Featured Application
The computational approaches hereby shown can be used in the rational design of biologic drugs.
Abstract
Factor VIII belongs to the coagulation cascade and is expressed as a long pre-protein (mature form, 2351 amino acids long). FVIII is deficient or defective in hemophilic A patients, who need to be treated with hemoderivatives or recombinant FVIII substitutes, i.e., biologic drugs. The interaction between FVIII and von Willebrand factor (VWF) influences the pharmacokinetics of FVIII medications. In vivo, full-length FVIII (FL-FVIII) is secreted in a plasma-inactive form, which includes the B domain, which is then proteolyzed by thrombin protease activity, leading to an inactive plasma intermediate. In this work, we analyzed through a computational approach the binding of VWF with two structure models of FVIII (secreted full-length with B domain, and B domain-deleted FVIII). We included in our analysis the atomic model of efanesoctocog alfa, a novel and investigational recombinant FVIII medication, in which the VWF is covalently linked to FVIII. We carried out a structural analysis of VWF/FVIII interfaces by means of protein–protein docking, PISA (Proteins, Interfaces, Structures and Assemblies), and protein contact networks (PCN) analyses. Accordingly, our computational approaches to previously published experimental data demonstrated that the domains A3-C1 of B domain-deleted FVIII (BDD-FVIII) is the preferential binding site for VWF. Overall, our computational approach applied to topological analysis of protein–protein interface can be aimed at the rational design of biologic drugs other than FVIII medications.
1. Introduction
Hemophilic A patients are treated periodically with the coagulation factor FVIII substitutes, such as purified hemoderivatives or FVIII biological drugs, which are currently biological advanced therapies approved for treatment of hemophilia A. This class of drugs includes recombinant full-length FVIII and B domain-deleted FVIII [1]. B domain-deleted FVIII (BDD-FVIII) biologics have been developed to improve the biotechnological production of these proteins. Furthermore, BDD-FVIII conjugated with Fc immunoglobulin fragment was reported to have the highest plasma half-life, providing the opportunity of scheduling a low number of infusions [2]. Therefore, BDD-FVIII biologics (products with extended half-life, EHL) have been claimed to decrease the number of infusions in hemophilic patients. EHL products may allow less frequent dosing; however, due to inter-patient differences in FVIII plasma stability and clearance, less frequent infusions may cause longer time periods with relatively low FVIII plasma levels, which could increase the risk of bleeding. The safety cutoff for plasma FVIII levels was set to >1%, and patients with levels >12% were subjected to less bleeding events, especially at joints [2,3]. Specifically, clinical trials carried out so far highlighted a similar capability of different FVIII substitutes on bleeding prevention, measured as annualized bleeding rate [2]. Interestingly, a pharmacokinetic (PK) modeling study predicted that mean values of FVIII plasma levels were similar in patients treated with either BDD-FVIII or full-length recombinant FVIII [1]. Furthermore, this study predicted that patients treated with full-length FVIII (infusions every 48 h) spent more time with FVIII above the 10 IU dL-1 than patients treated with BDD-FVIII product infusions every 72 h. Therefore, full-length FVIII could be characterized by higher plasma stability than B domain-deleted FVIII substitutes compared to BDD-FVIII [1]. This data raised a controversy [4], but the high plasma stability of FL-FVIII was confirmed at pre-clinical level [5].
FVIII half-life is strongly influenced by von Willebrand Factor (VWF), which is reported to bind with FVIII [6,7,8]. Several studies reported that the binding sites of VWF at FVIII are in the A3 and C1 FVIII domains [9,10,11,12] Therefore, with the aim to improve BDD-FVIII plasma stability, a novel FVIII substitute has been developed, efanesoctocog alfa (BIVV001). Efanesoctocog alfa is an investigational biologic drug where VWF D’-D3 domain is covalently bound (through a cleavable XTEN polypeptide linker) to an engineered BDD-FVIII [13]. The atomic model of cryo-electron microscopy (Cryo-EM) of efanesoctocog alfa was built and deposited on Protein Data Bank (PDB:7KWO); in this atomic model VWF binding to A3 and C1 domains of FVIII is confirmed. Mature FVIII is a 2351 a.a pre-protein that, during processing in the endoplasmic reticulum, is cleaved and reassembled in a heavy and a light chain, interacting by means of Van der Waals interactions, and through a metal complex interaction with a divalent cation (Figure 1).
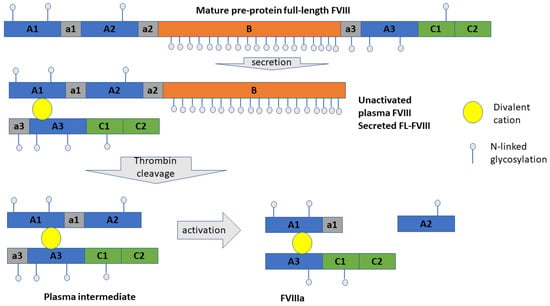
Figure 1.
FVIII processing. Capital letters refer to protein domains, while lowercase letters refer to loops linking two protein domains (modified from Pipe SW. Haemophilia 2009 [14]).
After secretion, the processed full-length FVIII (FL-FVIII) is cleaved mainly at a.a 1313, corresponding to the secreted inactivated form of FVIII, containing about 572 amino acids of B domain (secreted full-length FVIII) (Figure 1) [14,15]. However, several heterogenous forms of secreted FL-FVIII were found in plasma [16]. The domain B function has been linked to protection of FVIII against premature proteolysis. Moreover, B domain can inhibit FVIII binding to activated platelets, decreasing overall the inactivation rate of FVIII. B domain has also been involved in modulation of FVIII clearance through binding to the asialoglycoprotein receptor [14]. Finally, FVIII stability to aggregation events is driven by the content of B domain, since FVIII aggregation rate increases with shortening of B domain length [16]. Protein aggregation should generally be avoided for biologics, due to the increased risk of immunogenicity, adverse drug reactions, and modification of pharmacokinetics and pharmacodynamic properties, which then affect overall drug efficacy [17,18,19]. FVIII medication immunogenicity has been accounted to formation of anti-FVIII antibodies in patients (i.e., inhibitor formation); however, to date, no significant differences in FVIII inhibitor formation have been found in patients treated with full length or BDD-FVIII [20]. Since the stoichiometry of VWF binding to FVIII has not been univocally defined [21,22,23,24], and the VWF increases FL-FVIII stability [5,20], we explored through structural computational approach the binding of VWF to modeled B domain of FVIII. To date, the structure of B domain of FVIII has not been solved, although this domain has been identified in magnified Cryo-EM images [25]. Moreover, we reported a structural analysis of protein–protein interactions through molecular docking and structural analysis of protein—protein interfaces [26,27]. We also applied the Protein Contact Networks (PCN) methodology to analyze the topology of the VWF/FVIII complexes, which was validated on more than 1000 protein systems [28]. This methodology, applied to the prediction of interface binding energy, can depict the structure–function relationship in protein–protein complexes along with identification of allosteric binding sites [29,30,31,32,33].
