based Emotion Aware Literature Study
We summarize the following shortcomings of the ontology and IoT-based emotion aware state of the art analysis:
- Recommender systems use contextual information but do not exploit the emotions of the users. Recommender systems for enhancing people’s emotion and mental health are still lacking.
- There are emotion-aware recommendation systems for music but not for mental health nor using IoT (Abdul et al. [53]).
- There is no emotion knowledge graph considering neurotransmitters, hormones and relationships to physiological data (e.g., heart rate).
- There is no standard emotion ontology.
- Reviewing the state of the art is a time-consuming task. There is a need to share the literature analysis innovatively (e.g., an emotion knowledge repository supported by tools) to ease the work of other researchers. The ontology-based emotion projects are classified in Table 1.
- Few ontologies can be semi-automatically analyzed, since numerous ontologies are not accessible online. There is a need to disseminate ontology best practices such as FAIR principles [14] for better ontology analysis to semi-automatically extract emotional-based domain knowledge.
- There is a lack of research tools shared as open-source (e.g., web service, web application) that can be easily reused to analyze the strengths and weaknesses of the applications illustrating the research use cases.
- NLP techniques are applied on texts for sentiment analysis (e.g., Twitter, comments on blogs, etc.) rather than emotion ontologies or scientific publications describing ontologies.
- Explicit descriptions of food that boost emotion are missing within ontology-based emotion-aware systems.
- There is no naturopathy recommender system to boost emotion and mental health.
3. Interconnecting Interdisciplinary Emotional Knowledge
Interconnecting interdisciplinary emotional knowledge is described in Section 3.1. Competency questions are introduced in Section 3.2. An ontology-based IoT project catalog for emotion is presented in Section 3.3. Knowledge extraction from emotion ontologies is described in Section 3.4. Mapping to existing knowledge bases such as SNOMEDCT, FMA, RXNORM, MedDRA, LOINC, etc. and Emotion Ontologies is explained in Section 3.5. Provenance of the data is explained in Section 3.6. FAIR principles are introduced in Section 3.7. Finally, lessons learnt from automatic extraction or mapping are summarized in Section 3.8.
3.1. Interconnecting Interdisciplinary Emotional Knowledge
We investigated emotional-related knowledge from various interdisciplinary domains, as shown in Figure 1 and listed below. Note that we cited numerous books (in French); this is due to the availability of resources to investigate them. Moreover, we mainly focus on chapters related to emotion within those books.
- Affective Computing (Picard et al. [54] with a focus on emotion recognition by robots and wearable computers).
- –
- IoT for Emotions (in our past publication [55], see table on Well-being and IoT-based emotion applications (positive and negative)).
- –
- Internet of Robotic Things for Emotions (see our past publication on the Agile Co-Creation of Robots for Ageing (ACCRA) H2020 European project [49]).
- Artificial Intelligence with a focus on Knowledge Engineering (e.g., emotion ontologies) (Ontology Catalog for Emotion in Section 3.3).
- Biology
- –
- Human Physiology (Silverthorn et al. [56]).
- –
- Psychobiology (Breedlove et al. [57]).
- –
- Neurosciences (Purves et al. [58]—chapter on emotions and chapter on neurotransmitters), brain and behavior (Kolb et al. [59]). We focused on neurosciences to understand better neurotransmitters relevant for emotions.
- –
- Neurobiology of emotions (Belzung et al. [60,61]), brain chemistry with hormones (Loretta Graziano Breuning et al. [62] focuses on happy brain with serotonin, dopamine, oxytocin, and endorphin hormones).
- –
- Epigenetics [63]: Impact of emotions on DNA (Zammatteo et al. [64]).
- –
- Endocrinology to describe hormones. Endocrinology chapters can be found in physiology books (Silverthorn et al. [56]).
- Psychology:
- –
- Brain, Chemistry and Psychology relationships (Virol et al. [65]).
- –
- Affective Sciences (Hastings et al. [22] design an emotion ontology, Lisa Feldman Barrett et al. [66,67] focus also on the physiology of emotions).
- –
- Understanding human emotions from facial expressions (Ekman et al. [68]).
- –
- Cognitive Psychology (Lieury et al. [69]).
- –
- Psychophysiology (Morange-Majoux et al. [70]).
- –
- Neuropsychology (Roger Gil et al. [71]—chapter on neuropsychology of emotions).
- –
- Positive Psychology (Seligman et al. [72,73], Lecomte et al. [74], Palazzolo et al. [75]. We also have an interest in positive psychology at work (Arnaud et al. [76], Chief Happiness Officer from Motte et al. [77]).
- –
- Emotion Regulation (Mikolajczak et al. [78]).
- –
- Emotional Intelligence (Goleman et al. [79], Couzon et al. [80]).
3.2. Competency Questions
We defined a set of Competency Questions (CQ) that will be answered within prototypes described in Section 4.1:
- CQ: What are the basic emotions according to Eckman et al. [81]?
- CQ: What are the neurotransmitters relevant for emotions?
- CQ: What are the hormones relevant for emotions?
- CQ: What are the hormones related to stress?
- CQ: What are the hormones related to happiness?
- CQ: What are the physiological parameters/sensors relevant to deduce (basic) emotions?
- CQ: How to deduce meaningful information such as (basic) emotions from physiological data produced by sensors?
- CQ: What are the ontologies describing emotions?
3.3. Ontology-Based IoT Project Catalog for Emotion: LOV4IoT-Emotion
We design the LOV4IoT-Emotion, which is an ontology-based IoT emotion project knowledge base (summarized in Table 1) to release our state of the art analysis as an innovative and maintained tool. We are aware of Systematic Literature Review (SLR) guidelines such as [82]. Other ontology catalogs such as BioPortal [20] and Linked Open Vocabularies (LOV) [21] are not focused on IoT-based emotions applications. LOV4IoT-Emotion is more focused on the ontologies and scientific publications, sensors, and reasoning mechanisms employed as detailed in Table 1 which are not covered by other ontology catalogs. We continuously enrich the LOV4IoT ontology catalog [83] and address more and more keywords such as affective science, well-being, etc. LOV4IoT-Emotion provides a dump of ontology code, web services, and web-based ontology catalog, which were released for the Knowledge Extraction for the Web of Things Challenge. The IoT-based emotion ontologies analysis is also employed within the reasoning discovery explained hereafter. To extract knowledge, manual and semi-automatic analysis are performed [84,85] (see Section 3.4).
3.4. Knowledge Extraction from Emotion Ontologies
We semi-automatically analyzed the ontologies with Natural Language Processing techniques (NLP): (1) ontology code (RDF/XML) and (2) scientific publications describing the ontologies. When the ontologies can be processed, we selected a set of specific keywords (e.g., joy) to compare knowledge provided by ontologies.
From a set of publications on emotion ontology-based projects, we extract knowledge such as which sensors are employed, which reasoning mechanisms are used to interpret sensor measurements, is there an ontology available and reusable, etc. (as depicted in Figure 2).
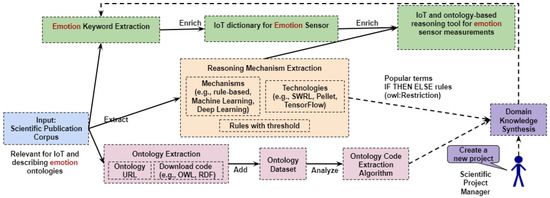
Figure 2.
Knowledge extraction architecture from emotion ontologies.
Ontologies are compared with each other to extract a common pattern to later generate a unified and federated emotional knowledge graph. As an example, we automatically retrieve key phrases (see Figure 3) if they mention emotion-related keywords such as: “fear”, “terror”, “anxiety”, “stress”, “stressed”, “despair”, “crying”, “anger”, “angry”, “irritation”, “joy”, “happy”, “happiness”, “pleasure”, “excited”, “calm”, “tired”, “bored”, “sad”, “disgust”, “love”, “surprise”, “hate”, “jealous”, “emotion”, “apraisal”, “guilt”, “sad”, “valence”, “face”. Since there is no corpus for such tasks, we have semi-manually built the gold dataset, as depicted in Figure 3. Each column corresponds to an ontology, and each row represents an emotion-related term.
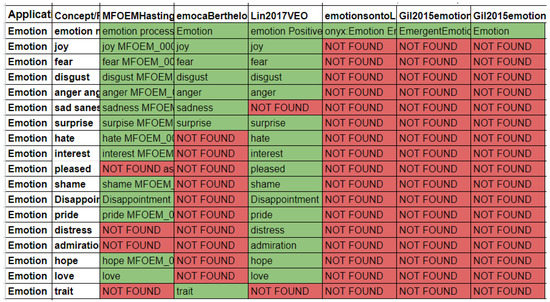
Figure 3.
Emotion gold dataset: knowledge extraction from emotion ontologies that highlight the need to collect and integrate knowledge from complementary emotion ontologies.
3.5. Mapping to Existing Knowledge Bases Such as SNOMED-CT, FMA, RXNORM, MedDRA, LOINC, MESH, GALEN, ChEBI, DBpedia, and Emotion Ontologies
We first describe key concepts related to hormones and neurotransmitters. We search on the Bioportal ontology catalog for key ontologies to be mapped with our Emotion Knowledge Graph. We found ontologies such as SNOMED-CT, Mapping Foundational Model of Anatomy (FMA), RXNORM, MedDRA, Logical Observation Identifier Names and Code (LOINC), Medical Subject Headings (MESH), GALEN, and Chemical Entities of Biological Interest Ontology (ChEBI). The mappings of hormones and neurotransmitters are summarized in Table 2 and Table 3. We also use DBpedia due to its popularity and link emotion-related concepts to existing emotion ontologies when available online. Most of the emotion ontologies cannot be found on BioPortal; only the Hastings’s ontology [22] is referenced on BioPortal.

