Abstract
Medicinal plants extracts are a rich natural source of bioactive phytochemicals (mainly polyphenols). This study aims at determining the total polyphenols content (TPC) of nine medicinal plants extracted using the UV-visible (UV-Vis) spectroscopic method, along with the Orange Data Mining Tool (ODMT). The TPC for the selected medicinal plant extracts (i.e., Daucus carota L. root, Ruta Chalepensis L. Leaves, Anisosciadium DC. Leaves, Thymus vulgaris L. Leaves, Senna alexandrina leaves, Myrtus communis L. leaves, Silybum Marianum L. Flower, Silybum marianum L. Leaves, and Rosa moschata Flower) was measured using gallic acid (GA) as a standard. The intended method requires a maximum of 1 mg of GA and only 1 mg of the plant extract. The wavelength range of the maximum absorption in the UV-vis spectrum was about 270 nm. For polyphenols, the purposed method linear dynamic concertation range (44.67 to 334.7 mg GA equivalent (GAE)/g dry weight (DW)) with a recovery percentage range of 95.3% to 104.3%, and the good regression value, was found to be R2 = 0.999. This method was easy, fast, accurate, and less expensive than the conventional Folin–Ciocalteu method.
1. Introduction
Phytochemicals originate from the Greek word (Phyto) meaning plant. They are biologically active chemical compounds that are naturally present in plant fruits, vegetables, whole grains, nuts, seeds, legumes, and other parts. They protect plant cells from pollution, dehydration, exposure to ultraviolet rays, and toxic substances as well as diseases, insects, and exposure to the ultraviolet rays [1,2]. They are responsible for the color, smell, and flavor of every plant. They also play a great role in protecting human health from many diseases; they provide the human body with more energy than micronutrients in industrial supplements [3]. Polyphenols are the main components of plants. They can be divided into several classes, including flavonoids, phenolic acids, tannins, stilbenes, and lignins. They can have various simple and complex structures such as simple phenolic acids (e.g., vanillin, GA, caffeic acid), polyphenols such as stilbenes, flavonoids, and polymers derived from these different groups [4]. Phenolic phytochemicals are an essential component of our food as they are responsible for the color and taste of fruits and vegetables. In particular, as polyphenols precipitate salivary proteins, this property could somewhat participate in the defense against their anti-nutritional effects [5].
UV-Vis spectrophotometric methods assessing TPC in plant extracts are cheaper, faster, and thus more accessible methods than analytical chromatography techniques, such as high-performance liquid chromatography (HPLC) [6]. Moreover, spectrophotometric assays identify compound categories rather than individual compounds. Among different spectrophotometric techniques, UV-Vis spectroscopy appears to be suitable for the quantification of phenolic contents in the plant extract [7]. Phenolic compounds contain π-conjugated systems with hydroxyl-phenolic groups. They can strongly absorb UV light where π type molecular orbitals electronic transitions of phenolic groups provide the UV-vis spectrum. UV-Vis spectroscopy for phenolic analysis is reported to quantify anthocyanins [8], phenolic acids [9], stilbenes [10], flavanols [11], and tannins [12]. However, the main limitations of UV-Vis spectroscopy are overestimating the specificity of the assays and assigning results to specific phenolic compounds instead of compound categories.
TPC is widely accepted as a key measure of quality for plants, and can be commonly assessed by the Folin–Ciocalteu method [13] or by ODMT method [14,15,16]. The Folin–Ciocalteu method is broadly used to decide complete polyphenols. This reaction happens by the phosphotungstic corrosive reduction, shaping a blue chromophore comprised of a phosphotungstic-phosphomolybdenum complex [17,18]. The maximum absorption of chromophores mainly depends on the alkaline solution and the concentration of phenolic derivatives [18]. However, this reagent decomposes quickly and easily in alkaline solutions, so a large surplus of reagents must be used to obtain a complete reaction but it is worth noting that a reverse reaction can occur because this excess can result in precipitates and high turbidity, making spectrophotometric analysis impossible [19]. Thus, many modifications of this method are found in many pharmacopeias and laboratory procedures, including the amount of the Folin–Ciocalteu reagents, concentration, wavelength, standard used, reaction time, and temperature [20].