Recently, this approach has also been applied to the analysis of the molecular mechanism behind the SARS-CoV2 infection, analyzing the protein–protein interactions of spike protein/ACE2 complex, providing insight also in the development of new therapeutic strategies [34,35,36,37]. Specifically, in our study we included computed novel PCN descriptors of protein–protein interfaces, which identified the key residues involved in the protein–protein interactions of FVIII/VWF complexes.
2. Materials and Methods
Structures have been retrieved from the Protein DataBank as PDB files: 2R7E (B domain-deleted FVIII), 6N29 (D’D3 von Willebrand factor binding domain to FVIII), 7KWO (D’D3 VWF bound to B-domainless FVIII, atomic model of efanesoctocog alfa).
2.1. Structure Modeling
The structure of secreted full-length FVIII was predicted through a two-step modeling approach: (i) the B domain was modeled with the I-Tasser web server; (ii) full-length FVIII model was built with the Advanced Molecular Modeling task of Schrodinger Maestro, by sequence alignment of available structure of B domain-deleted FVIII and modeled B domain, using as input primary sequences of heavy and light chains as reported in Figure 1. Five models of B domain structure were generated with I-Tasser. These models of B domain were used to build 5 models of secreted full-length FVIII, with the Advanced Molecular Modeling task of Schrodinger Maestro. However, 4 models of B domain were automatically excluded by the Advanced Molecular Modeling task due to steric clashes between other protein domains, and just 1 model (Figure S1, secondary structure plot–Supplementary Materials) was further optimized with automated energy minimization steps in the Advanced Molecular Modeling task of Schrodinger Maestro. After energy minimization steps, the optimized models of full-length FVIII (secreted FL-FVIII) and B domain-deleted FVIII (BDD-FVIII) were then subjected to protein–protein docking with a the D’D3 domain of von Willebrand Factor (PDB: 6N29) through PyDock, which provided the prediction of binding free energy of protein complexes (https://life.bsc.es/pid/pydock/ from 2016 to 2022) [38]. Pydock output also includes the scoring of predicted complexes. Rescoring of predicted complexes was also carried out with Prodigy web server (http://milou.science.uu.nl/services/PRODIGY/ from 2016 to 2022) [39].
2.2. Protein Contact Networks
We built the protein contact networks (PCNs) on FVIII complexes starting from the .pdb files, as previously described [27]. In a PCN, protein residues are the network nodes. Links between nodes are active contacts between residues, e.g., when the inter-residue distance lies between 4 and 8 Å, to account for non-covalent residue–residue interactions.
The mathematical representation of the PCN is given by the adjacency matrix, defined as:
where is the Euclidean distance between the i-th and j-th residue, defined as:
where and are the coordinates in a cartesian space of the i-th and j-th residues, respectively (represented by the coordinates of their -carbons).
Once the PCN was built, we computed the node degree for the i-th node, defined as the number of its links with other nodes, computed as the sum of the elements on the i-th row of the adjacency matrix A:
In order to characterize the topology of the protein–protein interactions, for each protein–protein interface we identified links between nodes belonging to the different interfacing chains and, accordingly, we introduced the inter-chain degree of each node as the number of links it shares with residues belonging different protein chains.
Nodes (residues) endowed with high inter-chain degree are defined as network hotspots of the protein complex interface, addressing their significant role in protein–protein interactions.
The energy of a graph E is defined as the sum of the absolute values of the adjacency matrix A eigenvalues. Although this is a purely topological descriptor, it captures some physical energy properties of the protein molecular structures [40], particularly oligomers interactions [41].
Focusing on a given interface between two chains, and , the overall inter-chain degree for a given interface is computed as the sum of the inter-chain degree of residues belonging to a single chain, characterizing the overall interface strength. We defined the average inter-chain degree as the average inter-chain degree value over the number of residues participating in the interface.
We adopted the geometrical descriptors of protein interfaces according to the method of Mei et al. in [42] for each interface between two chains in the complex: 1. the total number of residues for each chain in the interface; this number is in general lower than the total interface degree, due to multiple links between residues participating to the interface; 2. the length of the chain involved in the interface R; 3. the interface “roughness” Q/R (previously introduced [9]); 4. the interface amino acid range, IAR = R/N being N the total number of residues in the chain.
For a given interface between two chains, and , the average value of the inter-chain degree is simply given as:
We introduced energy descriptors, including the topological description provided by the PCNs method. Considering that the interaction energy is higher if the contact distance is smaller, we introduced a weight for each contact:
which is also the generic element of the interface energy matrix E, defined as:
For each interface, we sorted out the corresponding minor of the interface energy matrix E (corresponding as indices to the rectangular minors of the adjacency matrix) and we introduced the overall interface energy as the sum of for each of the active links at the interface, and the average value over the whole number of residues at the interface. We also analyzed the single residue contribution to the interface energy, defining the:
The overall value of energy of interaction is then computed as the sum of all contributions given by Equation (7) for all contacts between chain Ai and Aj. The average value of the interaction energy is given by:
Finally, we can define the graph energy of the interface as the difference between the graph energy of the complex minus the graph energy computed for the single chains (that is, considering the eigenvalues of the adjacency matrix minors corresponding to the single chains).
Furthermore, we completed the analysis through a thermodynamic analysis of the protein complexes via the PISA web server [8] (https://www.ebi.ac.uk/pdbe/pisa/ from 2016 to 2022), reporting from the analysis the following properties: for monomers, a. number of residues exposed at the surface; b. solvent-accessible surface area (ASA) in Å2; c. solvation free energy of folding of the corresponding structures in kcal/mol; for interfaces, a. number of residues exposed at the interface (not accessible to solvent); b. interface area in Å2 for each monomer (surface area, accessible to solvent in the monomer and no more accessible upon interface formation); c. solvation free energy gain upon formation of the interface in kcal/mol; the value was calculated as difference in the total solvation energies of isolated and interfacing structures; negative corresponds to hydrophobic interfaces, or positive protein affinity.