Table 2.
Subset of mapping hormones and neurotransmitters to existing knowledge bases to demonstrate the difficulty to reuse only one knowledge base. Legend: * means that we use rdfs:seeAlso since the concepts are similar but might not be an equivalent class.

Table 3.
Subset of mapping hormones and neurotransmitters to existing knowledge bases to demonstrate the difficulty to reuse only one knowledge base. Legend: * means that we use rdfs:seeAlso since the concepts are similar but might not be an equivalent class.
3.5.1. Mapping to SNOMED-CT
Systematized Nomenclature of Medicine for Clinical Terms (SNOMED-CT) (https://www.snomed.org/, accessed on 17 September 2022), designed by the College of American Pathologists, is the biggest biomedical ontology with more than 370,000 concepts. SNOMED-CT describes clinical terminology for patient electronic health records such as operations, diseases, devices, symptoms, treatments, and drugs. SNOMED-CT requires the payment of an annual fee to be used by health care organizations. SNOMED-CT is now part of UMLS.
We mapped Hormone, Neurotransmitter, Dopamine, Serotonin, Oxytocin, etc. classes to the ones from SNOMED-CT using
owl:equivalentClass property, since the URL names do not have an explicit name (they are numbers such as &SNOMEDCT;734617007). Adrenaline is mapped to SNOMED-CT via rdfs:seeAlso since the concept is “Epinephrine”.
Noradrenaline is mapped to SNOMED-CT via rdfs:seeAlso since the concept is “noradrenaline hydrochloride” and to “Norepinephrine”.
3.5.2. Mapping Foundational Model of Anatomy (FMA)
Foundational Model of Anatomy (FMA) (http://si.washington.edu/projects/fma, accessed on 17 September 2022) is an ontology-based knowledge base for biomedical informatics which defines the structural organization of the human body.
We mapped Hormone, Neurotransmitter, Dopamine, Serotonin, Oxytocin to the ones from FMA using owl:equivalentClass property, since the URL names do not have an explicit name (they are numbers such as &fma;fma12278). As an example, Glutamate was within FMA. Adrenaline is mapped to FMA via rdfs:seeAlso since the concept is “Epinephrine”. Noradrenaline is mapped to FMA
via rdfs:seeAlso since the concept is “Norepinephrine”.
3.5.3. Mapping to RXNORM
RxNorm (https://www.nlm.nih.gov/research/umls/rxnorm/index.html, accessed on 17 September 2022), part of UMLS, is maintained the by United States National Library of Medicine. It is a medicine terminology that describes clinical drugs. It is used to study drug-to-drug interactions, since a medical doctor cannot have a visibility of side effects on more than three medicines given to a patient. It is also used in personal health records applications.
We mapped Dopamine, Serotonin, Oxytocin, Glutamate to the concepts from RXNORM using the owl:equivalentClass property, since the URL names do not have an explicit name (they are numbers such as &RXNORM;7824). When the same terms are not found, we map the terms using the rdfs:seeAlso property: Cortisol is “cortisol succinate”, Endorphin is “beta-endorphin human”, Insulin is “insulin aspart, human/insulin degludec”, Noradrenaline is “Norepinephrine”.
3.5.4. Mapping to MedDRA
Medical Dictionary for Regulatory Activities (MedDRA) (https://www.meddra.org/, accessed on 17 September 2022) is an internationally used standardized medical terminology that describes medical conditions, medicines and medical devices. MedDRA assists regulators for sharing medical products information used by humans.
We mapped concepts such as Dopamine, Oxytocin, and Glutamate to the concepts from MedDRA using the owl:equivalentClass property, since the URL names do not have an explicit name (they are numbers such as &MEDDRA;10033329). Other concepts are mapped using the rdfs:seeAlso property, since the terms are not exactly the same such as: Serotonin is “Serotonin syndrome”, Dopamine is “Dopamine urine”, Glucocorticoid is “Glucocorticoid decreased”, and Noradrenaline is “Serum noradrenaline increased”.
3.5.5. Mapping to Logical Observation Identifier Names and Codes (LOINC)
Logical Observation Identifier Names and Codes (LOINC) is a common language for identifying health measurements, observations, and clinical documents, but it is not used for diagnosis and diseases.
We mapped Dopamine, Serotonin, and Glutamate to the ones from LOINC using owl:equivalentClass property, since the URL names do not have an explicit name (they are numbers such as &LOINC;MTHU013002). Endorphin is mapped to LOINC via rdfs:seeAlso since the concept is “Beta endorphin”. Glucocorticoid is mapped to LOINC via rdfs:seeAlso since the concept is “Synthetic glucocorticoid drug”. Noradrenaline is mapped to LOINC via rdfs:seeAlso since the concept is “Norepinephrine”.
3.5.6. Mapping to Medical Subject Headings (MESH)
Medical Subject Headings (MeSH) (https://www.nlm.nih.gov/mesh/meshhome.html accessed on 17 September 2022), part of the US NIH National Library of Medicine, is a thesaurus and vocabulary used for indexing, cataloging, and searching of biomedical and health-related information from MEDLINE/PubMed, the NLM Catalog, and other NLM databases.
We mapped concepts such as Dopamine, Serotonin, Oxytocin, Cortisol,
Prolactin, Insulin, Aldosterone, and Testosterone, etc. to the concepts from Medical Subject Headings (MESH) using the owl:equivalentClass property. The MESH ontology does not provide an explicit name within URLs (e.g., &mesh;D000450). Oestrogen is found within MESH as “estrogens”. Other concepts are mapped to MESH using the rdfs:seeAlso property, since we do not find an exact match of the term: Noradrenaline is “noradrenaline sulfate”, Adrenaline is adrenaline sulfate, Endorphin is “endorphin, humoral”, Glutamate is “Glutamic Acid”, Neurotransmitter is “Neurotransmitter Agents”. We did not find the concept Hormone with Mesh.
3.5.7. Mapping to GALEN
Generalised Architecture for Languages, Encyclopaedias, and Nomenclatures in medicine (GALEN) (https://opengalen.org/, accessed on 17 September 2022) was a European Union project (1992–1999) which provides an ontology that describes medical concepts for clinical systems. It became the OpenGALEN foundation, and 2020 marks OpenGALEN’s 30th anniversary. GALEN provides more than 25,000 concepts. GALEN can be used to describe medical procedures.
We mapped Hormone, Neurotransmitter, Serotonin Dopamine, cortisol, etc. to the ceoncepts from GALEN using owl:equivalentClass property. The GALEN ontology provides an explicit name (e.g., galen:Hormone). Oxytocin and glutamate were not found.
3.5.8. Mapping to Chemical Entities of Biological Interest Ontology (ChEBI)
Chemical Entities of Biological Interest (ChEBI) (https://www.ebi.ac.uk/ChEBI/, accessed on 17 September 2022) is a chemical ontology from the Open Biomedical Ontologies effort at the European Bioinformatics Institute.
We mapped concepts such as Neurotransmitter, Dopamine, Serotonin, Oxytocin, Cortisol, Adrenaline, Insulin, Prolactin, Noradrenaline, Aldosterone,
Testosterone, etc. to the concepts from ChEBI using the owl:equivalentClass property. The ChEBI ontology does not provide an explicit name within URLs (e.g., ChEBI:ChEBI_50114). Other concepts do not have the exact same term such as: Oestrogen is estrogen, Endorphin is “beta-endorphin”, and Glutamate is “glutamate(2)”.
3.5.9. Mapping to DBpedia
DBpedia is the semantic Wikipedia. It covers various domains but is less specific to the biomedical domain. DBpedia is not well known in the biomedical domain (not referenced within the Bioportal ontology catalog). However, it is intensively used as it is proven by the Linked Open Data cloud (https://lod-cloud.net/, accessed on 17 September 2022) or the Linked Open Vocabularies ontology catalog (https://lov.linkeddata.es/dataset/lov/vocabs/dbpedia-owl, accessed on 17 September 2022), which shows that DBpedia ontology has been used in at least 18 ontologies, uses 19 external ontologies, and it is used in at least 15 datasets.
Due to its popularity and numerous tools using DBpedia, we decided to map to DBpedia. We mainly used rdfs:seeAlso to map to terms such as Emotion Neurotransmitter. For instance, <rdfs:seeAlso rdf:resource="https://dbpedia.org/page/Emotion"/>, accessed on 17 September 2022). DBpedia uses the term Glutamic_acid for glutamate. Noradrenaline is mapped to Norepinephrine from DBpedia. Oestrogen is mapped to Estrogen from DBpedia.
3.5.10. Mapping to Emotion Ontologies
Our emotional knowledge graph adds explicit links to existing emotion ontologies:
- Emotion Ontology (EmOCA) for Context Awareness (Berthelon et al. [28] (http://ns.inria.fr/emoca/emoca.rdfs, accessed on 17 September 2022)) since it references six basic emotions: joy, fear, disgust, anger, sadness, surprise, and other concepts such as valence and arousal.
- Emotion Ontology (EMO) and mental health and disease ontologies (MFOEM) (Hasting et al. [22]) (https://raw.githubusercontent.com/jannahastings/emotion-ontology/master/ontology/MFOEM.owl, accessed on 17 September 2022).
- Visualized Emotion Ontology (VEO) (Lin et al. [24,25]) (https://bioportal.bioontology.org/ontologies/VEO accessed on 17 September 2022) for concepts such as Emotion, Fear, Surprise, Anger, Disgust, Pride, Interest, Pleased, Joy, Hope, Admiration, Disappointment, Distress, Hate, Shame, etc.
- Onyx Ontology (Lopez, Sanchez-Rada et al. [27,30]) (http://www.gsi.dit.upm.es/ontologies/onyx/1.5/ns accessed on 17 September 2022) for concepts such as Emotion, Appraisal, etc.
- Emotion and Cognition ontology (Gil et al. [26] (http://rhizomik.net/ontologies/2010/11/emotionsonto.owl accessed on 17 September 2022) for concepts such as Emotion, etc. We did not find a taxonomy of emotion in this ontology.
We explicitly added links such as <owl:equivalentClass rdf:resource = “obo;MFOEM_ 000053”/> or <rdfs:seeAlso rdf:resource = “obo;MFOEM_000196”/>
When the ontology code is not available, we enrich our emotional knowledge graph with more knowledge while reading the scientific papers. For instance, the taxonomy of mood states (see Figure A9) is based on Zenonos et al. [86] that references eight mood states that can be recognized: Excited, Happy, Calm, Tired, Bored, Sad, Stressed, Angry. We explicitly write the source of the knowledge within our knowledge graph to keep track of the provenance of data.
3.6. Keeping Track of Provenance Metadata
To keep track of the source of knowledge, we explicitly add this information within the rule RDF dataset and LOV4IoT RDF dataset as shown in Listing 1 by referring to the scientific publication in rdfs:comment and the link to the scientific paper with prov:hadPrimarySource from the W3C PROV-O ontology. For instance, prov:hadPrimarySource keeps track of the source by providing the URL of the scientific publication mentioning the rule or reasoning mechanism.