Recently, an attempt has been made based on the application of data prediction techniques for quality modeling of various products [21,22,23]. Due to the small amount of the natural extract of the plants and depletion of reagents used in the analysis of the phenolic compounds, the development of a new method consuming fewer amounts of samples and reagents remains of a great importance. In this study, the TPC of plant extracts using UV-Vis spectroscopy combined with ODMT is suggested, and a proof-of-concept (POC) and validation studies are reported. Importantly, this method is found to consume a small amount of reagent and avoid possible side reactions contrarily to the Folin–Ciocalteu method. Potential environmental factors, such as heat or cold, did not affect the reaction. The method developed proved to be reliable for the TPC determination of the crude extract of a bunch of plants (GAE in mg/g of DW). Furthermore, the UV-Vis spectrometry–ODMT method is suitable for different kinds of users, from data mining beginners to programmers who prefer a scripting interface. Therefore, the purposed method ends up being direct, precise, reproducible, and simple to perform [24,25]. It showed excellent results, compared to those obtained directly from the Folin–Ciocalteu method.
2. Materials and Methods
2.1. Materials and Instrumentation
All chemicals used were of analytical quality; ethanol (C2H5OH, 99.9%, Sigma-Aldrich, St. Louis, MI, USA), hexane (C6H14, 97%, Sigma-Aldrich), reagent Folin–Ciocalteu (3H2O·P2O5·13WO3·5MoO3·10H2O and 3H2O·P2O5·14WO3·4MoO3·10H2O)) produced by PROLABO, sodium carbonate (Na2CO3, ≥99.5%, Sigma-Aldrich), and GA (C7H6O5, 99%, PROLABO). All plants were collected in spring 2021 from El-Oued, southeastern Algeria. Analytical balance (Shanghai Suisse Instrument precision 0.0001 g). Rota evaporator branded (B.U.C.H.I) model R-210. equipped with a top cooler. A Shimadzu UV-Vis 1800 spectrophotometer characterized by high resolution and an error of less than 0.01 nm. This device is linked with a microcomputer to facilitate the processing of results. Whatman® cellulose chromatography papers 1 Chr sheets. (20 × 20) cm (GE Healthcare Life Sciences, Amersham, UK) were used for paper chromatography. The absorption spectrum was measured on a UV-Vis. Spectrophotometer (UV-Vis. Cary 4000. Agilent, Craven Arms, UK) controlled by Agilent Scan software.
2.2. Preparation of the Plant Extract (Maceration)
The plant samples (Daucus carota L. root, Ruta Chalepensis L. Leaves, Anisosciadium DC. leaves, Thymus vulgaris L. Leaves, Senna alexandrina leaves, Myrtus communis L. leaves, Silybum marianum L. Flower, Silybum marianum L. Leaves, and Rosa moschata Flower). Randomly, selected samples were taken to the laboratory for analysis. In the laboratory, only samples with no visible malformation or bacterial damage were carefully selected. Then, the samples were washed with distilled water to remove the dirt deposited on the surface of the samples. These plant samples were then naturally dried after washing with distilled water. The oven-dried samples were ground to a powder using a mortar and pestle and then sieved using a mesh sieve of 2 mm diameter. The process was left for 24 h and the solids were filtered out using a Whatman No. 1 filter. To extract the phenolic compounds, 10 mg of each sample were taken in a conical flask and extracted with organic 100 mL solvents ethanol in a mechanical shaker with temperature control (70 °C) at a constant stirring rate at 200 rpm and repeat the process three times. In the end, the solvents were separated from the extract [26]. To date, different solvents have been reported for polyphenol extraction such as water, methanol, ethanol, propanol, chloroform, n-hexane, ethyl acetate, and acetone. These solvents differ in their polarity; and thus, they have different influences on the efficiency of the extraction process. However, ethanol has been selected as a general solvent, as it is a safe option for plant extraction due to the fact that it leaves behind a safe to use, non-toxic oil product [27].