2.3. Network Clustering and Participation Coefficient Calculations
Finally, we applied a network spectral clustering algorithm to identify functional domains in all different conformations [43]; the methodology is based on the spectral decomposition of the network Laplacian, defined as:
where D is the degree matrix, a diagonal matrix whose diagonal is the degree vector, and A is the network adjacency matrix, as defined in Equation (1). Cluster partition is based on the value of the Fiedler vector v2 (the eigenvector corresponding to the second minor eigenvalue of L): the cluster number nc is user-defined. The v2 components interval is divided into nc subintervals, so that nodes (residues) are parted in clusters according to which subinterval their v2 components fall into.
On the basis of network clustering, the participation coefficient P is defined as:
where is the number of links the i-th node shares with nodes belonging to its own cluster.
The participation coefficient is able to identify residues’ role in transmitting signals between functional protein regions (protein network clusters) [30,34,40,44].
The PCN methodology is now implemented in open-source software [45].
We projected values of participation coefficient as b-factor and colored the ribbon structures of the complexes by means of an in-house Python script, according to the method previously described [27].
3. Results
3.1. Protein Docking
Protein–protein docking studies were carried out with PyDock on the model of full-length secreted form of FVIII, to predict VWF/FL-FVIII complex (Figure 2). To validate the protein–protein docking and the computational structure interface analysis approaches, we also docked the BDD-FVIII (PDB: 2R7E) with the fragment of von Willebrand factor (PDB: 6N29) (Figure 3). Pydock predicted that von Willebrand factor interacts with the B domain of secreted full-length FVIII with slightly more negative predicted binding free energy, compared to the BDD-FVIII/VWF complex (Table 1).
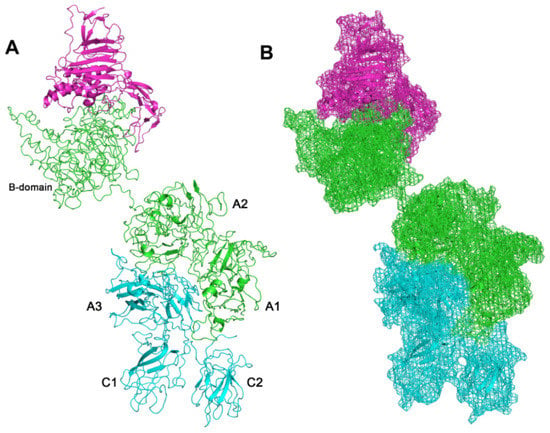
Figure 2.
VWF/FL-FVIII complex. (A) FL-FVIII is represented in cyan (light-chain) and green cartoons (heavy-chain, including domain (B)), VWF is represented with magenta cartoon. (B) Surfaces representation as mesh. Capital letters refer to protein domains of FVIII.
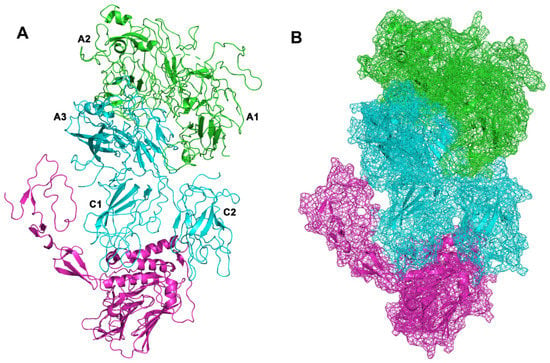
Figure 3.
VWF/BDD-FVIII complex. (A) BDD-FVIII is represented in cyan (light-chain) and green cartoon, VWF is represented with magenta cartoon. (B) Surfaces representation as mesh. Capital letters refer to protein domains of FVIII.

Table 1.
Pydock output. Predicted interactions between FVIII structure models and von Willebrand factor fragment (PyDock).
Shiltagh et al. in 2014 identified the D’D3 (PDB: 6N29) as the domain of von Willebrand factor (VWF) that interacts with FVIII [46]. Chiu et al. in 2015 found that the VWF interacts with the A3 and C1 domain in light chain of the B domain-deleted FVIII [47]. The Pydock output for VWF/BDD-FVIII complex is in accordance with the study results from Chiu et al. 2015 and Fuller et al. 2021 (Figure 3 and Figure 4) [13,47].
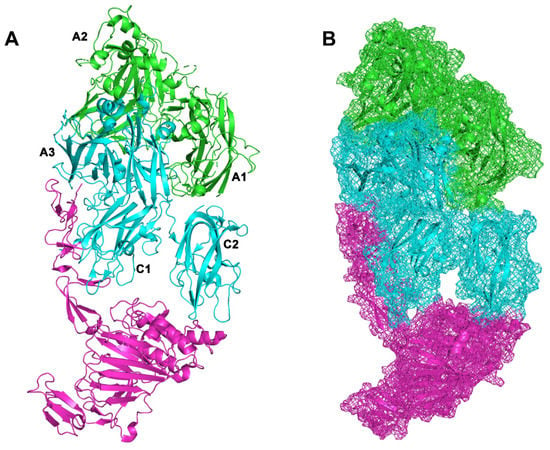
Figure 4.
Efanesoctocog alfa (A) FVIII is represented in green cartoon (heavy chain) and cyan cartoon (light chain). VWF is represented with magenta cartoon. (B) Surfaces representation as mesh. Capital letters refer to protein domains of FVIII.
Fuller et al., 2021, have solved the structure of the bioengineered clinical-stage FVIII substitute, BIVV001 or efanesoctocog alfa (PDB: 7KWO). In efanesoctocog alfa, the VWF-D′D3 is covalently linked to an Fc domain of a BDD-FVIII, resulting in a stabilized VWF-D′D3/BDD-FVIII complex (Figure 4).
