Development of a Convolutional Neural Network Model to Predict Coronary Artery Disease Based on Single-Lead and Twelve-Lead ECG Signals
Abstract
1. Introduction
1.1. Digitization and Feature Extraction
1.2. Neural Network
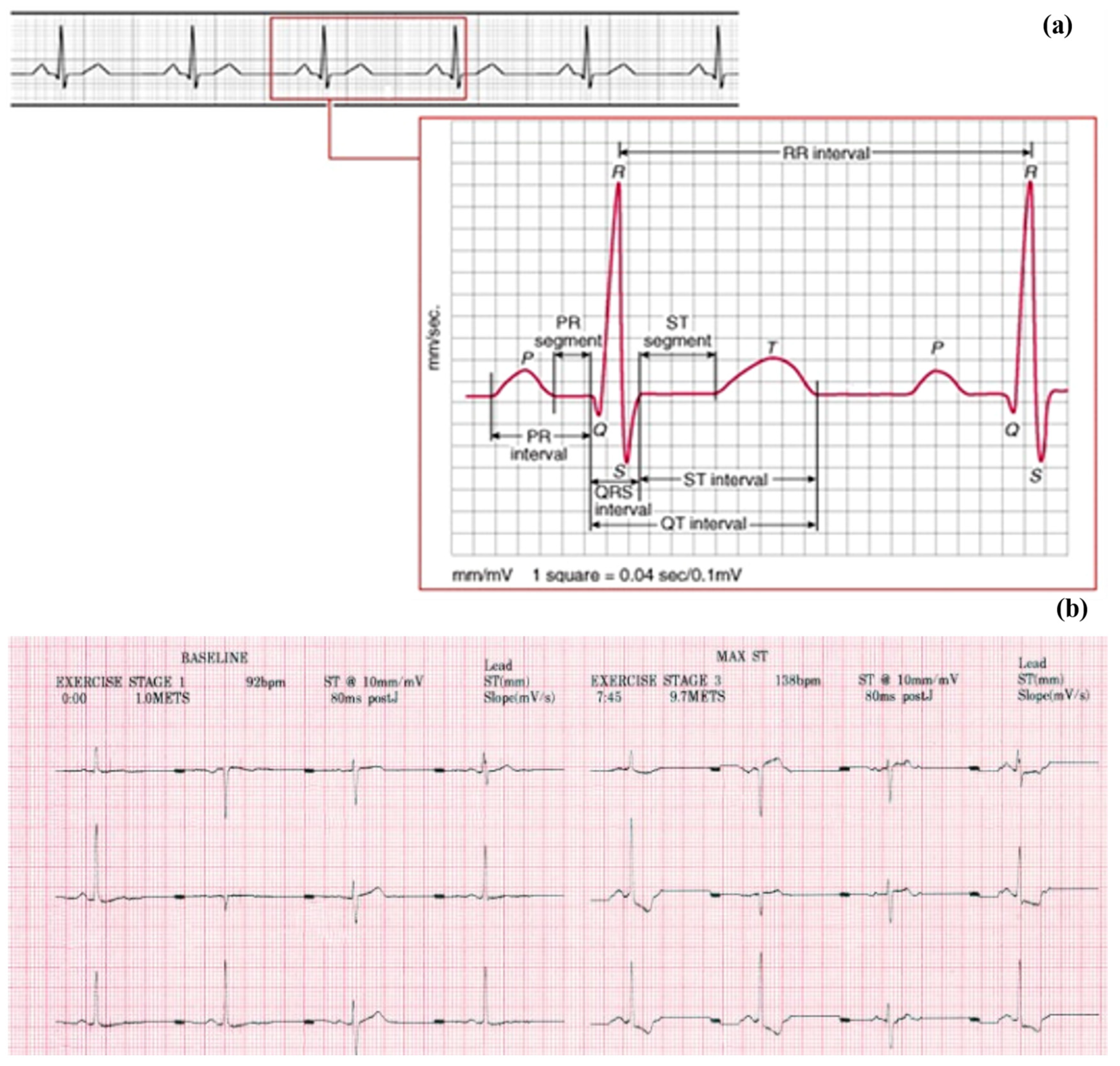
2. Methodology
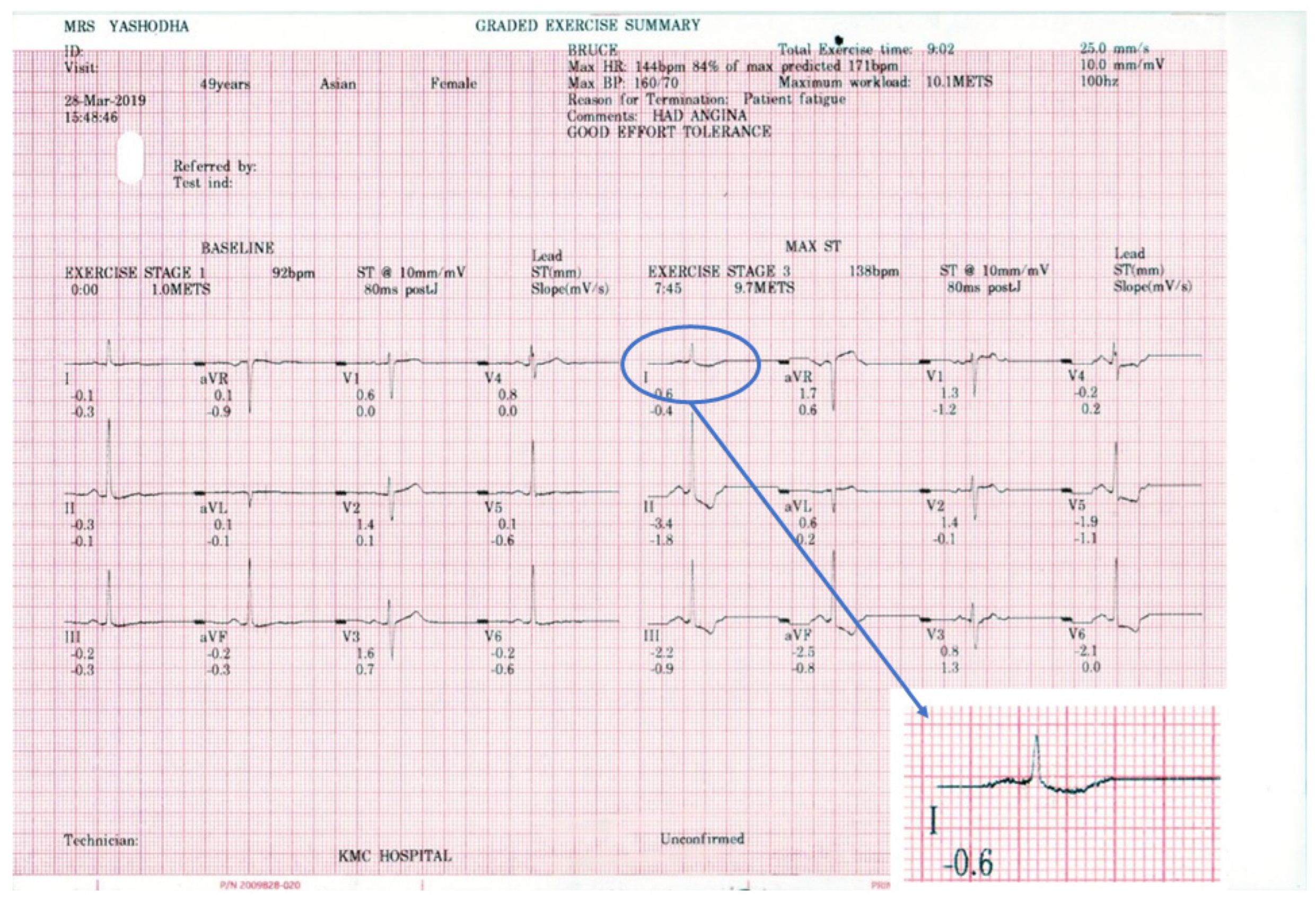