2.3. Determination of Total Polyphenol Content (TPC)
Measurements of TPC in various extracts are performed by the Singleton and Rossi method using the Folin–Ciocalteu method (GAE in mg/g of DW) [13,28]. Volumes of 1000 µL at different concentrations of the various phytoextracts were carried and added to 200 µL Na2CO3 (10%). Approximately, 1000 µL of Folin–Ciocalteu reagent diluted ten times was added to the reaction medium. After incubating in the dark at room temperature (RT) for 40 min., Folin–Ciocalteu’s phenol reagent does not contain phenol. Rather, the reagent will react with phenols and nonphenolic reducing substances to form chromogens that can be detected spectrophotometrically. The absorbance was measured a λmax nm. The calibration curve of GA at various concentrations as standard [29]. Therefore, the result is expressed in mg GAE/g DW. All data were presented as the mean of three separate experiments and error bars are displayed with standard error (mean ± standard deviation/STD).
2.4. Software and Tools
ODMT is a data-mining tool that is useful for visual programming and exploratory data analysis that can be written in Python. Orange has many components known as widgets. Each widget includes some tasks of data retrieval, preprocessing, visualization, modeling, or evaluation. The combination of different user interface elements in a workflow allows users to create comprehensive data analysis charts on the go with a large library of tools [30,31,32,33]. This study proposes a comparison of the Folin–Ciocalteu method [13], and prediction methods in the ODMT are used in this study for analysis and prediction of TPC in the plant extracts. The methodology flow used in this study is illustrated in Figure 1. In this Figure, the program was designed using prophecy models that are present in Orange software to extract the desired results, which are the TPC from a bunch of plant extracts.
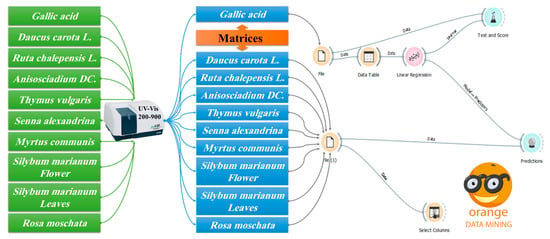
Figure 1.
Flow chart showing the method steps and the workflow in Orange software to determine quantitatively the TPC in a bunch of plant extracts.
The purposed method consumes a small amount of reagent, estimated at 1 mg, for one single time. As shown in Figure 1. First, using a new file function. The data whose concentration is known are entered in this file, and these data are represented in gallic acid with different concentrations in the Orange program, and these data are arrays of different concentrations of gallic acid in the absorption range of 200–900 and then the concentrations of the studied samples will be predicted. A spreadsheet is a table in which data are placed, organized, and arrays stored in a tabular form to facilitate the work of the program to predict the desired result. Linear regression function is used for finding the relationship between the GA concentration and the studied samples. Test and score function were used to extract the correction factor and calculate the success rate of the prediction model, and the result associated with the prediction models previously used. The second new file function is used to enter the plant extract information (sample of unknown concentration), where the second select column function is used for adding the UV-vis data (spreadsheet) of plant extracts. Eventually, the matrix function is used to link the concentrations of GA (standard) and the plant extracts (unknown samples).
3. Results and Discussion
TPC in the plants were estimated in two ways, whereas the closeness ratio between the two results was observed. The first result (GAE in mg/g of DW) was obtained by the method of Singleton and Rossi [28]. The second result was obtained by the ODMT method.
3.1. Prediction of Various Concentrations of GA
The predictions are to extract the concentrations obtained from the program for the three studied plant samples, of unknown concentrations, which is the concentration of polyphenols. Various concentrations are prepared of gallic acid in ethanol at 70 °C, and these concentrations are shown in the Figure 2.

Figure 2.
UV-Vis spectrophotometric analysis of GA: (a) UV-absorbance curve at different concentrations from 0.00538 to 0.02727 mg/mL; (b) Standard solutions calibration curve.