The two structures of predicted VWF/BDD-FVIII and of efanesoctocog alfa have been superimposed, and RMSD of alignment was 1.4 Å. PRODIGY rescoring (Table 2) provided binding energy and dissociation constants for analyzed complexes. The binding energy (−14.1 kcal/mol, pKd 10.4) of the predicted complex VWF/FL-FVIII, characterized by the binding of VWF with B domain, was higher (less favorable), although comparable to the binding energy of VWF bound to the A3-C1 domains of BDD-FVIII (−15.3 kcal/mol, pKd 11.3), resembling the slight differences predicted by PyDock. We included the prediction of binding free energy of VWF in efanesoctocog alfa. In the efanesoctocog alfa atomic model, the predicted binding free energy of protein–protein interactions between VWF and FVIII was more negative (~4 logs), compared to binding free energy of the other complexes predicted through protein–protein docking (VWF/FL-FVIII and VWF/BDD-FVIII) (Table 2). Therefore, our computational approach is in accordance with the experimental findings reporting highly stable VWF and FVIII interactions in efanesoctocog alfa or BIVV001.

Table 2.
PRODIGY rescoring of PYDOCK-predicted complexes.
3.2. Interface Analysis
Since binding free energy prediction with PyDock and PRODIGY, generally, provides an indication of binding driving forces, we further focused our calculations on specific protein–protein interface descriptors. We analyzed the protein–protein interfaces of VWF/FVIII complexes: B domain-deleted FVIII (Table 3), secreted full-length FVIII (Table 4), and efanesoctocog alfa (Table 5). Two interfaces were analyzed: (i) one that involves the heavy and the light chains interactions; (ii) and the VWF/FVIII interface. BDD-FVIII and the FL-FVIII showed similar heavy/light chain interfaces (Table 3 and Table 4) in terms of ΔΔGSOLV, which was slightly lower in the VWF/FL-FVIII (more favorable) compared to BDD-FVIII complex. These results are coherent with PyDock and Prodigy predictions. Furthermore, according to prediction of binding free energy (PRODIGY calculations) the interactions in the VWF/FVIIII interface of the efanesoctocog alfa were characterized by the most favorable (lowest) predicted free energy ΔΔGsolv, kcal/mol (Table 5), compared to the other analyzed complexes. This lower ΔΔGsolv, kcal/mol (more favorable) corresponded to higher ΔΔGsolv, kcal/mol at interface between heavy and light chains in efanesoctocog alfa, compared to other analyzed complexes. Therefore, it is likely that more stable interactions between VWF/FVIII correspond to a destabilization of FVIII heavy/light chain interface. The results reported in Table 3, Table 4 and Table 5 indicated that the efanesoctocog alfa is the most stable VWF/FVIII complex, in terms of specific stability of monomers and interface interaction, compared to the VWF/BDD-FVIII and VWF/FL-FVIII complexes, according to findings of Fuller et al. [13].

Table 3.
PISA results for the VWF/BDD-FVIII complex. Values within brackets refer to the value specific per residue. Accessible surface area (ASA) of interface (Å2).

Table 4.
PISA results for the VWF/FL-FVIII complex. Values within brackets refer to the value specific per residue. Accessible surface area (ASA) of interface (Å2).

Table 5.
PISA results for efanesoctocog alfa. Values within brackets refer to the value specific per residue. Accessible surface area (ASA) of interface (Å2).
To obtain further insight in the protein–protein interactions, we carried out PCN analysis of the three complexes: VWF/BDD-FVIII, efanesoctocog alfa, and VWF/FL-FVIII. Results are shown in Table 6, according to the description provided in Materials and Methods.

Table 6.
Topological descriptors for the complex interfaces. is the total number of residues for each chain in the interface. R is the length of the chain involved in the interface. Q/R is the interface “roughness.” IAR = R/N is the interface amino acid range, where N is the total number of residues in the chain. is the inter-chain degree. is the average value of the inter-chain degree. is the energy of interface interaction, computed as the sum of all contributions for all contacts between chain Ai and Aj. is the average value of the interaction energy. H is the label for heavy chain of FVIII, L is the label for light chain of FVIII, and V is the label for VWF. In the VWF/FL-FVIII complex, the VWF interacts with B domain in the heavy chain of FVIII.
PCN analysis (Table 6) has shown that the interface roughness, (Q/R) was very similar in analyzed FVIII structure models. The highest Q/R value was associated to heavy chain of FL-FVIII, because FL-FVIII includes the aminoacids of B domain. In fact, the differences in IAR can be accounted to different length of protein chains in contact with the ligand, the VWF.
The PCN analysis, as regards the topology of heavy/light chain interface (H:L), shows that the number of residues involved into active links between heavy and light chains was higher for the FL-FVIII, compared to the BDD-FVIII and the atomic model of efanesoctocog alfa. This result diverges from PISA output, but PCN and PISA reside on two different methods. However, the “absolute interface energy” at heavy/light (H:L) chain interface was higher (more favorable) in the VWF/FL-FVIII (81.07 a.u.), compared to values of the VWF/BDD-FVIII (40.49 a.u.) and efanesoctocog alfa (42.89 a.u.) complexes. This trend of energy values, calculated with PCN, are in accordance with interface energy values calculated with PISA. Moreover, the “absolute interface energy” is proportional to the average inter-chain degree, which was greater at H:L interface of FL-FVIII, compared to other VWF/FVIII complexes.
Looking at FVIII /VWF interface, the values of “absolute interface energy” were higher (more favorable) for the efanesoctocog alfa (31.20 a.u.) and BDD-FVIII (28.92 a.u.), compared to FL-FVIII (22.66 a.u.); indeed, PCN parameters have been in accordance with experimental data and PISA calculations. These differences were mirrored by other topological PCN parameters, i.e., the average value of the interaction energy.
Particularly in the comparison between efanesoctocog alfa and BDD-FVIII, we found subtle differences in graph energy parameters regarding topology of protein–protein interfaces. Therefore, we carried out PCN clustering and participation coefficient (P) calculation, in order to identify through a quantitative approach (see method section, i.e., 2.3 Network clustering and participation coefficient calculations) residues involved in allosteric modulation of protein structure, i.e., allosteric residues (Figure 5, Figure 6 and Figure 7). The VWF/FL-FVIII (around 1%, Figure 5) complexes showed the lowest number of allosteric residues, compared to the VWF/BDD-FVIII (about 4%, Figure 6) and the efanesoctocog alfa (more than 5%, Figure 7).

Figure 5.