Listing 1.
Adding provenance metadata using the W3C PROV Ontology.
Listing 1.
Adding provenance metadata using the W3C PROV Ontology.
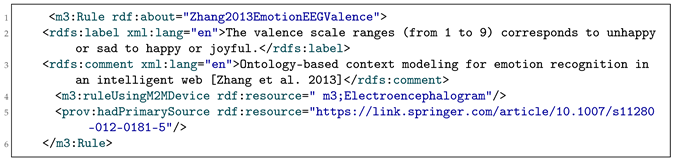 |
To describe the rule dataset itself, we use the Data Catalog Vocabulary (DCAT) as shown in Listing 2.

Listing 2.
Describing the rule dataset using the Data Catalog Vocabulary (DCAT).
Listing 2.
Describing the rule dataset using the Data Catalog Vocabulary (DCAT).
 |
Table 4 reminds of the namespaces used: the first column for the prefix name, the second column for the namespace description, and the third column for the ontology namespace URL.

Table 4.
Ontology Namespace. URLs accessed on 17 September 2022.
3.7. FAIR Principles
Findable, Accesssible, Interoperable, Resuable (FAIR) principles [14] encourage researchers to share their reproducible experiments by publishing online the resources such as ontologies, datasets, rules, etc. The set of ontology code available online can be automatically processed; if the ontology code is not available, the scientific publications describing the emotion ontologies are semi-automatically processed with Natural Language Processing (NLP) techniques to feed the emotion reasoning engine to build the recommender system. As we mentioned earlier, the LOV4IoT-Emotion ontology catalog (Section 3.3) encourages FAIR principles.
3.8. Lessons Learnt from Automatic Extraction or Mapping
We encountered a set of errors while conducting automatic extraction or mapping:
- Dead ontology URL. The ontology URL was mentioned in the scientific paper but is not available anymore. For this reason, it is important to encourage better FAIR principles (e.g., findable resources).
- The ontology cannot be loaded. Errors such as Unable to complete the HTTP request are encountered. There are also issues when dealing with ontology code generated with various ontology editors, libraries, etc. in various formats such as RDF/XML, RDF/Turtle.
- The ontology can be loaded but no terms for automatic extraction can be found. It can happen when there are no label or comments within the ontology or when the ontology URL was automatically built (e.g., MEDDRA;10033329).
- To map the ontologies, we have to deal with synonyms as well.
- Picking the right ontology fitting our need is challenging; numerous ontologies can cover the same terms, but all terms that we need are not covered in only one ontology.
4. Emotional Recommender System: Knowledge Discovery and Reasoning for Emotion
The implementation of competency questions is described in Section 4.1. The sensor dictionary for emotion is defined in Section 4.2. The emotion reasoner is introduced in Section 4.3. Additional emotional use cases are summarized in Section 4.4.
4.1. Implementation: Emotional Knowledge Graph and Prototypes Answering Competency Questions
The set of implemented prototypes answers the competency questions introduced in Section 3.2:
- CQ: What are the basic emotions according to Ekman et al. [81] (see Figure A1). Ekman et al. defines six basic emotions: happiness/enjoyment, sadness, anger, fear, surprise, and disgust.
- CQ: What are the hormones relevant for emotions? (Figure A4). The source of knowledge is extracted for instance from the Brain, Chemistry and Psychology relationships’s book (Virol et al. [65]), Kolb et al. [59], among others).
- CQ: What are the hormones related to happiness? (Figure A6). For instance, serotonin dopamine, oxytocin, and endorphin are described within Breuning et al. [62], among others.
- CQ: What are the hormones related to stress? (Figure A5). The answer is based on Kolb et al. [59], among others.
- CQ: What are the neurotransmitters relevant for emotions? (Figure A3). The answer is based on Kolb et al. [59], Brain, Chemistry and Psychology relationships’s book (Virol et al. [65]), among others).
- CQ: What are the physiological parameters/sensors relevant to deduce (basic) emotions? (Figure A8 and see Section 4.2)
- CQ: What are the reasoning mechanisms to analyze data from physiological parameters/sensors to deduce (basic) emotions? (Figure A7 and Section 4.3)
4.2. Semantic Sensor Emotion Dictionary
We extended our sensor dictionary and applied it to the emotion domain. The sensor dictionary is compliant with the ETSI SmartM2M SAREF standard. The sensor dictionary classifies sensors, measurements, and units for the emotion domain. We also take into consideration synonyms. For each sensor, we keep the provenance of the source of knowledge using it (e.g., past projects referenced within the emotion IoT-based ontology catalog (see Section 3.3 and Table 1, and reasoning employed to infer additional knowledge from emotion sensor data (see the SLOR-Emotion reasoning discovery project in Section 4.3).
The sensor emotion dictionary is provided as a web service: (http://sensormeasurement.appspot.com/emotion/getAllEmotionSensors/?nameClass=EmotionM2MDevice&format=xml, accessed on 17 September 2022) (see Figure 4) or can be hidden within the Graphical User Interface (GUI) (see Section 4.3 and Figure A8).
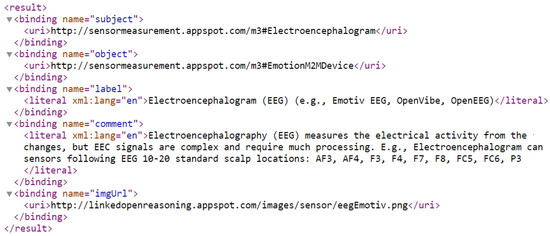
Figure 4.
Web service to retrieve all emotion sensors.
Semantic Annotation: We design an SAREF-compliant sensor dictionary to map common terms. Emotion datasets describe sensor measurement; its value, its unit, and its timestamp are represented in JSON or XML and follow the SenML format (https://tools.ietf.org/html/rfc8428 accessed on 18 September 2022). Semantic annotation requires a rule repository to explicitly describe the context of the measurement. For instance, “t” or “temp” or “temperature” can represent a room temperature or a body temperature and will be annotated following the dictionary which is implemented as an ontology.
4.3. Knowledge Discovery and Reasoning for Emotion with Sensor-Based Linked Open Reasoning (S-LOR Emotion)
The sensor emotion dictionary described in Section 4.2 is used within the emotion rule discovery. We classified emotion sensors within a dictionary (Figure A7), where each sensor is automatically retrieved using SPARQL queries.
The reasoning engine uses data produced by sensors (e.g., heartbeat). The Semantic Annotator API component explicitly annotates the data (e.g., unit of the measurement, context such as body temperature or room temperature) and unifies the data when needed. The semantic annotation uses ontologies that can be found through our LOV4IoT-Emntion ontology catalogs (Section 3.5.10). The ontology chosen must be compliant with a set of rules to infer additional information. The Reasoning Engine API (inspired from [55,87] and supported by the AIOTI group on semantic interoperability [19]) deduces additional knowledge from the data (e.g., abnormal heartbeat) using rule-based reasoning. The IF THEN ELSE rules executed by the reasoning engine will add new data in the data storage. The enriched data can be later used within end-user applications (e.g., call the family or doctor when the heartbeat is abnormal since it might be an emergency, or deducing the fear emotion).
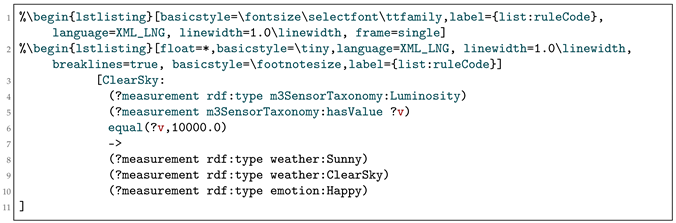
To implement the inference engine, we used the Jena framework; a rule code example is shown in Listing 3. The rule is compliant with our Machine-to-Machine-Measurement (M3) ontology which classifies sensor type, measurement type, units, etc. to ensure an interoperable knowledge-based reasoning. Our web services (Table 5) are employed to later code rules as depicted in Listing 3 for the scenarios according to the applications’ needs where the scenarios provide GUIs.