2.1. Dataset
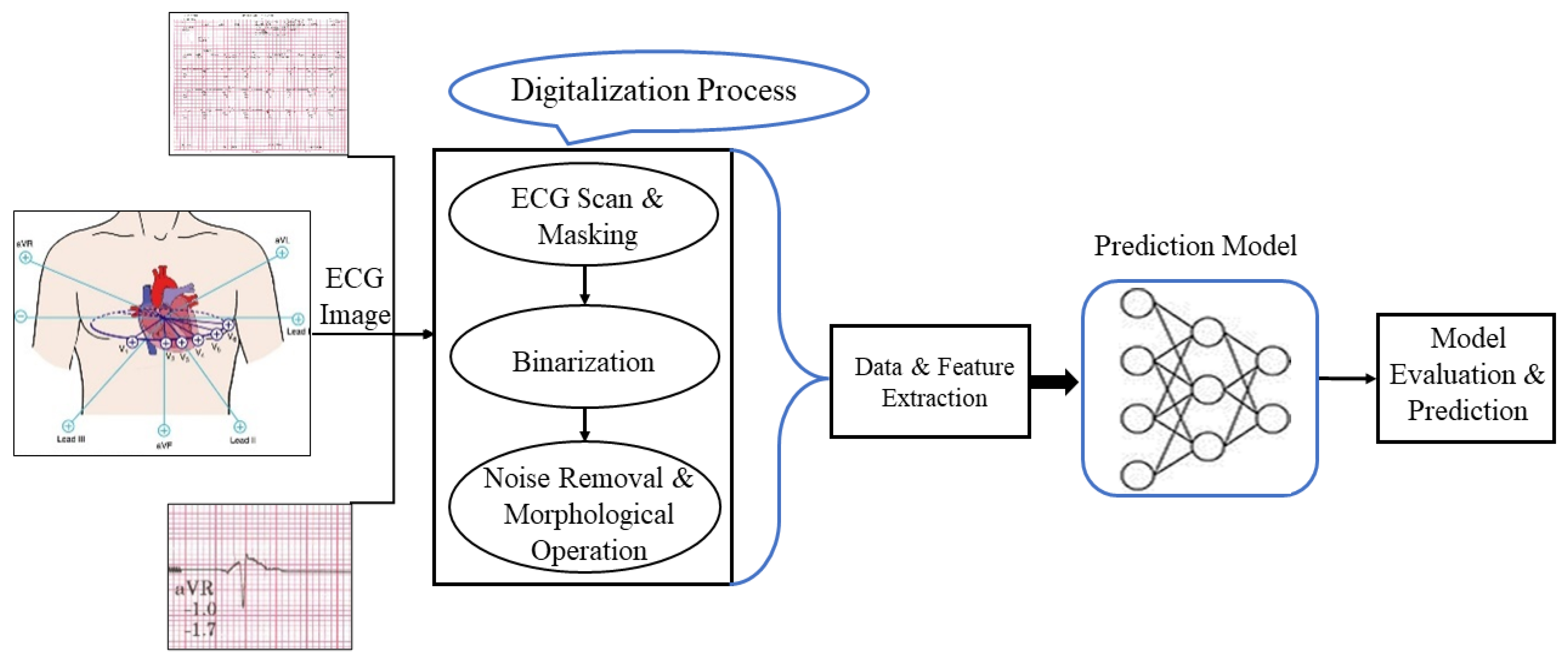
2.2. Data Extraction
2.2.1. Image Scanning
2.2.2. Image Masking
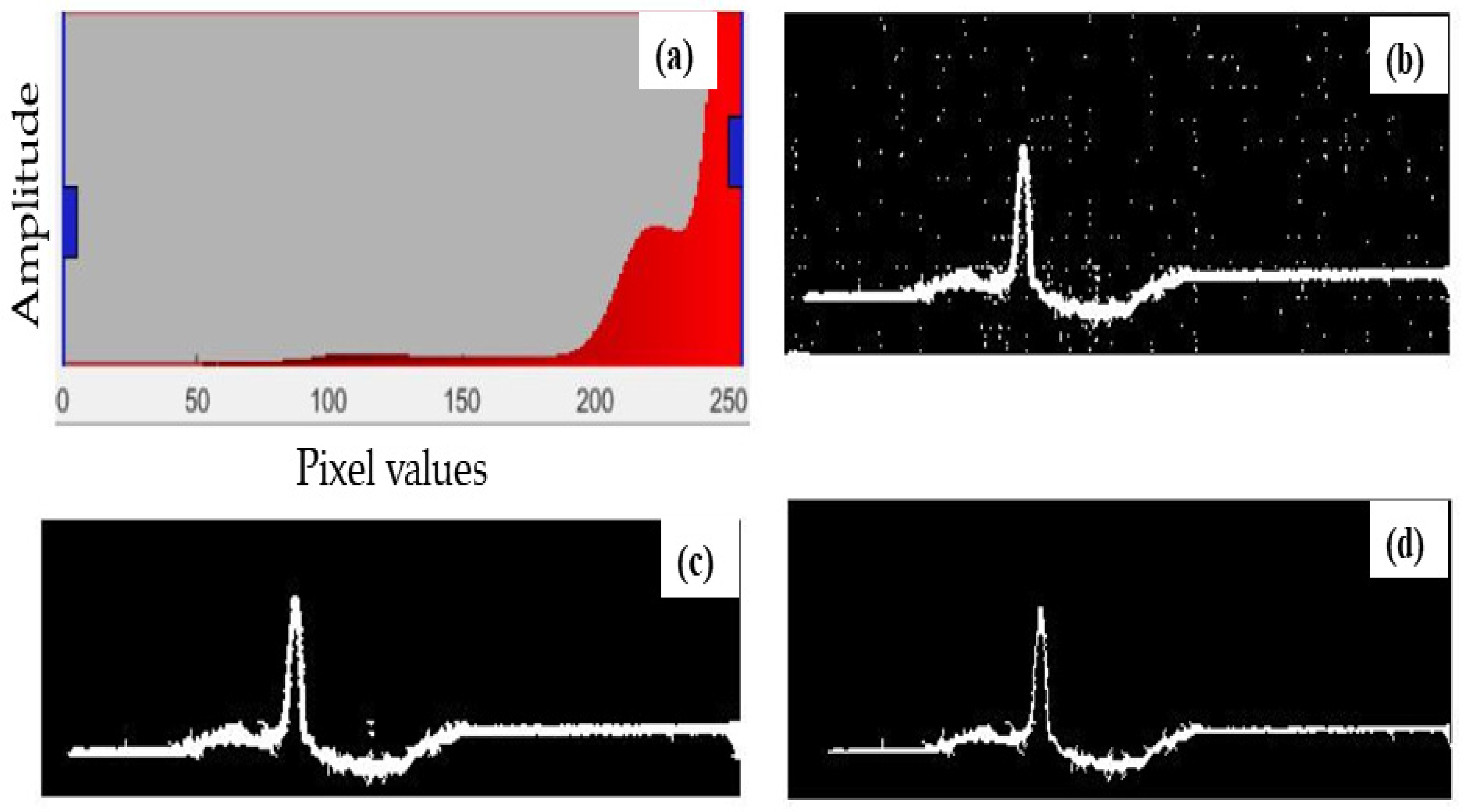