Then the samples are read in the UV-vis device from the range 200–500 of the gallic acid where a large matrix is obtained that links the concentrations and absorption for each sample separately. Figure 2a shows the calibration curve of the absorbance (200–900) versus the concertation of the GA, with a correction factor (R2 = 0.9998) (Figure 2b). Eventually, the resulting UV-Vis spectrometric data were entered into the ODMT, to be considered as the database to predict the concentration of unknown TPC in different plants.
3.2. Determination of TPC in Plant Extracts
The TPC for a group of plant extracts (Daucus carota L. root, Ruta Chalepensis L. Leaves, Anisosciadium DC. Leaves, Thymus vulgaris L. Leaves, Senna alexandrina leaves, Myrtus communis L. leaves, Silybum marianum L. Flower, Silybum marianum L. Leaves, and Rosa moschata Flower) was measured by using GA as a standard. The TPC in the different plant extracts samples ranges from 44.67 to 334.7 mg GAE/g DW (Table 1).

Table 1.
Total polyphenols content (TPC) in the different plant extracts measured using the Folin–Ciocalteu method as a reference method. Data are expressed in gallic acid equivalent (GAE) of dry weight (DW) extract.
3.3. Mathematical Calculations
Equation (1) was used to estimate TPC in different plant extracts using data (Figure 2a) stored in the ODMT.
where,
- : Concentration of polyphenol of plant extract (mg GA/g DW);
- : Concentration calculated by Orange Data Mining Tool in ;
- : Concentration of plant extract (mg/mL);
- : Sample absorbance in λmax in (nm), it varies from one plant to another plant;
- λmax: Larger absorption of the sample in (nm).
3.4. Spectroscopic Prediction of Total Polyphenols Content
A series of dilutions were prepared for each plant extract, as shown in Table 2. Then, the UV-Vis spectrum of each sample and their series of dilutions were recorded separately (Figure 3 and Figure 4). The data’s matrix that connects the concentrations and absorption of each plant is then entered for each of the TPC in the plant extracts (dilution series) in the second part of the program, and the following concentrations from the ODMT were then obtained. Table 3 indicates that the results obtained in the reaction method are very close to the results obtained from the ODMT, with a recovery percentage range of 95.3% to 104.3%.

Table 2.
Two comparison methods for total polyphenol content (TPC) in the indicated plant extract.
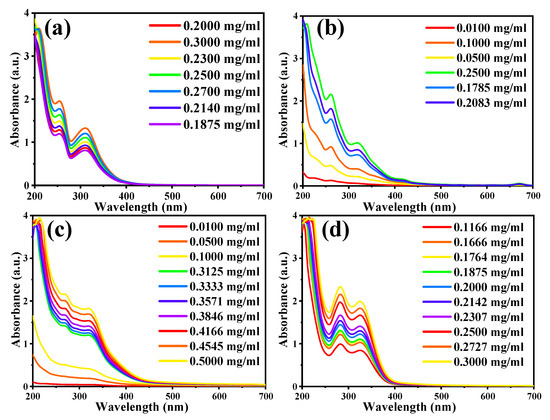
Figure 3.
UV-Vis absorbance curve of the plant extracts at different indicated concentrations: (a) Daucus carota L. (root), (b) Ruta Chalepensis L. (Leaf), (c) Anisosciadium DC. (Leaf), and (d) Thymus vulgaris L. (Leaf).
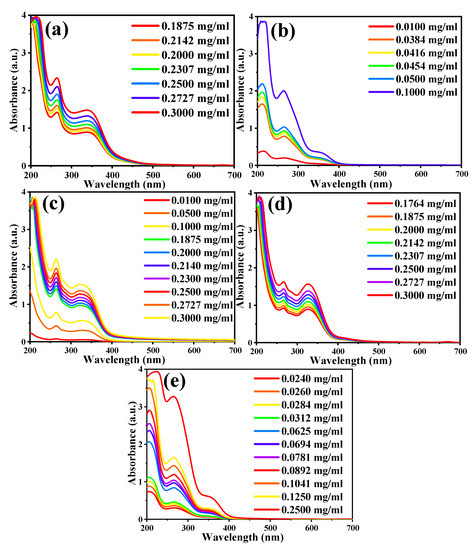
Figure 4.