Table 5.
Specification draft of the S-LOR Knowlege Reasoning API.

Listing 3.
A rule example.
Listing 3.
A rule example.
 |
Various rules can be provided by the Sensor-based Linked Open Rules (S-LOR) tool (Figure A7) which references rules per domain and per sensor. A drop-down list is shown with a set of IoT sub-domains, such as emotion, that we are looking for. Once the domain is selected, the list of sensors relevant for this domain (e.g., skin conductance) is depicted. The developer clicks on the button “Get Project” to retrieve existing projects already using such sensors or the “Get rule” button to retrieve existing rules relevant for this sensor to deduce meaningful information from sensor data.
Demos provide tooltips to provide the veracity of the facts. The facts come, most of the time, from scientific publications referenced within the LOV4IoT catalog and the Sensor-based Linked Open Rules (S-LOR) tool. Such technical AI solutions can be later embedded within the robots to better understand users’ emotions from sensor devices.
A future work is to consider visual [88] and audio data [89] from more complex sensors such as a microphone and camera to detect emotions.
4.4. Emotional Use Cases
We reference in this section how the emotional knowledge graph and related tools are employed: (1) ACCRA H2020 European Project in Section 4.4.1, (2) AIOTI for Health and Urban Living in Section 4.4.2, (3) AI4EU H2020 European Project–Knowledge Extraction for the Web of Things (KE4WoT) Challenge in Section 4.4.3, and (4) other emotion use cases in Section 4.4.4 such as the naturopathy recommender system to boost mental health or the use cases answering the competency question from Section 3.2.
4.4.1. Emotional Robots to Reduce Social Isolation for Ageing People: ACCRA H2020 European Project in Collaboration with Japan
How can Internet of Robotic Things (IoRT) technology and co-creation methodologies help to design emotional-based robotic applications? To respond to this research question, the ACCRA (Agile Co-Creation of Robots for Ageing) project [49] designs advanced social companion robot-based applications to support active and healthy aging, so elderly people can stay independent at home and decrease their social isolation. Applications are designed following co-creation methodologies involving multidisciplinary stakeholders and researchers (e.g., elderly people, medical professionals, and computer scientists such as roboticists or IoT engineers). The ACCRA project provides three robots, Buddy, ASTRO, and RoboHon, used for daily life, mobility, and conversation, to understand and convey emotions in real time using IoT and AI technologies (e.g., knowledge-based reasoning).
4.4.2. AIOTI for Health and Urban Living
AIOTI Health WG and Urban Living WG (https://aiotieu.github.io/urbansociety/catalogue.html accessed on 18 September 2022) are collaborating to release a white paper in September 2022 for the AIOTI Signature Event 2022. AIOTI has been developed in collaboration with the Urban Society, Buildings, Innovation Ecosystems, Energy and Health communities a report that is covering topics of improving healthy urban lifestyle, value-based health care and mitigating the risk and social impact of isolation and loneliness in urban societies.
This report focuses on how IoT can be deployed to foster mental and physical health, thereby putting the weight on disease prevention instead of treatment. Furthermore, cultural and behavioral factors, including the degree of trust in IoT and other technical applications, digital data, governments and the industry itself, are explicitly included and covered by the report. We will present the results of the report and have an open discussion between contributors and experts on how IoT and technology can support to identify and mitigate diseases and increase prevention, thus supporting broader societal goals of benefiting people’s health and well-being.
4.4.3. AI4EU H2020 European Project-Knowledge Extraction for the Web of Things (KE4WoT) Challenge
To encourage research collaborations to automatically extract the knowledge from ontologies and scientific publication describing the ontology, we designed the Knowledge Extraction for the Web of Things (KE4WoT) Challenge supported by the AI4EU project and by the Web Conference Conference (WWW 2018) [90]. The KE4WoT challenge encourages processing the expertise published by domain experts and making the domain knowledge usable, interoperable, and integrated by machines. We released the set of ontologies in various domains (including emotion) as dumps, web services, and tutorials and made them available for the challenge.
Furthermore, the Linked Open Reasoning tool, introduced in Section 4.3, is referenced on the AI4EU platform (https://www.ai4europe.eu/research/ai-catalog/linked-open-reasoning accessed on 18 September 2022).
4.4.4. Other Emotional Use Cases
- Taxonomy of mood states (see Figure A9) (e.g., based on Zenonos et al. [86] references eight mood states: Excited, Happy, Calm, Tired, Bored, Sad, Stressed, Angry).
- IAMHAPPY: an IoT knowledge-based well-being recommendation system to encourage happiness [55].
- Naturopathy Recommender System extended for Emotion: We extended our naturopathy knowledge graph to cover better emotion-related knowledge and to enhance mental health, for instance:
- –
- Food that contains magnesium is recommended for depression (e.g., chocolate) (Figure A10).
- –
- Food that contains Vitamin D is recommended for depression (Figure A11).
- Heartbeat data to deduce fear emotion (Figure A12).
- Skin conductance data to deduce anxiety emotion (Figure A13).
- Luminosity data to deduce weather (cloud cover) (Figure A14).
5. Evaluation: Applying Semantic Web Best Practices to Enhance the Emotional Knowledge Graph
We employed our designed PerfecO methodology [91] that references a set of semantic web best practices and collection of tools providing web services to evaluate ontologies to enhance our emotional knowledge graph: (1) Syntactic Ontology Evaluation in Section 5.1, (2) ontology design evaluation in Section 5.2, (3) LOV4IoT-Emotion Ontology Catalog statistics in Section 5.3, and (4) the emotional knowledge graph evaluated with emotion scenarios in Section 5.4.
5.1. Syntactic Ontology Evaluation
The ontology can be loaded with Jena version 2.11, Protege 4.3 (Figure 5 and Figure 6), and GraphDB 9.10 (Figure 7). We evaluated our emotion ontology with the TripleChecker syntax validator in May 2022. Mistakes found such as “Literal value does not match datatype” (data format and decimal format) have been fixed.
Figure 5.
Subset of the emotion ontology (visualization under the Protege ontology editor).