2.2.3. Defining a Threshold Value
2.2.4. Binarization
2.2.5. Open Area Deletion
2.2.6. Morphological Operation
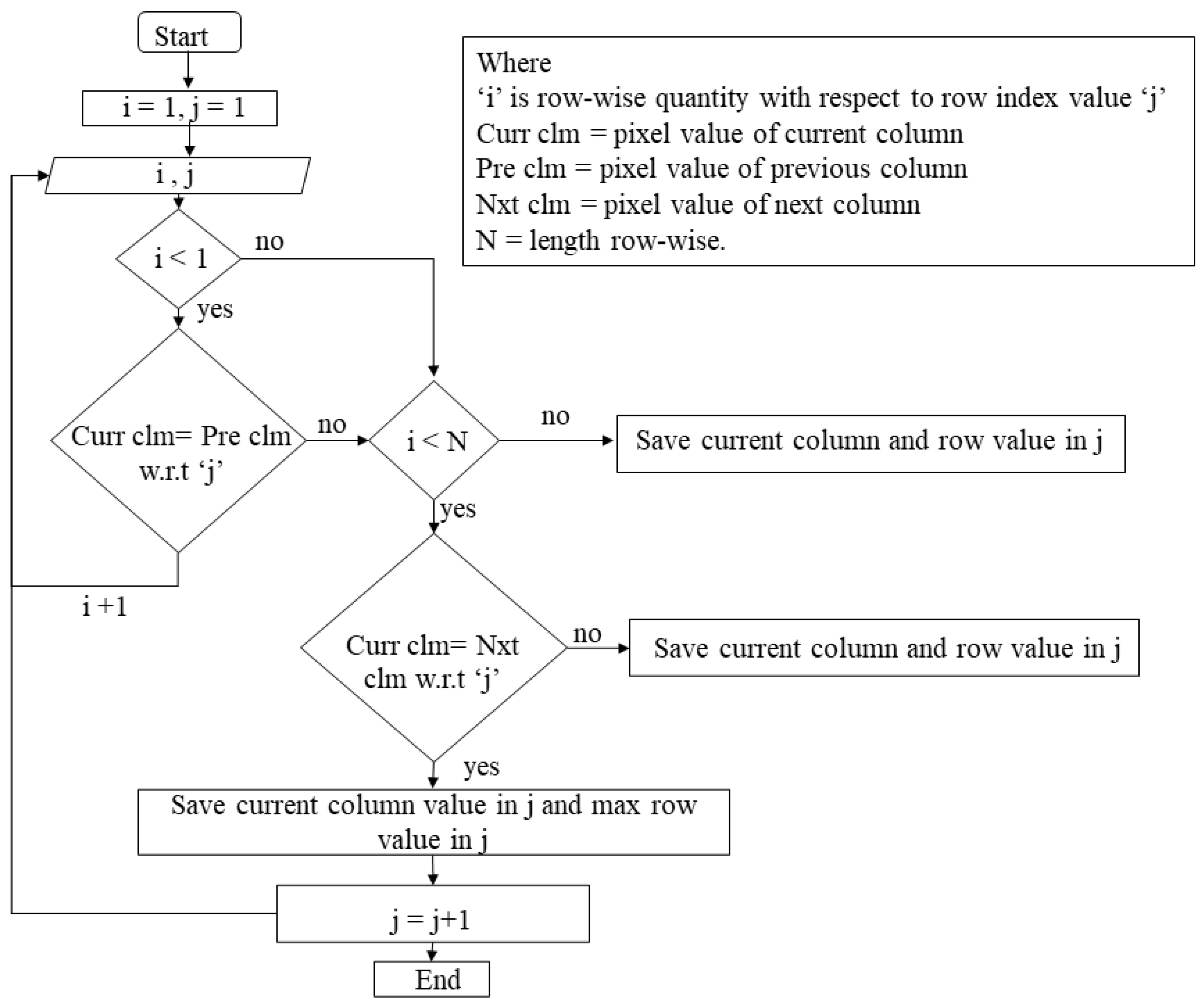