VWF/FL-FVIII complex allosteric residues. Participation coefficient P heat map in color scale (blue to red, increasing values of P). Blue residues have a P = 0. Capital letters refer to protein domains of FVIII.
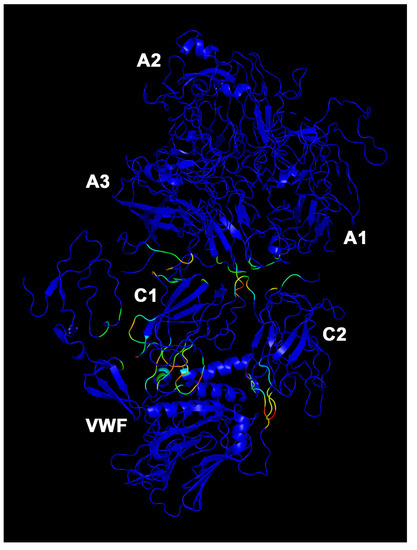
Figure 6.
VWF/BDD-FVIII complex allosteric residues. Participation coefficient P heat map in color scale (blue to red, increasing values of P). Blue residues have a P = 0. Capital letters refer to protein domains of FVIII.
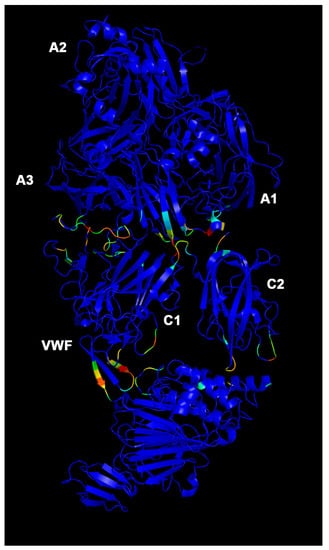
Figure 7.
Efanesoctocog alfa allosteric residues. Participation coefficient P heat map in color scale (blue to red, increasing values of P). Blue residues have a P = 0. Capital letters refer to protein domains of FVIII.
The few allosteric residues (high P values) in the FL-FVIII/VWF complex (Figure 5) are localized at domain-B/VWF interface, domain B, and A3-C1 interface. The main difference between VWF/BDD-FVIII (Figure 6) and efanesoctocog alfa (Figure 7) stands in the localization and number of allosteric residues. Particularly in VWF/BDD-FVIII (Figure 6), allosteric residues (P > 0) are more distributed in the C1 domain of heavy chain at interface with VWF, compared to efanesoctocog alfa (Figure 7), where most of the allosteric residues (high coefficient P value) are located at A3-C1 and A3-C2 domain interfaces.
These results support the validity and accordance of different computational methods hereby applied: protein–protein docking, docking rescoring, PISA, and PCN analysis of protein and protein domains interfaces.
4. Discussion
FVIII is a key protein involved in the coagulation cascade, and genetic defects in the FVIII gene (F8) cause hemophilia A, an x-linked recessive inherited disease. Hemophilia A is a rare, life-threatening disease affecting 1 in 6000 males, which causes spontaneous and prolonged hemorrhages due to FVIII deficiency. FVIII replacement therapy is the major therapeutic strategy for treatment of hemophilia A, and FVIII medications are listed by the World Health Organization (WHO) as essential medicines [48]. FVIII substitutes consist of full-length FVIII extracted and purified from human plasma, along with several biologic drugs, such as recombinant full-length FVIII (rFL-FVIII) and recombinant B domain-deleted FVIII (rBDD-FVIII). BDD-FVIII products have been developed to improve production yield and standardize recombinant protein production processes, but rBDD-FVIII were also found to be associated to increased FVIII plasma half-life; therefore, these products are also denominated as extended half-life (EHL) FVIII [49]. Interaction between von Willebrand Factor (VWF) and FVIII has been reported to improve FVIII plasma stability and pharmacokinetics properties, without modification of FVIII pharmacodynamics [8,9,50]. From this perspective, a novel FVIII investigational replacement medication has been developed, i.e., the BIVV001 or efanesoctocog alfa, which is a recombinant fusion protein in which the B domain-deleted FVIII is covalently linked to VWF, through a XTEN polypeptide linker [13]. VWF binds with most favorable energy to A3-C1 domains of FVIII light chain, however, binding of VWF to other domains of FVIII should not be excluded, since FVIII and VWF stoichiometry has not been univocally identified [21,22]. Moreover, the VWF binding to full-length FVIII has been linked to increased FVIII stability and decreased FVIII immunogenicity (i.e., inhibitor formation). This explains the trend of FL-FVIII medication to show a longer half-life over time and a similar or lower rate of inhibitor formation in treatment-naïve patients, compared to BDD-FVIII medications [5,8,20]. These data can also be attributed to binding of B domain to other plasma proteins, such as albumin [20]. Our in silico study could be considered as a small step to translational investigation, because it outlined how B domain in FL-FVIII/VWF would be an additional, but not the most favorable, binding site for VWF, thus putatively contributing to high plasma stability of FL-FVIII medication.
Our in silico study provided a structural insight on the binding of von Willebrand Factor to FVIII, and our computational data are in accordance with the experimental findings, i.e., FVIII A3-C1 domain is the most stable binding site of VWF. Our computational approach suggested that one of the driving forces of VWF binding at this preferential binding site could be related to conformational modifications of FVIII, through modulation of allosteric residues.
The PISA and PCN analysis of topological parameters suggested an unfavorable binding of VWF at B domain of FVIII, due to an increased stability of the interface between the heavy and the light chain, compared to VWF/BDD-FVIII and efanesoctocog alfa. In fact, the most stable VWF/FVIII complex (efanesoctocog alfa) was characterized by highest (less favorable) interaction energy between heavy and light chains of FVIII. Differences between the modeled VWF/BDD-FVIII complex and the atomic model of efanesoctocog alfa are not likely to be attributed to the lower resolution of BDD-FVIII X-ray structure (PDB:2R7E), compared to the atomic model of efanesoctocog alfa (PDB: 7KWO). Furthermore, the X-ray structure of BDD-FVIII showed 4% of Phi and Psi angles in disallowed regions of the Ramachandran plot, but these residues are located in disordered loops of A1 and A2 domains of heavy chain, a region not involved in the binding with VWF [51].