UV-Vis absorbance curve of the plant extracts at the different indicated concentrations: (a) Senna alexandrina, (b) Myrtus communis L. (Leaf), (c) Silybum marianum L. (Flower), (d) Silybum marianum L. (Leaf), and (e) Rosa moschata L. (Flower).

Table 3.
Comparison of total polyphenol content (TPC) in different plants by Orange Data Mining Tool (ODMT) and Folin–Ciocalteu method (FC) used as the reference method. Data are expressed in terms of the gallic acid equivalent (GAE) in mg/g dry weight (DW) of the plant extract.
4. Conclusions
UV-Vis spectroscopic methods for the prediction of the TPC of medicinal plants extracts can be improved by adjusting the reagent concentration, pH, reaction time, temperature, and absorption wavelength. Herein, we developed an accurate UV-Vis spectrophotometric-ODMT method for the determination of TPC in nine medicinal plant extracts. The method used very small amounts of reagents (required a maximum of 1 mg of GA reagent and only 1 mg of the plant extract). The determination of TPC by the newly suggested method is based on creating a database (standard calibration curve) associated with the GA reagent and ODMT. Interestingly, TPC obtained by the standard calibration curve obtained using the Folin–Ciocalteu method (GAE) (in mg/g of the DW plant extract) are in agreement with the results predicted by the ODMT. The proposed ODMT method linear dynamic concentration ranges from 44.67 to 334.7 mg GAE/g of DW plant extract with a recovery percentage range of 95.3% to 104.3%. Therefore, from this research, it can be said that the value of polyphenols can be predicted in any plant in the future, and as mentioned in the introduction, the TPC can be predicted and apply to all medicinal plants using the new ODMT method.
Author Contributions
Conceptualization. S.E.L. F.G., S.M., and A.B. (Abderrhmane Bouafia) and A.B. (Ahmed Barhoum); methodology. A.B. (Abderrhmane Bouafia), A.B. (Ahmed Barhoum), S.M., and F.M.; software. F.G. and A.B. (Abderrhmane Bouafia); validation. A.B. (Abderrhmane Bouafia), F.G, A.T., K.A.M., S.A.A., F.M., and A.B. (Ahmed Barhoum); formal analysis. S.E.L., A.R., A.T., K.A.M., S.A.A., F.M., and F.G.; investigation. A.B. (Abderrhmane Bouafia), and F.M.; resources. A.R., and F.M.; data curation. S.E.L., A.T. K.A.M., S.A.A., and F.M.; writing—original draft preparation. F.G., A.B. (Abderrhmane Bouafia), S.M., and A.B. (Ahmed Barhoum); writing—review, and editing. F.G., A.B. (Abderrhmane Bouafia). A.B. (Ahmed Barhoum), S.M., A.T., K.A.M., S.A.A., and F.M.; supervision. S.E.L., A.R., and F.M. Submission. F.M. All authors have read and agreed to the published version of the manuscript.
Funding
The authors did not receive any external funds for this project.