2.2.7. Pixel Indexing
2.2.8. Data Storage
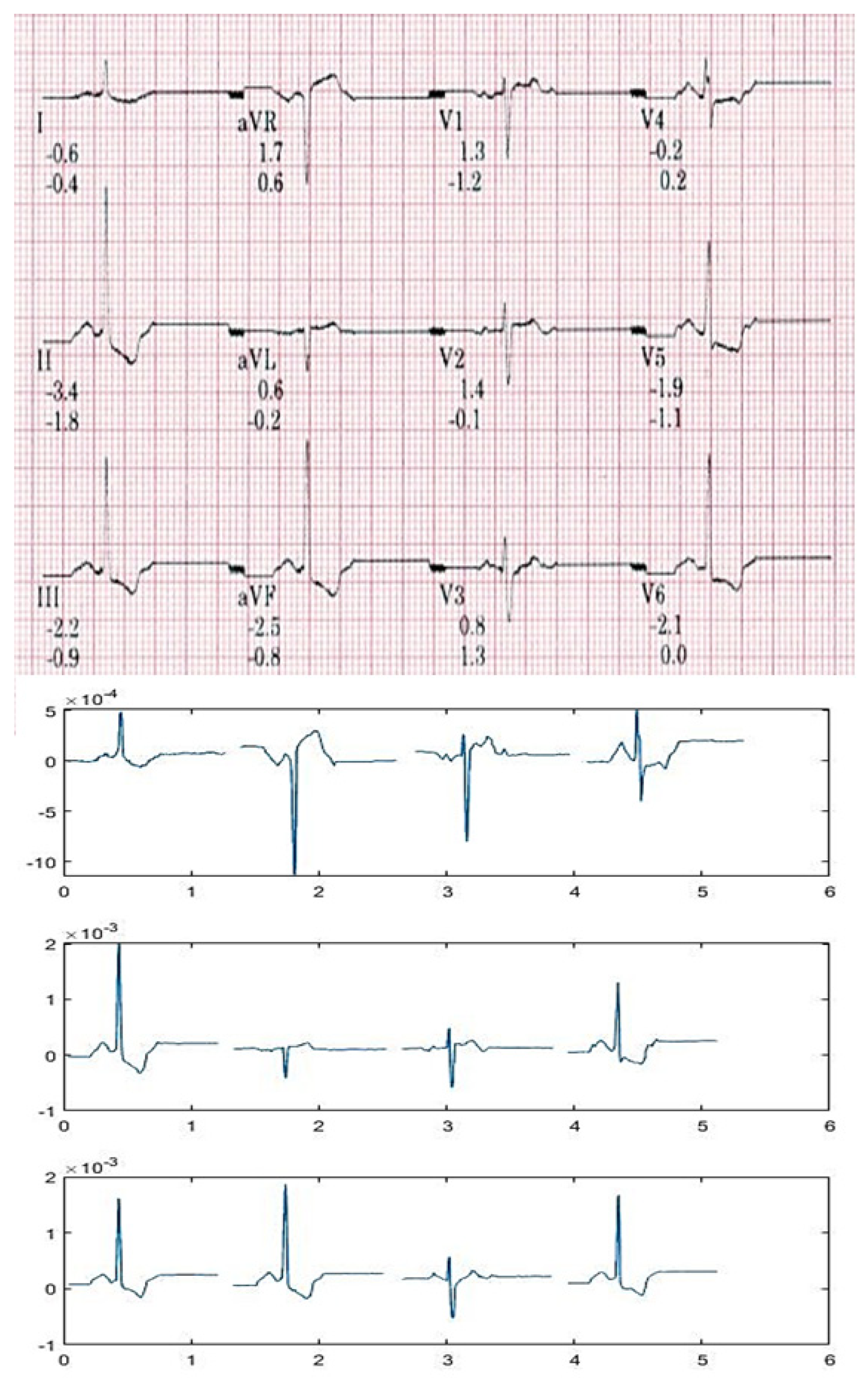
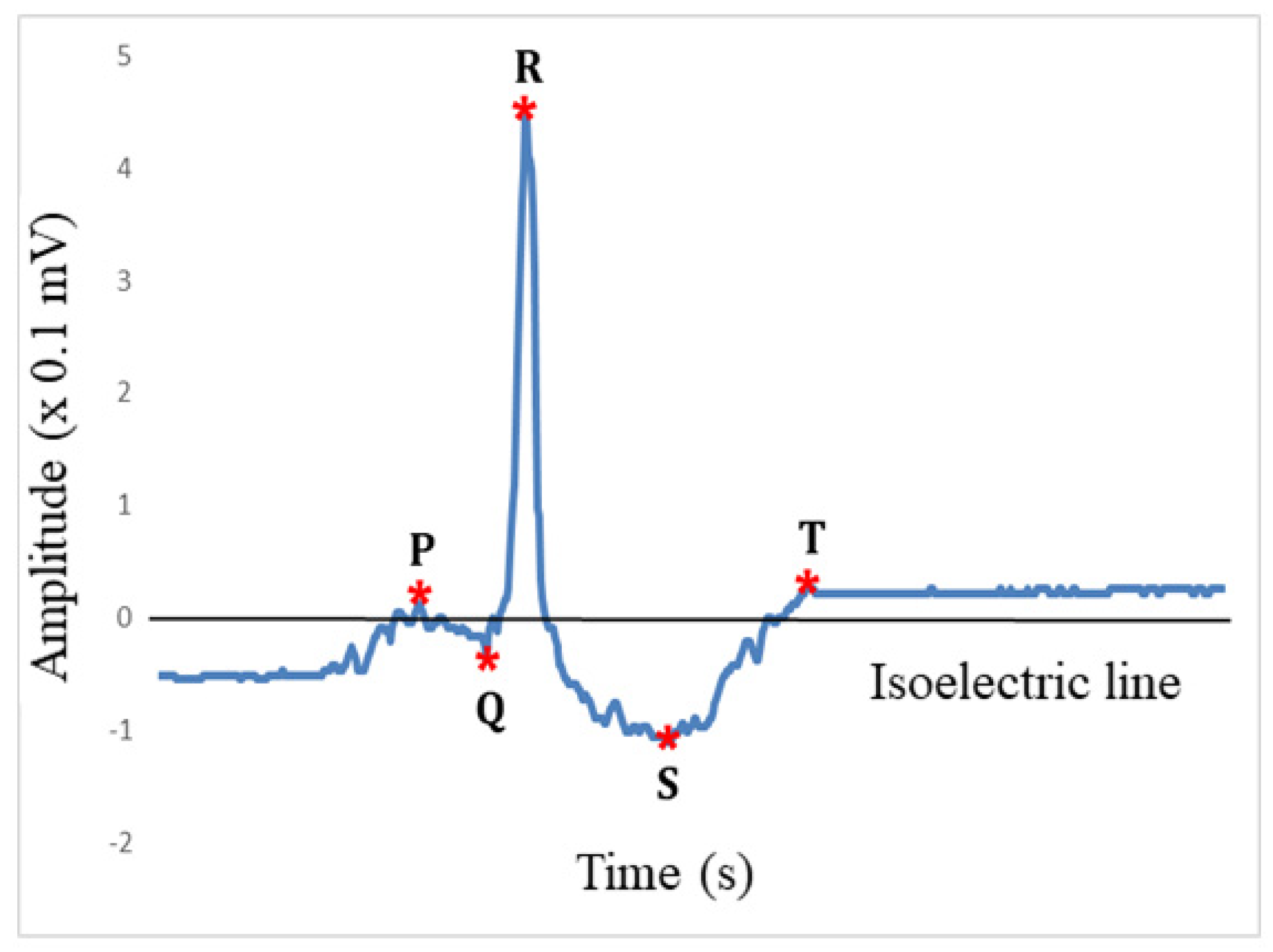