Moreover, the PCN clustering and participation coefficient P calculations revealed that VWF binding to the B domain of FL-FVIII was characterized by lower number of residues with participation coefficient P > 0 (allosteric residues), compared to VWF/BDD-FVIII and efanesoctocog alfa. The participation coefficient is an output parameter of PCN analysis, and has been found useful for identification of allosteric residues in different protein system and protein domains, according to the methodological approach hereby used and previously applied [31,40]. The topological parameters, coming from PCN analysis, also provided information on the local contribution to interface energy, which is useful for identification of key residues (e.g., allosteric amino acids) in protein–protein complex formation. Therefore, we can hypothesize that one of the driving forces in VWF binding to FVIII is attributed to allosteric modulation of protein structure.
Further investigations can shed light on putative allosteric and cooperative protein–protein interaction, e.g., by simulating FVIII structures with several replicas of VWF. These studies may provide new hints about the structural role of different domains of FVIII, along with interaction with other plasma proteins, such as the serum albumin. One of the main limitations of our study is attributed to the intrinsic approximation structural modeling of B domain, whose structure has never been solved, with the exclusion of Cryo-EM density images [25]. Another limitation is related to approximation of intrinsic and essential FVIII post-translational modifications, such as protein glycosylation (Figure 1), and the most glycosylated domain of FVIII is the B domain.
5. Conclusions
In conclusion, we hereby carried out an integrated computational approach which provided outputs that are in accordance with experimental data: i.e., most favorable binding site for VWF in the FVIII (A3-C1 domains in the light chain). Our computational approach provided new hints about the involvement of domain B of FVIII as another putative, although less favorable, binding site for VWF. Additionally, we can hypothesize, given the accordance between different computational methods, that the most stable VWF/FVIII complex (efanesoctocog alfa) is characterized by most unfavorable interface energy between heavy and light chains of FVIII, paralleled by the most favorable VWF/FVIII interface, likely due to the involvement of the highest number of residues with high participation coefficient (i.e., allosteric residues). Overall, our computational approaches provided new hints on interdomain allosteric communication in proteins or protein–protein complexes, which are considered as one of the driving forces in the protein–protein binding stability. Thereby, our integrated computational approach will be helpful in the rational structure design of biologic drugs.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/app12157855/s1, Figure S1: Secondary structure plot of secreted full-length FVIII.
Author Contributions
Conceptualization, C.B.M.P. and V.D.; methodology, C.B.M.P. and L.D.P.; software, C.B.M.P., L.D.P., and C.B.; formal analysis, C.B.M.P.; C.L., and L.D.P.; Investigation, V.D., C.B.M.P., C.L., and L.D.P.; resources, C.B., V.D.; data curation, V.D., C.B.M.P., and R.B.; writing—original, draft preparation, C.B.M.P.; writing—review and editing, C.B.M.P., L.D.P., C.L., V.D., R.B., and C.B.; visualization, C.B.M.P.; supervision, C.B.M.P., V.D.; project administration, C.B.M.P., V.D.; funding acquisition, C.B.M.P., C.B. All authors have read and agreed to the published version of the manuscript.
Funding
C.B.M.P. has been supported by the funding PON Ricerca e Innovazione D.M. 1062/21–Contratti di ricerca, from the Italian Ministry of University (MUR). Contract #: 08-I-17629-2.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Structural models will be provided by the corresponding author upon request of the readers.
Acknowledgments
The manuscript is completed in memory of Salvatore Asero (BIOVIIIx), who contributed to conceptualization of the project. Authors wish to thank the company BIOVIIIx (Via Alessandro Manzoni 1, 80128 Napoli, Italy) for unrestricted support and fruitful scientific discussion. The manuscript is dedicated to Salvatore (Totò) Salomone, who spent his career in the field of cardiovascular pharmacology, and unfortunately departed too early from the Pharmacology section of the BIOMETEC department-University of Catania.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gringeri, A.; Wolfsegger, M.; Steinitz, K.N.; Reininger, A.J. Recombinant full-length factor VIII (FVIII) and extended half-life FVIII products in prophylaxis-new insight provided by pharmacokinetic modelling. Haemophilia 2015, 21, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Lieuw, K. Many factor VIII products available in the treatment of hemophilia a: An embarrassment of riches? J. Blood Med. 2017, 8, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Den Uijl, I.E.M.; Fischer, K.; Van Der Bom, J.G.; Grobbee, D.E.; Rosendaal, F.R.; Plug, I. Analysis of low frequency bleeding data: The association of joint bleeds according to baseline FVIII activity levels. Haemophilia 2011, 17, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.D.; Li, S. Response to Gringeri et al.: “Recombinant full-length factor VIII (FVIII) and extended half-life FVIII products in prophylaxis--new insight provided by pharmacokinetic modelling”. Haemophilia 2015, 21, e489–e492. [Google Scholar] [CrossRef]