Institutional Review Board Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
F.M. thank Fluorotronics, Inc/California Innovations Corporation, a pioneered company in optical and nanooptical devices and applications, for its technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gibson, E.L.; Wardle, J.; Watts, C.J. Fruit and Vegetable Consumption, Nutritional Knowledge and Beliefs in Mothers and Children. Appetite 1998, 31, 205–228. [Google Scholar] [CrossRef] [PubMed]
- Mathai, K. Nutrition in the adult years. In Krause’s Food, Nutrition, & Diet Therapy, 10th ed.; Mahan, L.K., Escott-Stump, S., Eds.; Saunders: Philadelphia, PA, USA, 2000; Volume 271, pp. 274–275. [Google Scholar]
- Hasler, C.M.; Blumberg, J.B. Phytochemicals: Biochemistry and physiology. Introduction. J. Nutr. 1999, 129, 756S–757S. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.M.; Markham, K.R. Flavonoids: Chemistry, Biochemistry and Applications; CRC Press: Boca Raton, FL, USA, 2005; ISBN 142003944X. [Google Scholar]
- Cheynier, V. Phenolic compounds: From plants to foods. Phytochem. Rev. 2012, 11, 153–177. [Google Scholar] [CrossRef]
- Gaurav, D.; Malik, A.K.; Rai, P.K. High-Performance Liquid Chromatographic Methods for the Analysis of Explosives. Crit. Rev. Anal. Chem. 2007, 37, 227–268. [Google Scholar] [CrossRef]
- Aleixandre-Tudo, J.L. The Role of UV-Visible Spectroscopy for Phenolic Compounds Quantification in Winemaking. In Frontiers and New Trends in the Science of Fermented Food and Beverages; Solís-Oviedo, R.L., De La Cruz Pech-Canul, Á, Eds.; IntechOpen: Rijeka, Croatia, 2019; Chapter 3; ISBN 978-1-78985-496-1. [Google Scholar]
- Giusti, M.M.; Wrolstad, R.E. Characterization and Measurement of Anthocyanins by UV-Visible Spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, F1.2.1–F1.2.13. [Google Scholar] [CrossRef]
- Kowalski, R.; Kowalska, G. Phenolic acid contents in fruits of aubergine (Solanum melongena L.). Pol. J. Food Nutr. Sci. 2005, 14, 37–41. [Google Scholar]
- Ish-Shalom, M.; Fitzpatrick, J.D.; Orchin, M. Quantitative analysis by ultraviolet spectrophotometry: The cis-trans-stilbene system. J. Chem. Educ. 1957, 34, 496. [Google Scholar] [CrossRef]
- Ramos, R.T.M.; Bezerra, I.C.F.; Ferreira, M.R.A.; Soares, L.A.L. Spectrophotometric Quantification of Flavonoids in Herbal Material, Crude Extract, and Fractions from Leaves of Eugenia uniflora Linn. Pharmacogn. Res. 2017, 9, 253–260. [Google Scholar] [CrossRef]
- Loum, J.; Byamukama, R.; Wanyama, P.A.G. UV–Vis Spectrometry for Quantitative Study of Tannin and Flavonoid Rich Dyes from Plant Sources. Chem. Afr. 2020, 3, 449–455. [Google Scholar] [CrossRef]
- Chavan, Y.; Singhal, R.S. Ultrasound-assisted extraction (UAE) of bioactives from arecanut (Areca catechu L.) and optimization study using response surface methodology. Innov. Food Sci. Emerg. Technol. 2013, 17, 106–113. [Google Scholar] [CrossRef]
- Devi, S.K.; Krishnapriya, S.; Kalita, D. Prediction of heart disease using data mining techniques. Indian J. Sci. Technol. 2016, 9, 1–5. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Tardaguila, J.; Fernández-Novales, J.; Diago, M.P. Data Mining and NIR Spectroscopy in Viticulture: Applications for Plant Phenotyping under Field Conditions. Sensors 2016, 16, 236. [Google Scholar] [CrossRef]
- Dey, U.K.; Masud, A.H.; Uddin, M.N. Rice yield prediction model using data mining. In Proceedings of the 2017 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 16–18 February 2017; pp. 321–326. [Google Scholar]