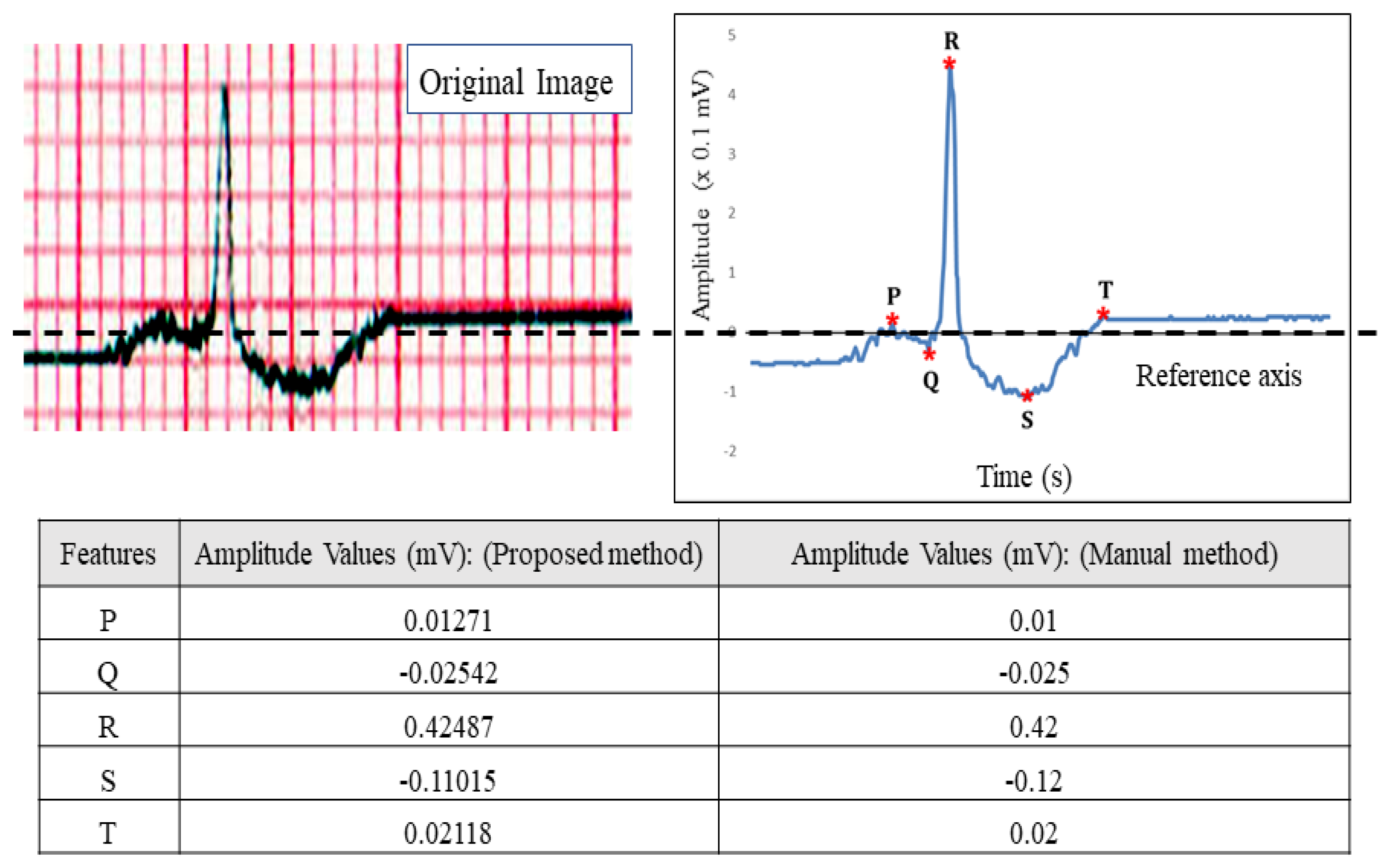
2.2.9. Feature Extraction
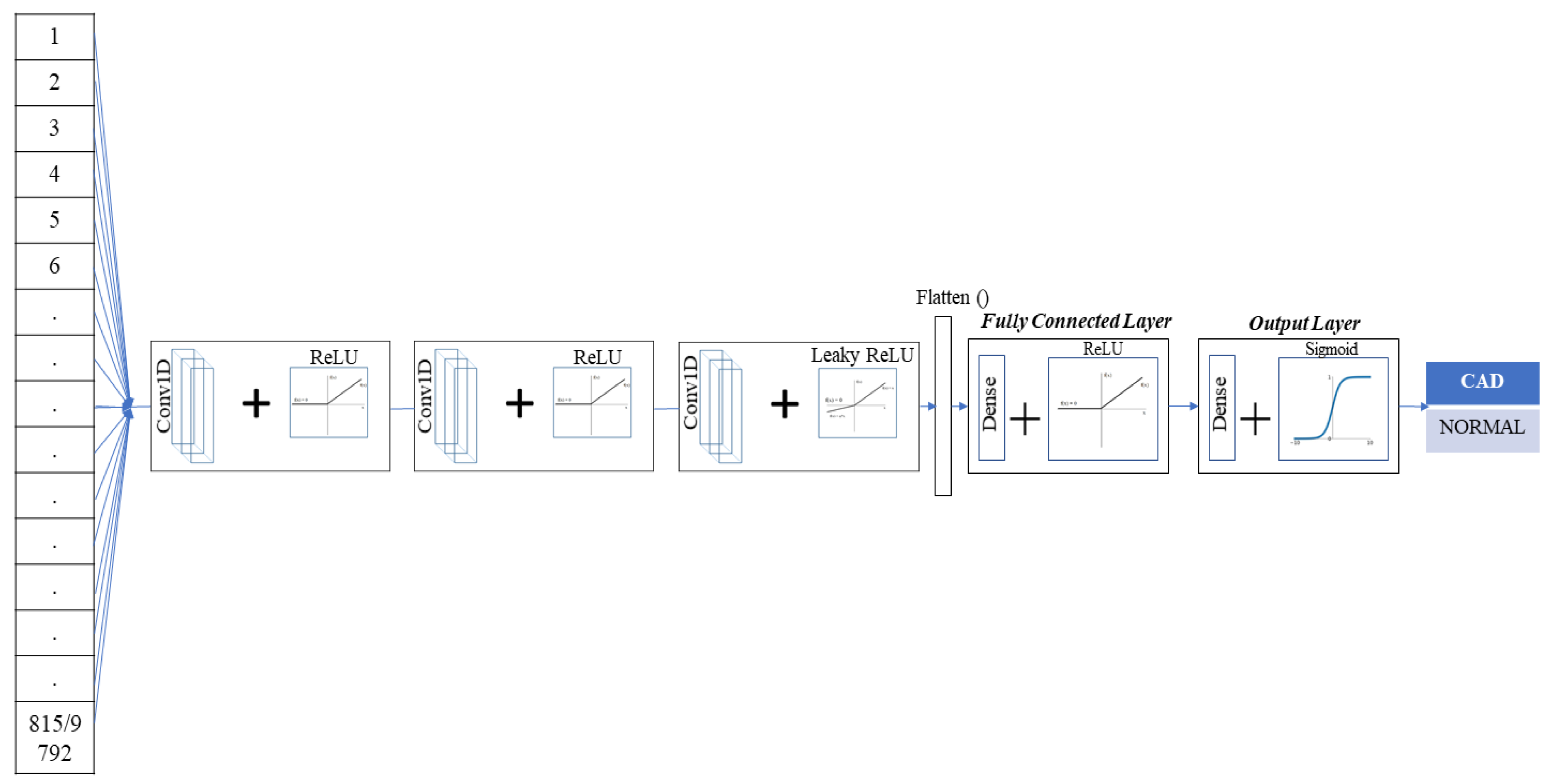
2.3. Prediction System Architecture