- Bloem, E.; Karpf, D.M.; Nørby, P.L.; Johansen, P.B.; Loftager, M.; Rahbek-Nielsen, H.; Petersen, H.H.; Blouse, G.E.; Thim, L.; Kjalke, M.; et al. Factor VIII with a 237 amino acid B-domain has an extended half-life in F8-knockout mice. J. Thromb. Haemost. 2019, 17, 350–360. [Google Scholar] [CrossRef]
- Pipe, S.W.; Montgomery, R.R.; Pratt, K.P.; Lenting, P.J.; Lillicrap, D. Life in the shadow of a dominant partner: The FVIII-VWF association and its clinical implications for hemophilia A. Blood 2016, 128, 2007–2016. [Google Scholar] [CrossRef]
- Shi, Q.; Kuether, E.L.; Schroeder, J.A.; Perry, C.L.; Fahs, S.A.; Gill, J.C.; Montgomery, R.R. Factor VIII inhibitors: Von Willebrand factor makes a difference in vitro and in vivo. J. Thromb. Haemost. 2012, 10, 2328–2337. [Google Scholar] [CrossRef]
- Vollack-Hesse, N.; Oleshko, O.; Werwitzke, S.; Solecka-Witulska, B.; Kannicht, C.; Tiede, A. Recombinant VWF Fragments Improve Bioavailability of Subcutaneous Factor VIII in Hemophilia A Mice. Blood 2020, 137, 1072–1081. [Google Scholar] [CrossRef]
- Yee, A.; Gildersleeve, R.D.; Gu, S.; Kretz, C.A.; McGee, B.M.; Carr, K.M.; Pipe, S.W.; Ginsburg, D. A von Willebrand factor fragment containing the D′D3 domains is sufficient to stabilize coagulation factor VIII in mice. Blood 2014, 124, 445–452. [Google Scholar] [CrossRef]
- Saenko, E.L.; Scandella, D. The acidic region of the factor VIII light chain and the C2 domain together form the high affinity binding site for von Willebrand factor. J. Biol. Chem. 1997, 272, 18007–18014. [Google Scholar] [CrossRef]
- Dagil, L.; Troelsen, K.S.; Bolt, G.; Thim, L.; Wu, B.; Zhao, X.; Tuddenham, E.G.; Nielsen, T.E.; Tanner, D.A.; Faber, J.H.; et al. Interaction Between the a3 Region of Factor VIII and the TIL’E’ Domains of the von Willebrand Factor. Biophys. J. 2019, 117, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, G.E.; Kaufman, R.J.; Arena, A.A.; Miao, H.; Pipe, S.W. Four hydrophobic amino acids of the factor VIII C2 domain are constituents of both the membrane-binding and von Willebrand factor-binding motifs. J. Biol. Chem. 2002, 277, 6374–6381. [Google Scholar] [CrossRef] [PubMed]
- Fuller, J.R.; Knockenhauer, K.E.; Leksa, N.C.; Peters, R.T.; Batchelor, J.D. Molecular determinants of the factor VIII/von Willebrand factor complex revealed by BIVV001 cryo-electron microscopy. Blood 2021, 137, 2970–2980. [Google Scholar] [CrossRef]
- Pipe, S.W. Functional roles of the factor VIII B domain. Haemophilia 2009, 15, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, M.A.; Patel, H.; Rouse, J.C.; Marzilli, L.A.; Weston, S.B.; Sharpe, P.J. Defining “full-length” recombinant factor VIII: A comparative structural analysis. Haemophilia 2007, 13, 30–37. [Google Scholar] [CrossRef]
- Anzengruber, J.; Feichtinger, M.; Bärnthaler, P.; Haider, N.; Ilas, J.; Pruckner, N.; Benamara, K.; Scheiflinger, F.; Reipert, B.M.; Malisauskas, M. How Full-Length FVIII Benefits from Its Heterogeneity–Insights into the Role of the B-Domain. Pharm. Res. 2019, 36, 77. [Google Scholar] [CrossRef]
- Moussa, E.M.; Panchal, J.P.; Moorthy, B.S.; Blum, J.S.; Joubert, M.K.; Narhi, L.O.; Topp, E.M. Immunogenicity of Therapeutic Protein Aggregates. J. Pharm. Sci. 2016, 105, 417–430. [Google Scholar] [CrossRef]
- Roberts, C.J. Therapeutic protein aggregation: Mechanisms, design, and control. Trends Biotechnol. 2014, 32, 372–380. [Google Scholar] [CrossRef]
- Joubert, M.K.; Luo, Q.; Nashed-Samuel, Y.; Wypych, J.; Narhi, L.O. Classification and characterization of therapeutic antibody aggregates. J. Biol. Chem. 2011, 286, 25118–25133. [Google Scholar] [CrossRef]
- Schiavoni, M.; Napolitano, M.; Giuffrida, G.; Coluccia, A.; Siragusa, S.; Calafiore, V.; Lassandro, G.; Giordano, P. Status of Recombinant Factor VIII Concentrate Treatment for Hemophilia a in Italy: Characteristics and Clinical Benefits. Front. Med. 2019, 6, 261. [Google Scholar] [CrossRef]
- Vlot, A.; Koppelman, S.; Berg, M.V.D.; Bouma, B.; Sixma, J. The affinity and stoichiometry of binding of human factor VIII to von Willebrand factor. Blood 1995, 85, 3150–3157. [Google Scholar] [CrossRef] [PubMed]
- Lollar, P.; Parker, C.G. Stoichiometry of the porcine factor VIII-von Willebrand factor association. J. Biol. Chem. 1987, 262, 17572–17576. [Google Scholar] [CrossRef]
- Fischer, B.E.; Kramer, G.; Mitterer, A.; Grillberger, L.; Reiter, M.; Mundt, W.; Dorner, F.; Eibl, J. Effect of multimerization of human and recombinant von Willebrand factor on platelet aggregation, binding to collagen and binding of coagulation factor VIII. Thromb. Res. 1996, 84, 55–66. [Google Scholar] [CrossRef]
- Fischer, B.E.; Schlokat, U.; Reiter, M.; Mundt, W.; Dorner, F. Biochemical and functional characterization of recombinant von Willebrand factor produced on a large scale. Cell. Mol. Life Sci. 1997, 53, 943–950. [Google Scholar] [CrossRef]
- Grushin, K.; Miller, J.; Dalm, D.; Parker, E.T.; Healey, J.F.; Lollar, P.; Stoilova-McPhie, S. Lack of recombinant factor VIII B-domain induces phospholipid vesicle aggregation: Implications for the immunogenicity of factor VIII. Haemophilia 2014, 20, 723–731. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Di Paola, L.; Platania, C.B.M.; Oliva, G.; Setola, R.; Pascucci, F.; Giuliani, A. Characterization of Protein-Protein Interfaces through a Protein Contact Network Approach. Front. Bioeng. Biotechnol. 2015, 3, 170. [Google Scholar] [CrossRef][Green Version]
- Di Paola, L.; Paci, P.; Santoni, D.; De Ruvo, M.; Giuliani, A. Proteins as sponges: A statistical journey along protein structure organization principles. J. Chem. Inf. Model. 2012, 52, 474–482. [Google Scholar] [CrossRef]
- Hu, G.; Di Paola, L.; Liang, Z.; Giuliani, A. Comparative Study of Elastic Network Model and Protein Contact Network for Protein Complexes: The Hemoglobin Case. BioMed Res. Int. 2017, 2017, 2483264. [Google Scholar] [CrossRef]