- Blainski, A.; Lopes, G.C.; De Mello, J.C. Application and Analysis of the Folin Ciocalteu Method for the Determination of the Total Phenolic Content from Limonium Brasiliense L. Molecules 2013, 18, 6852–6865. [Google Scholar] [CrossRef]
- Schofield, P.; Mbugua, D.M.; Pell, A.N. Analysis of condensed tannins: A review. Anim. Feed Sci. Technol. 2001, 91, 21–40. [Google Scholar] [CrossRef]
- Cicco, N.; Lanorte, M.T.; Paraggio, M.; Viggiano, M.; Lattanzio, V. A reproducible, rapid and inexpensive Folin–Ciocalteu micro-method in determining phenolics of plant methanol extracts. Microchem. J. 2009, 91, 107–110. [Google Scholar] [CrossRef]
- Bueno, F.G.; Machareth, M.A.D.; Panizzon, G.P.; Lopes, G.C.; Mello, J.C.P.; Leite-Mello, E.V.S. Development of a UV/Vis spectrophotometric method for analysis of total polyphenols from Caesalpinia peltophoroides Benth. Quim. Nova 2012, 35, 822–826. [Google Scholar] [CrossRef]
- Muzolf-Panek, M.; Kaczmarek, A.; Gliszczyńska-Świgło, A. A predictive approach to the antioxidant capacity assessment of green and black tea infusions. J. Food Meas. Charact. 2021, 15, 1422–1436. [Google Scholar] [CrossRef]
- Hosu, A.; Cristea, V.-M.; Cimpoiu, C. Analysis of total phenolic, flavonoids, anthocyanins and tannins content in Romanian red wines: Prediction of antioxidant activities and classification of wines using artificial neural networks. Food Chem. 2014, 150, 113–118. [Google Scholar] [CrossRef]
- Guiné, R.; Gonçalves, C.; Matos, S.; Gonçalves, F.; Costa, D.V.T.D.; Mendes, M. Modelling through artificial neural networks of the phenolic compounds and antioxidant activity of blueberries. Iran. J. Chem. Chem. Eng. 2018, 37, 193–212. [Google Scholar]
- Aleixandre-Tudo, J.L.; Buica, A.; Nieuwoudt, H.; Aleixandre, J.L.; du Toit, W. Spectrophotometric Analysis of Phenolic Compounds in Grapes and Wines. J. Agric. Food Chem. 2017, 65, 4009–4026. [Google Scholar] [CrossRef]
- Weber, F.; Passon, M. Chapter 7—Characterization and Quantification of Polyphenols in Fruits. In Polyphenols in Plants, 2nd ed.; Watson, R.R., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 111–121. ISBN 978-0-12-813768-0. [Google Scholar]
- Guemari, F.; Laouini, S.E.; Rebiai, A.; Bouafia, A. Phytochemical screening and Identification of Polyphenols, Evaluation of Antioxidant activity and study of Biological properties of extract Silybum marianum (L.). Asian J. Res. Chem. 2020, 13, 190. [Google Scholar] [CrossRef]
- Baümler, E.R.; Carrín, M.E.; Carelli, A.A. Extraction of sunflower oil using ethanol as solvent. J. Food Eng. 2016, 178, 190–197. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Waterhouse, A. Folin-ciocalteau micro method for total phenol in wine. Am. J. Enol. Vitic. 2006, 48, 357–363. [Google Scholar]
- Záková, M.; Podpecan, V.; Zelezný, F.; Lavrac, N. Advancing data mining workflow construction: A framework and cases using the orange toolkit. Proc. 2nd Intl. Wshop. Third Gener. Data Min. Towar. Serv. Knowl. Discov. 2009, 39–52. [Google Scholar]
- Vaishnav, D.; Rao, B.R. Comparison of Machine Learning Algorithms and Fruit Classification using Orange Data Mining Tool. In Proceedings of the 2018 3rd International Conference on Inventive Computation Technologies (ICICT), Coimbatore, India, 15–16 November 2018; pp. 603–607. [Google Scholar]
- Zupan, B.; Demsar, J. Open-Source Tools for Data Mining. Clin. Lab. Med. 2008, 28, 37–54. [Google Scholar] [CrossRef][Green Version]
- Jayawiguna, I.B.P. Comparison of Model Prediction for Tile Production in Tabanan Regency with Orange Data Mining Tool. Int. J. Eng. Emerg. Technol. 2020, 5, 72–76. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).