2.4. Single-Lead ECG/TMT-ECG-Based CNN Architecture
2.5. Multi (12)-Lead ECG/TMT-ECG with CNN Architecture
3. Results and Discussion
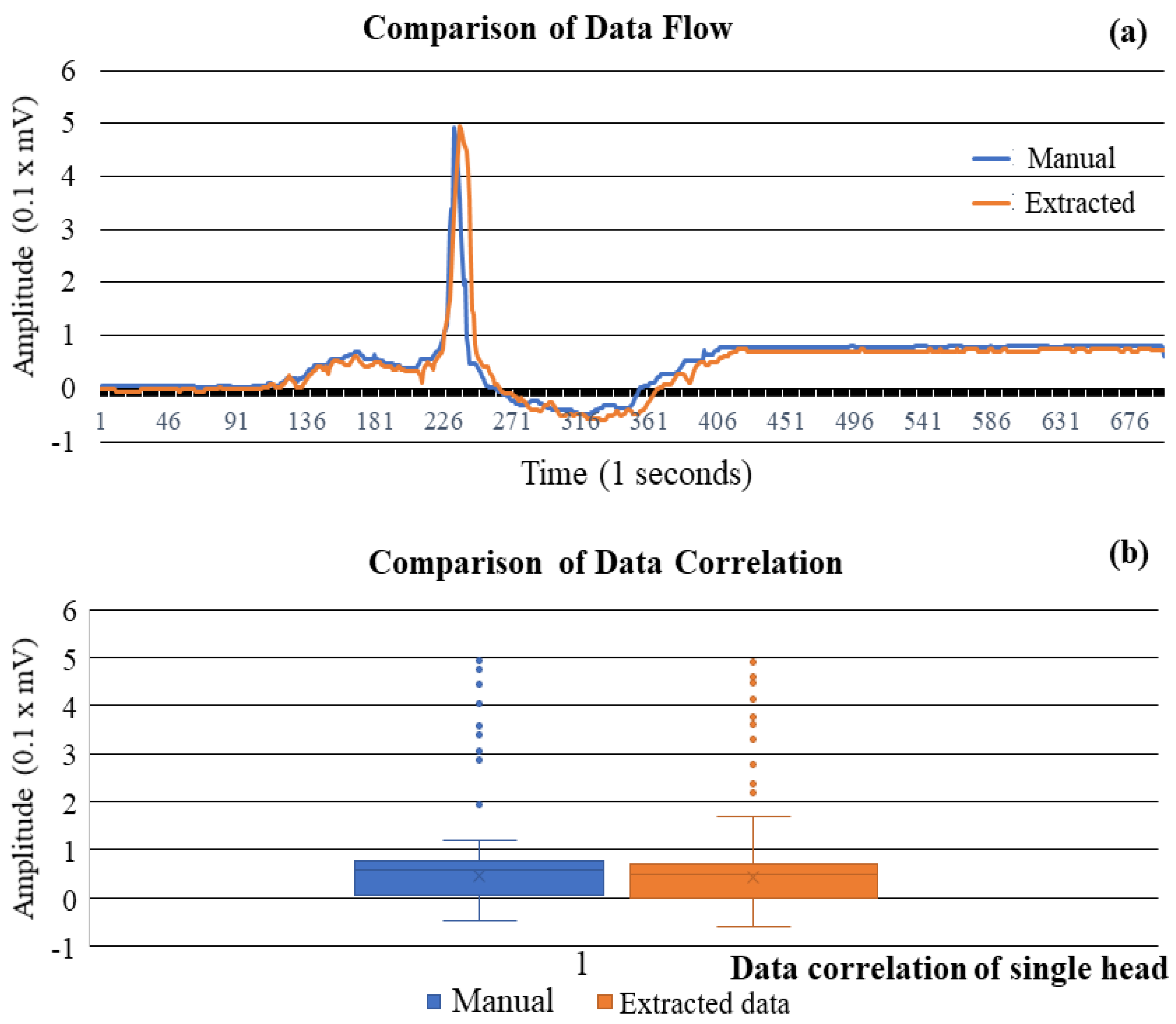