Figure 6.
Subset of the emotion ontology (visualization under the Protege ontology editor).
Figure 7.
Subset of the emotion ontology (visualization under GraphDB).
We also used the W3C RDF Validator (https://www.w3.org/RDF/Validator/rdfval accessed on 18 September 2022) and OWL Manchester Validator (http://mowl-power.cs.man.ac.uk:8080/validator/validate accessed on 18 September 2022), and no errors have been found.
5.2. Ontology Design Evaluation
We evaluated our emotion ontology with OOPS! OntOlogy Pitfall Scanner! [92], which detects pitfalls. We have identified eleven pitfalls, which are classified as critical, important or minor:
- One critical pitfall: P31 Defining wrong equivalent classes.
- Four important pitfalls: P10 Missing disjointness, P34 Untyped class, P38 No owl ontology declaration, and P30 Equivalent classes not explicitly declared.
- Six minor pitfalls: P04 Creating unconnected ontology elements, P08 Missing annotation, P20 Missing ontology annotations, P22 Using different naming conventions in the ontology, P32 Several classes with the same label, P36 URI contains file extension.
We are currently fixing the pitfalls. To fix P30 Equivalent classes not explicitly declared, we use rdfs:seeAlso instead of owl:equivalentClass.
Regarding P30 Equivalent classes not explicitly declared, we disagree, Hastings et al. distinguish Joy (MFOEM_000034) from Pleasure (MFOEM_000035). For this reason, we kept both as separate concepts.
For the P34 Untyped class pitfall, we need to import the relevant ontologies: Hastings et al. FMA, VOAF, emoca, SNOMEDCT, MedDRA, RXNORM, VEO, etc.
We also added ontology metadata (Figure 8 and Listing 4) to our emotion ontology as recommended by the Linked Open Vocabularies ontology catalog [21].
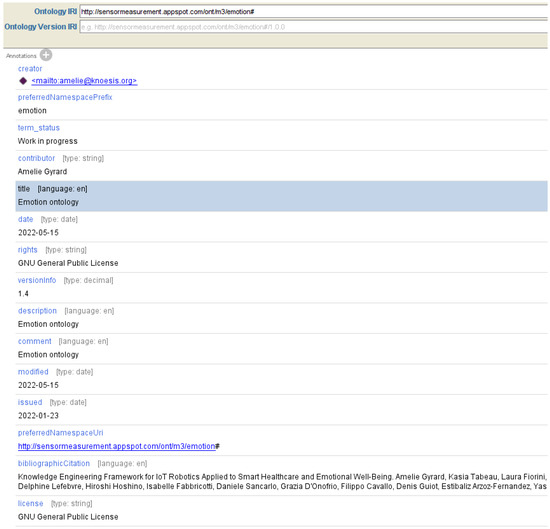
Figure 8.
Ontology metadata of our emotion ontology under the Protege Ontology Editor.

Listing 4.
Ontology Metadata Code Example of the Emotion Ontology.
Listing 4.
Ontology Metadata Code Example of the Emotion Ontology.
 |
To enhance clarity, we provide rdfs:label and rdfs:comment to explicitly describe concepts in natural language and even provide the source of the knowledge using the PROV-O ontology (as explained in Section 3.6).
To ensure deferencable URI, we tested the ontology with Vapour [93] to test the availability of the ontology on servers (e.g., deferencable URI). Deferencing Entity URI (without content negociation) passed (Figure 9). However, we can enhance availability by providing other sources such as HTML, JSON-LD, RDF/XML, and TURTLE. Moreover, http://sensormeasurement.appspot.com/ont/m3/emotion.owl (accessed on 17 September 2022) can be downloaded, but the server configuration must be improved. We are using Google Application Engine to host the application online, with web services running, and obtain the DNS name.
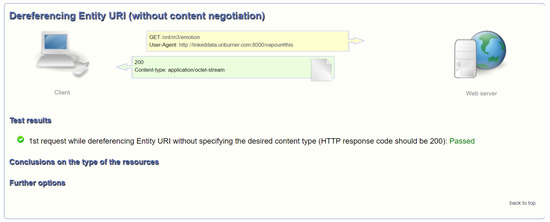
Figure 9.
Deferencable URI evaluated with Vapour.
5.3. LOV4IoT-Emotion Ontology Catalog: Page View Statistics
The LOV4IoT-Emotion web page has been visited 5511 times (3987 as unique views) between January 2018, and March 2022 according to Google Analytics (Figure 10). Visitors return to this dataset which demonstrates its usefulness. To make the GUI more appealing, there is still a need for front-end developers. The results are encouraging to update the dataset with additional domains and ontologies.
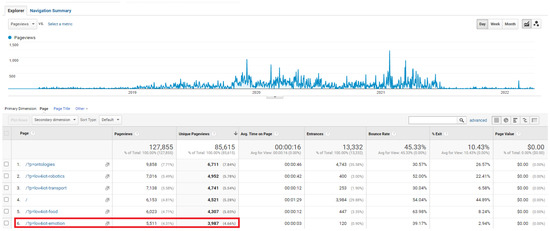
Figure 10.
LOV4IoT Google Analytics per IoT domain (e.g., emotion).
AIOTI IoT Ontology Landscape: LOV4IoT is considered with the AIOTI (The Alliance for the Internet of Things Innovation) IoT ontology landscape survey form (https://ec.europa.eu/eusurvey/runner/OntologyLandscapeTemplate accessed on 17 September 2022) that references and classifies ontologies (https://aioti.eu/wp-content/uploads/2022/02/AIOTI-Ontology-Landscape-Report-R1-Published-1.0.1.pdf accessed on 17 September 2022), which is led by the Standard WG–Semantic Interoperability Expert Group. The group helps industrial practitioners and non-experts find which ontologies are relevant in a certain domain, where to find them, how to choose the most suitable, and ontology maintenance-related questions.
StandICT.eu 2023 Standardized Ontology Landscape: Another similar action to collect standardized ontologies is under development within the StandICT.eu 2023 project. A landscape on standardized ontologies should be released before the end of 2022.
5.4. Emotional Knowledge Graph Evaluated with Emotion Scenarios
A subset of emotional-related scenarios has been introduced in Section 4.4. More and more scenarios are integrated.
Rules are implemented and employed within the emotion scenarios (see Table 6 for demonstrator’s URLs such as sensor discovery, rule discovery, emotion ontology, etc.).

Table 6.
Set of online demonstrators: ontology catalog, rule discovery, and full scenarios. accessed on 15 September 2022.
Technical Evaluation We summarize 16 rules that encourage ontology best practices in Table 7, so researchers will enhance the reusability and better interoperability of their emotion knowledge [91]. Each rule is associated with bad practices and best practice examples to ease the task of beginners in their learning journey (Step-by-step tutorial to improve the ontology quality, dissemination, reuse, etc. Semantic Web Best Practices: https://goo.gl/Rg4cGr, accessed on 18 September 2022) [94]. We also disseminated those best practices to ISO SC 42 Artificial Intelligence within ISO/IEC 5392 Knowledge Engineering Reference Architecture (KERA) and the Ontologies, Knowledge Engineering, and Representation (OKER) Report.

Table 7.
Ontology best practices: check list summary [91]. Legend: * means the level of complexity to achieve the rule. ** is secondary complexity. *** is the most complex one.
6. Conclusions and Future Work
Designing cross-domain emotion applications requires acquiring knowledge from different communities (e.g., affective sciences, affective computing, biology, neurosciences, psychology, physiology, psychophysiology, neuropsychology, etc.). We extended our sensor dictionary for the emotion domain. For each sensor, we discover related knowledge, extract rules, make rules compliant with our sensor dictionary, and integrate rules in a emotion-based scenario. Domain knowledge is also extracted from our LOV4IoT-Emotion IoT-based ontology catalog, referencing more than 49 projects (in March 2022). LOV4IoT reuses the domain expertise from past ontology-based emotion IoT projects. Integrating machine interpretable knowledge implemented within ontologies helps when domain experts are not available. We contributed to standards (e.g., we are editors of the ISO/IEC 21823-3 IoT semantic interoperability), Alliances for IoT such as AIOTI (Standard WG-IoT Semantic Interoperability sub-group, Health WG, and Urban WG), and (European) projects such as AI4EU, ACCRA and StandICT.eu 2023.
Mid-term challenges: There is not yet any emotion ontology that is standardized. The emotion knowledge graph could be disseminated more within standards such as W3C, ISO, NIST, IEEE, etc.
Long-term challenges: Deducing meaningful knowledge from IoT physiological data produced by sensors using more sophisticated AI technologies. Our SAREF-compliant semantic reasoning uses AI techniques such as knowledge and rule-based reasoning, but it could be enhanced addressing more sophisticated scenarios (e.g., dealing with ECG and EEG, which requires Machine Learning and Deep Learning). Encouraging more synergies among standards and communities (e.g., ETSI SmartM2M SAREF, W3C SOSA/SSN, W3C Web of Things, and iot.schema.org ) is partially addressed via the AIOTI Alliance and the StandICT project, and there is a need for more open-source tools, supports, etc.
Author Contributions
Conceptualization, A.G.; methodology, A.G.; software, A.G.; validation, A.G.; writ- 756 ing—original draft preparation, A.G.; Writing—review & editing, A.G. and K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. However, this research is related to the European Union’s 759 Horizon 2020 research and innovation program (ACCRA) under grant agreement No. 738251, StandICT.eu 760 2023 No. 951972 (open call), and AI4EU No. 825619 (challenge open call). The opinions expressed are those 761 of the authors and do not reflect those of the sponsors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
See Table 6 for the set of URLs, such as the emotion ontology, online demonstra- 765 tors (ontology catalog, rule discovery, and full scenarios).
Acknowledgments
We would like to acknowledge the Kno.e.sis research team (lead by Amit Sheth) from Wright State University, Ohio, USA for fruitful discussions about related topics such as “Semantic, Cognitive, and Perceptual Computing” during Gyrard’s post-doc in 2018–2019.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
See Table 6 for the set of URLs, such as the emotion ontology, online demonstrators (ontology catalog, rule discovery, and full scenarios).
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial Intelligence |
| ACCRA | Agile Co-Creation of Robots for Ageing |
| AIOTI | Alliance for the Internet of Things Innovation |
| BioPortal | Biomedical ontology catalog |
| CHEBI | Chemical Entities of Biological Interest Ontology |
| DBpedia | Semantic Wikipedia |
| FMA | Foundational Model of Anatomy |
| IoT | Internet of Things |
| KE4WoT | Knowledge Extraction for the Web of Things |
| KG | Knowledge Graph |
| FMA | Foundational Model of Anatomy |
| GALEN | Generalized Architecture for Languages, Encyclopedias, and Nomenclatures in medicine |
| LOINC | Logical Observation Identifier Names and Codes |
| LOV4IoT-Emotion | Linked Open Vocabularies for Internet of Things for Emotion |
| MedDRA | Medical Dictionary for Regulatory Activities |
| MESH | Medical Subject Headings |
| NLP | Natural Language Processing |
| RS | Recommender System |
| SNOMED-CT | Systematized Nomenclature of Medicine for Clinical Terms |
Appendix A. Implementation: Complementary Demos

Figure A1.
What are the basic emotions according to Ekman et al. [81]?
Figure A2.
LOV4IoT-Emotion Ontology Catalog.