- Minicozzi, V.; Di Venere, A.; Nicolai, E.; Giuliani, A.; Caccuri, A.M.; Di Paola, L.; Mei, G. Non-symmetrical structural behavior of a symmetric protein: The case of homo-trimeric TRAF2 (tumor necrosis factor-receptor associated factor 2). J. Biomol. Struct. Dyn. 2020, 39, 319–329. [Google Scholar] [CrossRef]
- Platania, C.B.M.; Di Paola, L.; Leggio, G.M.; Romano, G.L.; Drago, F.; Salomone, S.; Bucolo, C. Molecular features of interaction between VEGFA and anti-angiogenic drugs used in retinal diseases: A computational approach. Front. Pharmacol. 2015, 6, 248. [Google Scholar] [CrossRef] [PubMed]
- Platania, C.B.M.; Ronchetti, S.; Riccardi, C.; Migliorati, G.; Marchetti, M.C.; Di Paola, L.; Lazzara, F.; Drago, F.; Salomone, S.; Bucolo, C. Effects of protein-protein interface disruptors at the ligand of the glucocorticoid-induced tumor necrosis factor receptor-related gene (GITR). Biochem. Pharmacol. 2020, 178, 114110. [Google Scholar] [CrossRef] [PubMed]
- Platania, C.B.M.; Bucolo, C. Molecular Dynamics Simulation Techniques as Tools in Drug Discovery and Pharmacology: A Focus on Allosteric Drugs. Methods Mol. Biol. 2021, 2253, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Di Paola, L.; Hadi-Alijanvand, H.; Song, X.; Hu, G.; Giuliani, A. The Discovery of a Putative Allosteric Site in the SARS-CoV-2 Spike Protein Using an Integrated Structural/Dynamic Approach. J. Proteome Res. 2020, 19, 4576–4586. [Google Scholar] [CrossRef]
- Verkhivker, G.M.; Di Paola, L. Integrated Biophysical Modeling of the SARS-CoV-2 Spike Protein Binding and Allosteric Interactions with Antibodies. J. Phys. Chem. B 2021, 125, 4596–4619. [Google Scholar] [CrossRef]
- Verkhivker, G.M.; Di Paola, L. Dynamic Network Modeling of Allosteric Interactions and Communication Pathways in the SARS-CoV-2 Spike Trimer Mutants: Differential Modulation of Conformational Landscapes and Signal Transmission via Cascades of Regulatory Switches. J. Phys. Chem. B 2021, 125, 850–873. [Google Scholar] [CrossRef]
- Hadi-Alijanvand, H.; Di Paola, L.; Hu, G.; Leitner, D.M.; Verkhivker, G.M.; Sun, P.; Poudel, H.; Giuliani, A. Biophysical Insight into the SARS-CoV2 Spike-ACE2 Interaction and Its Modulation by Hepcidin through a Multifaceted Computational Approach. ACS Omega 2022, 7, 17024–17042. [Google Scholar] [CrossRef]
- Rosell, M.; Rodríguez-Lumbreras, L.A.; Romero-Durana, M.; Jiménez-García, B.; Díaz, L.; Fernández-Recio, J. Integrative modeling of protein-protein interactions with pyDock for the new docking challenges. Proteins Struct. Funct. Bioinform. 2020, 88, 999–1008. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, 32, 514–3678. [Google Scholar] [CrossRef]
- Cimini, S.; Di Paola, L.; Giuliani, A.; Ridolfi, A.; De Gara, L. GH32 family activity: A topological approach through protein contact networks. Plant Mol. Biol. 2016, 92, 401–410. [Google Scholar] [CrossRef]
- Di Paola, L.; Mei, G.; Di Venere, A.; Giuliani, A. Exploring the stability of dimers through protein structure topology. Curr. Protein Pept. Sci. 2016, 17, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Mei, G.; Di Venere, A.; Rosato, N.; Finazzi-Agrò, A. The importance of being dimeric. FEBS J. 2005, 272, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Tasdighian, S.; Di Paola, L.; De Ruvo, M.; Paci, P.; Santoni, D.; Palumbo, P.; Mei, G.; Di Venere, A.; Giuliani, A. Modules identification in protein structures: The topological and geometrical solutions. J. Chem. Inf. Model. 2014, 54, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Di Venere, A.; Nicolai, E.; Minicozzi, V.; Caccuri, A.; Di Paola, L.; Mei, G. The odd faces of oligomers: The case of traf2-c, a trimeric c-terminal domain of tnf receptor-associated factor. Int. J. Mol. Sci. 2021, 22, 5871. [Google Scholar] [CrossRef]
- Guzzi, P.H.; Di Paola, L.; Giuliani, A.; Veltri, P. PCN-Miner: An open-source extensible tool for the Analysis of Protein Contact Networks. Bioinformatics 2022, 7, btac450. [Google Scholar] [CrossRef]
- Shiltagh, N.; Kirkpatrick, J.; Cabrita, L.D.; McKinnon, T.A.J.; Thalassinos, K.; Tuddenham, E.G.D.; Hansen, D.F. Solution structure of the major factor VIII binding region on von Willebrand factor. Blood 2014, 123, 4143–4151. [Google Scholar] [CrossRef]
- Chiu, P.-L.; Bou-Assaf, G.M.; Chhabra, E.S.; Chambers, M.G.; Peters, R.T.; Kulman, J.D.; Walz, T. Mapping the interaction between factor VIII and von Willebrand factor by electron microscopy and mass spectrometry. Blood 2015, 126, 935–938. [Google Scholar] [CrossRef]
- World Health Organization. WHO Model List of Essential Medicines. 2021. Available online: https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2021.02 (accessed on 1 June 2022).
- Kessler, C.M.; Gill, J.C.; White, G.C.; Shapiro, A.; Arkin, S.; Roth, D.A.; Meng, X.; Lusher, J.M. B-domain deleted recombinant factor VIII preparations are bioequivalent to a monoclonal antibody purified plasma-derived factor VIII concentrate: A randomized, three-way crossover study. Haemophilia 2005, 11, 84–91. [Google Scholar] [CrossRef]
- Przeradzka, M.A.; Van Galen, J.; Ebberink, E.H.T.M.; Hoogendijk, A.J.; Van Der Zwaan, C.; Mertens, K.; Biggelaar, M.V.D.; Meijer, A.B. D’ domain region Arg782-Cys799 of von Willebrand factor contributes to factor VIII binding. Haematologica 2020, 105, 1695–1703. [Google Scholar] [CrossRef]
- Shen, B.W.; Spiegel, P.C.; Chang, C.-H.; Huh, J.-W.; Lee, J.-S.; Kim, J.; Kim, Y.-H.; Stoddard, B.L. The tertiary structure and domain organization of coagulation factor VIII. Blood 2008, 111, 1240–1247. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).