Figure A3.
Example of a subset of the taxonomy of neurotransmitters [65].

Figure A4.
Example of a subset of the taxonomy of hormones [59].

Figure A5.
Example of a subset of the taxonomy of hormones related to stress [59,65].
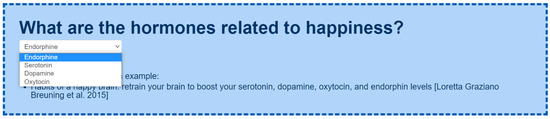
Figure A6.
Example of a subset of the taxonomy of hormones related to happiness [62].
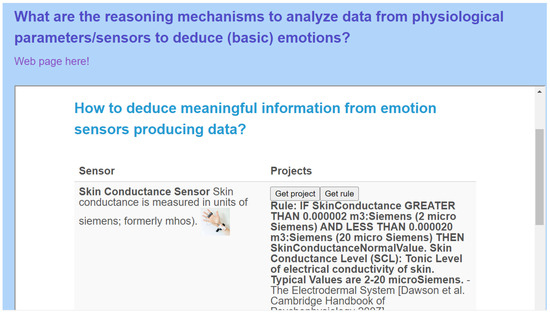
Figure A7.
Reasoning mechanisms relevant to analyze emotion data.

Figure A8.
Subset of the emotion sensor dictionary.
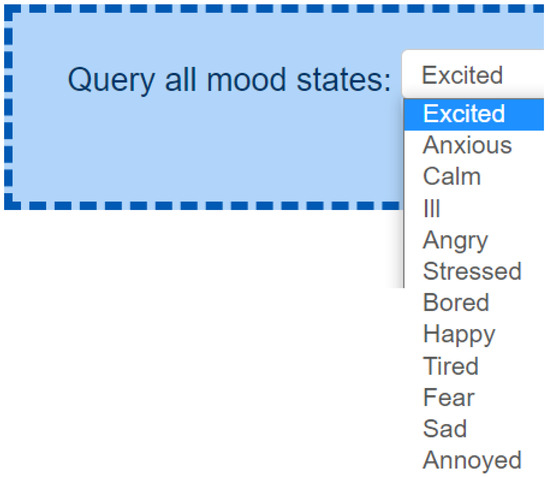
Figure A9.
Emotional State.

Figure A10.
Food that contains magnesium which is recommended for depression, anxiety, etc.
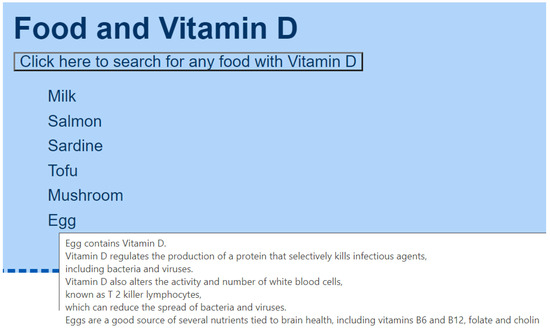
Figure A11.
Food that contains Vitamin D.

Figure A12.
Deducing fear from heartbeat data.

Figure A13.
Deducing anxiety from skin conductance.

Figure A14.
Luminosity and Emotion Scenario.
References
- Gruber, T.R. Toward principles for the design of ontologies used for knowledge sharing? Int. J.-Hum.-Comput. Stud. 1995, 43, 907–928. [Google Scholar] [CrossRef]
- Noy, N.F.; McGuinness, D.L. Ontology Development 101: A Guide to Creating YOUR First Ontology. 2001. Available online: https://www.researchgate.net/publication/243772462_Ontology_Development_101_A_Guide_to_Creating_Your_First_Ontology/link/00b495322175126bd7000000/download (accessed on 17 September 2022).
- Suarez-Figueroa, M.C.; Gomez-Perez, A.; Fernandez-Lopez, M. The NeOn Methodology for Ontology Engineering. In Ontology Engineering in a Networked World; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Paulheim, H. Knowledge graph refinement: A survey of approaches and evaluation methods. Semant. Web 2017, 8, 489–508. [Google Scholar] [CrossRef]
- Bonatti, P.A.; Decker, S.; Polleres, A.; Presutti, V. Knowledge Graphs: New Directions for Knowledge Representation on the Semantic Web (Dagstuhl Seminar 18371); Dagstuhl Reports; Schloss Dagstuhl-Leibniz-Zentrum fuer Informatik: Wadern, Germany, 2019. [Google Scholar]
- Jannach, D.; Zanker, M.; Felfernig, A.; Friedrich, G. Recommender Systems: An Introduction; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Aggarwal, C.C. Recommender Systems; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Portugal, I.; Alencar, P.; Cowan, D. The Use of Machine Learning Algorithms in Recommender Systems: A Systematic Review. Expert Syst. Appl. 2018, 97, 205–227. [Google Scholar] [CrossRef]
- Lu, J.; Wu, D.; Mao, M.; Wang, W.; Zhang, G. Recommender System Application Developments: A Survey. Decis. Support Syst. 2015, 74, 12–32. [Google Scholar] [CrossRef]
- Bobadilla, J.; Ortega, F.; Hernando, A.; Gutiérrez, A. Recommender Systems Survey. Knowl.-Based Syst. 2013, 46, 109–132. [Google Scholar] [CrossRef]
- Adomavicius, G.; Tuzhilin, A. Toward the Next Generation of Recommender Systems: A Survey of the State-of-the-Art and Possible Extensions. IEEE Trans. Knowl. Data Eng. 2005, 17, 734–749. [Google Scholar] [CrossRef]
- Nouh, R.M.; Lee, H.H.; Lee, W.J.; Lee, J.D. A Smart Recommender Based on Hybrid Learning Methods for Personal Well-Being Services. Sensors 2019, 19, 431. [Google Scholar] [CrossRef]
- Garcia-Ceja, E.; Riegler, M.; Nordgreen, T.; Jakobsen, P.; Oedegaard, K.J.; Tørresena, J. Mental Health Monitoring with Multimodal Sensing and Machine Learning: A survey. Pervasive Mob. Comput. 2018, 51, 1–26. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for Scientific DataManagement and Stewardship. Nat. Sci. Data J. 2016, 3, 60018. [Google Scholar]
- Poveda-Villalón, M.; Espinoza-Arias, P.; Garijo, D.; Corcho, O. Coming to Terms with FAIR Ontologies. In Proceedings of the International Conference on Knowledge Engineering and Knowledge Management, Bolzano, Italy, 16–20 September 2020. [Google Scholar]
- ISO/IEC 21823; Semantic Interoperability: Interoperability for Internet of Things Systems Part 3 Semantic Interoperability. ISO: Geneva, Switzerland, 2020.
- Bauer, M.; Baqa, H.; Bilbao, S.; Corchero, A.; Daniele, L.; Esnaola, I.; Fernandez, I.; Franberg, O.; Garcia-Castro, R.; Girod-Genet, M.; et al. Semantic IoT Solutions-A Developer Perspective (Semantic Interoperability White Paper Part I). 2019. Available online: https://www.researchgate.net/publication/336679022 (accessed on 17 September 2022).
- Bauer, M.; Baqa, H.; Bilbao, S.; Corchero, A.; Daniele, L.; Esnaola, I.; Fernandez, I.; Franberg, O.; Garcia-Castro, R.; Girod-Genet, M.; et al. Towards Semantic Interoperability Standards Based on Ontologies (Semantic Interoperability White Paper Part II). 2019. Available online: https://www.researchgate.net/publication/336677616 (accessed on 17 September 2022).
- Murdock, P.; Bassbouss, L.; Bauer, M.; Alaya, M.B.; Bhowmik, R.; Brett, P.; Chakraborty, R.N.; Dadas, M.; Davies, J.; Diab, W.; et al. Semantic Interoperability for the Web of Things (White Paper). 2016. Available online: https://www.researchgate.net/publication/307122744 (accessed on 17 September 2022).
- Noy, N.F.; Shah, N.H.; Whetzel, P.L.; Dai, B.; Dorf, M.; Griffith, N.; Jonquet, C.; Rubin, D.L.; Storey, M.A.; Chute, C.G.; et al. BioPortal: Ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009, 37 (Suppl. 2), W170–W173. [Google Scholar] [CrossRef]
- Vandenbussche, P.Y.; Atemezing, G.A.; Poveda-Villalón, M.; Vatant, B. Linked Open Vocabularies (LOV): A Gateway to Reusable Semantic Vocabularies on the Web. Semant. Web J. 2016, 8, 437–452. [Google Scholar] [CrossRef]
- Hastings, J.; Ceusters, W.; Smith, B.; Mulligan, K. The emotion ontology: Enabling interdisciplinary research in the affective sciences. In Proceedings of the Modeling and Using Context: 7th International and Interdisciplinary Conference, CONTEXT 2011, Karlsruhe, Germany, 26–30 September 2011. [Google Scholar]
- Larsen, R.R.; Hastings, J. From affective science to psychiatric disorder: Ontology as a semantic bridge. Front. Psychiatry 2018, 9, 487. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Amith, M.; Liang, C.; Duan, R.; Chen, Y.; Tao, C. Visualized Emotion Ontology: A Model for Representing Visual Cues of Emotions. BMC Med. Inform. Decis. Mak. 2018, 18, 101–113. [Google Scholar] [CrossRef]
- Lin, R.; Amith, M.T.; Liang, C.; Tao, C. Designing an Ontology for Emotion-Driven Visual Representations. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine, Kansas City, MO, USA, 13–16 November 2017. [Google Scholar]
- Gil, R.; Virgili-Gomá, J.; García, R.; Mason, C. Emotions Ontology for Collaborative Modelling and Learning of Emotional Responses. Comput. Hum. Behav. 2015, 51, 610–617. [Google Scholar] [CrossRef]
- López, J.M.; Gil, R.; García, R.; Cearreta, I.; Garay, N. Towards an Ontology for Describing Emotions. In Proceedings of the First World Summit on the Knowledge Society, WSKS 2008, Athens, Greece, 24–26 September 2008. [Google Scholar]
- Berthelon, F.; Sander, P. Emotion Ontology for Context Awareness. In Proceedings of the 2013 IEEE 4th International Conference on Cognitive Infocommunications (CogInfoCom), Budapest, Hungary, 2–5 December 2013. [Google Scholar]
- Berthelon, F. Modélisation et Détection des émotions à Partir de Données Expressives et Contextuelles. Ph.D. Thesis, Université Nice Sophia Antipolis, Nice, France, 2013. [Google Scholar]
- Sánchez-Rada, J.F.; Iglesias, C.A. Onyx: A Linked Data approach to Emotion Representation. Inf. Process. Manag. 2016, 52, 99–114. [Google Scholar] [CrossRef]
- Abaalkhail, R.; Guthier, B.; Alharthi, R.; El Saddik, A. Survey on Ontologies for Affective States and Their Influences. Semant. Web 2018, 9, 441–458. [Google Scholar] [CrossRef]
- Tabassum, H.; Ahmed, S. Emotion: An Ontology for Emotion Analysis. In Proceedings of the Conference on Emerging Trends and Innovations in Computing and Technology, Karachi, Pakistan, 15–16 December 2016. [Google Scholar]
- Chen, J.; Hu, B.; Moore, P.; Zhang, X.; Ma, X. Electroencephalogram-based emotion assessment system using ontology and data mining techniques. Appl. Soft Comput. 2015, 30, 663–674. [Google Scholar] [CrossRef]
- Arguedas, M.; Xhafa, F.; Daradoumis, T.; Caballe, S. An Ontology about Emotion Awareness and affective Feedback in Elearning. In Proceedings of the International Conference on Intelligent Networking and Collaborative Systems (INCOS), Taipei, Taiwan, 2–4 September 2015. [Google Scholar]
- Tapia, S.A.A.; Gomez, A.H.F.; Corbacho, J.B.; Ratte, S.; Torres-Diaz, J.; Torres-Carrion, P.V.; Garcia, J.M. A Contribution to the Method of Automatic Identification of Human Emotions by using Semantic Structures. In Proceedings of the Conference on Interactive Collaborative Learning, Dubai, United Arab Emirates, 3–6 December 2014. [Google Scholar]
- Sykora, M.D.; Jackson, T.W.; O’Brien, A.; Elayan, S. Emotive Ontology: Extracting Fine-Grained Emotions from Terse, Informal Messages. IADIS Int. J. Comput. Sci. Inf. Syst. 2013, 8, 106–118. [Google Scholar]
- Ptaszynski, M.; Rzepka, R.; Araki, K.; Momouchi, Y. A Robust Ontology of Emotion Objects. In Proceedings of the Annual Meeting of The Association for Natural Language Processing (NLP-2012), Kanazawa, Japan, 22–24 October 2012. [Google Scholar]
- Baldoni, M.; Baroglio, C.; Patti, V.; Rena, P. From Tags to Emotions: Ontology-driven Sentiment Analysis in the Social Semantic Web. Intell. Artif. 2012, 6, 41–54. [Google Scholar] [CrossRef]
- Honold, F.; Schüssel, F.; Panayotova, K.; Weber, M. The Nonverbal Toolkit: Towards a Framework for Automatic Integration of Nonverbal Communication into Virtual Environments. In Proceedings of the Conference on Intelligent Environments, Guanajuato, Mexico, 26–29 June 2012. [Google Scholar]
- Eyharabide, V.; Amandi, A.; Courgeon, M.; Clavel, C.; Zakaria, C.; Martin, J.C. An Ontology for Predicting Students’ Emotions During a Quiz. Comparison with Self-Reported Emotions. In Proceedings of the Workshop on Affective Computational Intelligence (WACI), Paris, France, 14 April 2011. [Google Scholar]
- Francisco, V.; Gervás, P.; Peinado, F. Ontological Reasoning to Configure Emotional Voice Synthesis. In Proceedings of the Conference on Web Reasoning and Rule Systems, Innsbruck, Austria, 7–8 June 2007. [Google Scholar]
- Grassi, M. Developing HEO Human Emotions Ontology. In Proceedings of the European Workshop on Biometrics and Identity Management, Madrid, Spain, 16–18 September 2009. [Google Scholar]
- Radulovic, F.; Milikic, N. Smiley Ontology. In Proceedings of the International Workshop On Social Networks Interoperability, Shanghai, China, 7–9 December 2009. [Google Scholar]
- Yan, J.; Bracewell, D.B.; Ren, F.; Kuroiwa, S. The Creation of a Chinese Emotion Ontology Based on HowNet. Eng. Lett. 2008, 16, EL_16_1_24. [Google Scholar]
- Benta, K.I.; Rarău, A.; Cremene, M. Ontology Based Affective Context Representation. In Proceedings of the Conference on Telematics and Information Systems, Faro, Portugal, 14–17 May 2007. [Google Scholar]
- Rojas, A.; Vexo, F.; Thalmann, D.; Raouzaiou, A.; Karpouzis, K.; Kollias, S. Emotional Body Expression Parameters In Virtual Human Ontology. In Proceedings of the 1st International Workshop on Shapes and Semantics, Athens, Greece, 6–8 December 2006. [Google Scholar]
- Obrenovic, Z.; Garay, N.; López, J.M.; Fajardo, I.; Cearreta, I. An Ontology for Description of Emotional Cues. In Proceedings of the Conference on Affective Computing and Intelligent Interaction (ACII, B-Rank Conference), Beijing, China, 22–24 October 2005. [Google Scholar]
- Hastings, J.; Smith, B.; Ceusters, W.; Jensen, M.; Mulligan, K. Representing Mental Functioning: Ontologies for Mental Health and Disease. In Proceedings of the International Conference on Biomedical Ontology (ICBO), Graz, Austria, 21–25 July 2012. [Google Scholar]
- Gyrard, A.; Tabeau, K.; Fiorini, L.; Kung, A.; Senges, E.; De Mul, M.; Giuliani, F.; Lefebvre, D.; Hoshino, H.; Fabbricotti, I.; et al. Knowledge Engineering Framework for IoT Robotics Applied to Smart Healthcare and Emotional Well-Being. Int. J. Soc. Robot. 2021, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.; Ali, S.I.; Ali, R.; Hussain, M.; Ali, T.; Khan, W.A.; Amin, B.A.; Kang, B.H.; Lee, S. Personalization of Wellness Recommendations using Contextual Interpretation. Expert Syst. Appl. 2018, 96, 506–521. [Google Scholar] [CrossRef]
- Lim, T.P.; Husain, W.; Zakaria, N. Recommender System for Personalized Wellness Therapy. Int. J. Adv. Comput. Sci. Appl. 2013, 4, 54–60. [Google Scholar]
- Lin, Y.; Jessurun, J.; De Vries, B.; Timmermans, H. Motivate: Towards Context-Aware Recommendation Mobile System for Healthy Living. In Proceedings of the Conference on Pervasive Computing Technologies for Healthcare (PervasiveHealth) and Workshops, Dublin, Ireland, 23–26 May 2011. [Google Scholar]
- Abdul, A.; Chen, J.; Liao, H.Y.; Chang, S.H. An emotion-aware personalized music recommendation system using a convolutional neural networks approach. Appl. Sci. 2018, 8, 1103. [Google Scholar] [CrossRef]
- Picard, R.W. Affective Computing; MIT Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Gyrard, A.; Sheth, A. IAMHAPPY: Towards An IoT Knowledge-Based Cross-Domain Well-Being Recommendation System for Everyday Happiness. In Proceedings of the IEEE/ACM Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE) Conference, Crystal City, VA, USA, 16–18 December 2020. [Google Scholar]
- Silverthorn, D.U. Physiologie Humaine: Une Approche Intégrée; Pearson Education: Paris, France, 2007. [Google Scholar]
- Rosenzweig, M.R.; Leiman, A.L.; Breedlove, S.M. Psychobiologie. Rev. Philos. Fr. 2001, 191, 131. [Google Scholar]
- Purves, D.; Augustine, G.J.; Fitzpatrick, D.; Hall, W.; LaMantia, A.S.; White, L. Neurosciences; De Boeck Supérieur: Louvain-la-Neuve, Belgique, 2019. [Google Scholar]
- Kolb, B.; Whishaw, I.Q.; Teskey, G.C. Cerveau et Comportement; De Boeck Supérieur: Louvain-la-Neuve, Belgique, 2019. [Google Scholar]
- Belzung, C. Biologie des émotions; De Boeck: Louvain-la-Neuve, Belgique, 2007. [Google Scholar]
- Belzung, C. Neurobiologie des émotions; UPPR Editions: Paris, France, 2017. [Google Scholar]
- Breuning, L.G. Habits of a Happy Brain: Retrain Your Brain to Boost Your Serotonin, Dopamine, Oxytocin, & Endorphin Levels; Simon and Schuster: New York, NY, USA, 2015. [Google Scholar]
- de Rosnay, J.; Gouyon, P.H.; Junien, C.; Khayat, D.; Ornish, D. La Révolution Épigénétique: Votre Mode de vie Compte plus que Votre Hérédité; Albin Michel: Paris, France, 2018. [Google Scholar]
- L’impact des Émotions sur l’ADN. Available online: https://www.amazon.fr/Limpact-%C3%A9motions-lADN-Nathalie-Zammatteo/dp/2358051292 (accessed on 17 September 2022).
- Virol, F. Cerveau, Chimie et Psychologie: Neurophysiologie; GRANCHER. 2015. Available online: https://www.amazon.fr/Cerveau-Chimie-Psychologie-Neurophysiologie-psychologie/dp/2733913352 (accessed on 17 September 2022).
- Barrett, L.F. How Emotions Are Made: The Secret Life of the Brain; Houghton Mifflin Harcourt: Boston, MA, USA, 2017. [Google Scholar]
- Barrett, L.F.; Lewis, M.; Haviland-Jones, J.M. Handbook of Emotions; Guilford Publications: New York, NY, USA, 2016. [Google Scholar]
- Paul, E. Emotions Revealed: Recognizing Faces and Feelings to Improve Communication and Emotional Life; OWL Books: New York, NY, USA, 2007. [Google Scholar]
- Lieury, A. Manuel Visuel de Psychologie Cognitive, 4th ed.; Dunod: Paris, France, 2020. [Google Scholar]
- Morange-Majoux, F. Manuel Visuel de Psychophysiologie, 2nd ed.; Dunod: Paris, France, 2017. [Google Scholar]
- Gil, R. Neuropsychologie, 7th ed.; Elsevier Masson: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Seligman, M.E.; Lecomte, J. Vivre la Psychologie Positive: Comment être Heureux au Quotidien par le Fondateur de la Psychologie Positive; InterEditions: Paris, France, 2011. [Google Scholar]
- Seligman, M. Flourish: A Visionary New Understanding of Happiness and Well-Being (Book); Simon and Schuster: New York, NY, USA, 2012. [Google Scholar]
- Lecomte, J. Introduction à la Psychologie Positive; Dunod: Paris, France, 2014. [Google Scholar]
- Palazzolo, J. La Psychologie Positive:«Que sais-je?» n° 4191 Que sais-je. 2020. Available online: https://www.amazon.fr/psychologie-positive-Que-sais-je-4191-ebook/dp/B08J48N446 (accessed on 17 September 2022).
- Arnaud, B.; Mellet, É. La Boîte à Outils de la Psychologie Positive au Travail; Dunod: Paris, France, 2019. [Google Scholar]
- Motte, A.; Larabi, S.; Boutet, S. La boîte à outils du Chief Happiness Officer; Dunod: Paris, France, 2018. [Google Scholar]
- Mikolajczak, M.; Desseilles, M. Traité de régulation des émotions; De Boeck Supérieur: Louvain-la-Neuve, Belgique, 2012. [Google Scholar]
- Goleman, D. Emotional Intelligence: Why It Can Matter More Than IQ; Bantam: New York, NY, USA, 2012. [Google Scholar]
- Couzon, E.; Dorn, F. Les émotions: Développer son Intelligence Émotionnelle; ESF. 2007. Available online: https://www.amazon.fr/%C3%A9motions-D%C3%A9velopper-son-intelligence-%C3%A9motionnelle/dp/2710122979 (accessed on 17 September 2022).
- Ekman, P. Emotions revealed. BMJ 2004, 328, 0405184. [Google Scholar] [CrossRef]
- Kitchenham, B.; Pretorius, R.; Budgen, D.; Brereton, O.P.; Turner, M.; Niazi, M.; Linkman, S. Systematic literature reviews in software engineering—A tertiary study. Inf. Softw. Technol. 2010, 52, 792–805. [Google Scholar] [CrossRef]
- Gyrard, A.; Bonnet, C.; Boudaoud, K.; Serrano, M. LOV4IoT: A Second Life for Ontology-based Domain Knowledge to build Semantic Web of Things Applications. In Proceedings of the IEEE International Conference on Future Internet of Things and Cloud, Vienna, Austria, 22–24 August 2016. [Google Scholar]
- Noura, M.; Gyrard, A.; Heil, S.; Gaedke, M. Automatic Knowledge Extraction to build Semantic Web of Things Applications. IEEE Internet Things J. 2019, 6, 8447–8454. [Google Scholar] [CrossRef]
- Noura, M.; Gyrard, A.; Heil, S.; Gaedke, M. Concept Extraction from the Web of Things Knowledge Bases. In International Conference WWW/Internet; Outstanding Paper Award; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Zenonos, A.; Khan, A.; Kalogridis, G.; Vatsikas, S.; Lewis, T.; Sooriyabandara, M. HealthyOffice: Mood recognition at work using smartphones and wearable sensors. In Proceedings of the IEEE International Conference on Pervasive Computing and Communication Workshops (PerCom Workshops), Sydney, Australia, 14–18 March 2016. [Google Scholar]
- Gyrard, A.; Serrano, M.; Datta, S.; Jares, J.; Intizar, A. Sensor-based Linked Open Rules (S-LOR): An Automated Rule Discovery Approach for IoT Applications and its use in Smart Cities. In Proceedings of the 3rd International ACM Smart City Workshop (AW4city) in conjunction with International World Wide Web Conference (WWW), Perth, Australia, 3–7 April 2017. [Google Scholar]
- Wu, C.H.; Lin, J.C.; Wei, W.L. Survey on audiovisual emotion recognition: Databases, features, and data fusion strategies. APSIPA Trans. Signal Inf. Process. 2014, 3, e12. [Google Scholar] [CrossRef]
- Atmaja, B.T.; Sasou, A.; Akagi, M. Survey on bimodal speech emotion recognition from acoustic and linguistic information fusion. Speech Commun. 2022, 140, 11–28. [Google Scholar] [CrossRef]
- Gyrard, A.; Gaur, M.; Padhee, S.; Sheth, A.; Juganaru-Mathieu, M. Knowledge Extraction for the Web of Things (KE4WoT): WWW 2018 Challenge Summary. In Companion Proceedings of the The Web Conference; ACM: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Gyrard, A.; Atemezing, G.; Serrano, M. PerfectO: An Online Toolkit for Improving Quality, Accessibility, and Classification of Domain-based Ontologies; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Poveda-Villalón, M.; Gómez-Pérez, A.; Suárez-Figueroa, M.C. Oops!(ontology pitfall scanner!): An on-line tool for ontology evaluation. Int. J. Semant. Web Inf. Syst. 2014, 10, 7–34. [Google Scholar] [CrossRef]
- Berrueta, D.; Fernández, S.; Frade, I. Cooking HTTP content negotiation with Vapour. In Proceedings of the Workshop on Scripting for the Semantic Web (SFSW), Tenerife, Canary Islands, 2 June 2008. [Google Scholar]
- Gyrard, A.; Serrano, M.; Atemezing, G. Semantic Web Methodologies, Best Practices and Ontology Engineering Applied to Internet of Things. In Proceedings of the IEEE World Forum on Internet of Things, Milan, Italy, 14–16 December 2015. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).