3.1. Correlation Study of Signal Data
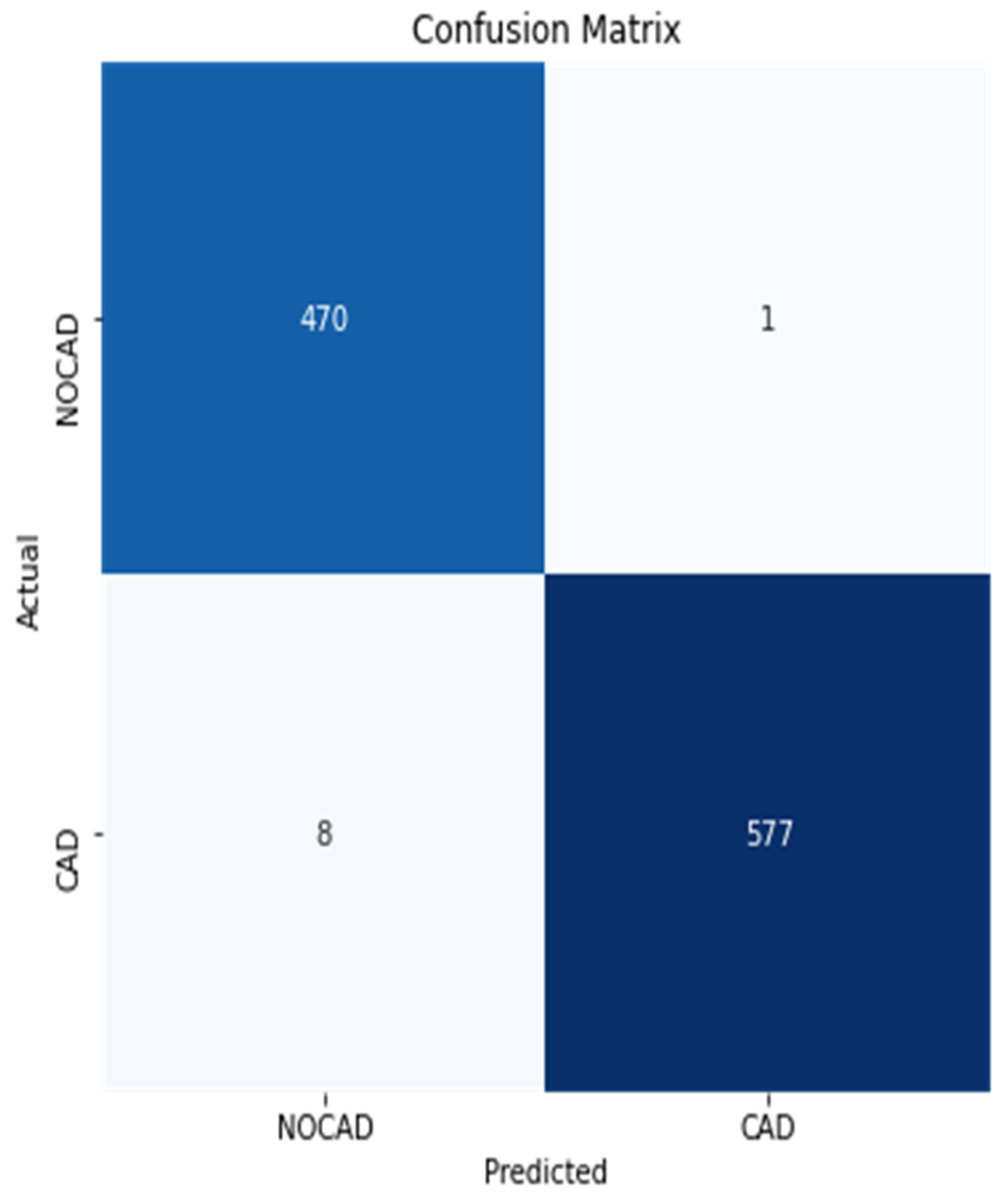
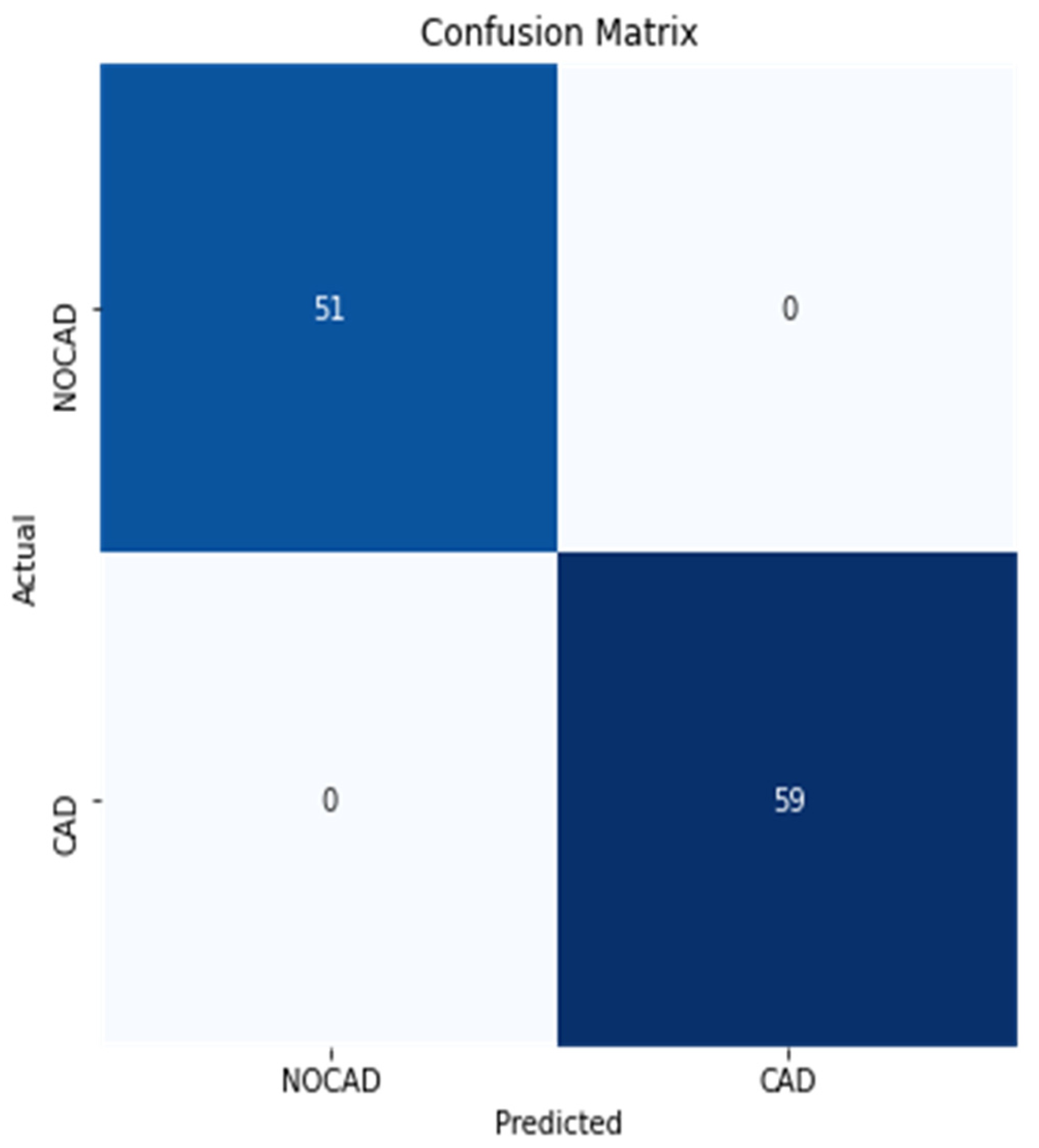
3.2. Performance Metrics Evaluation
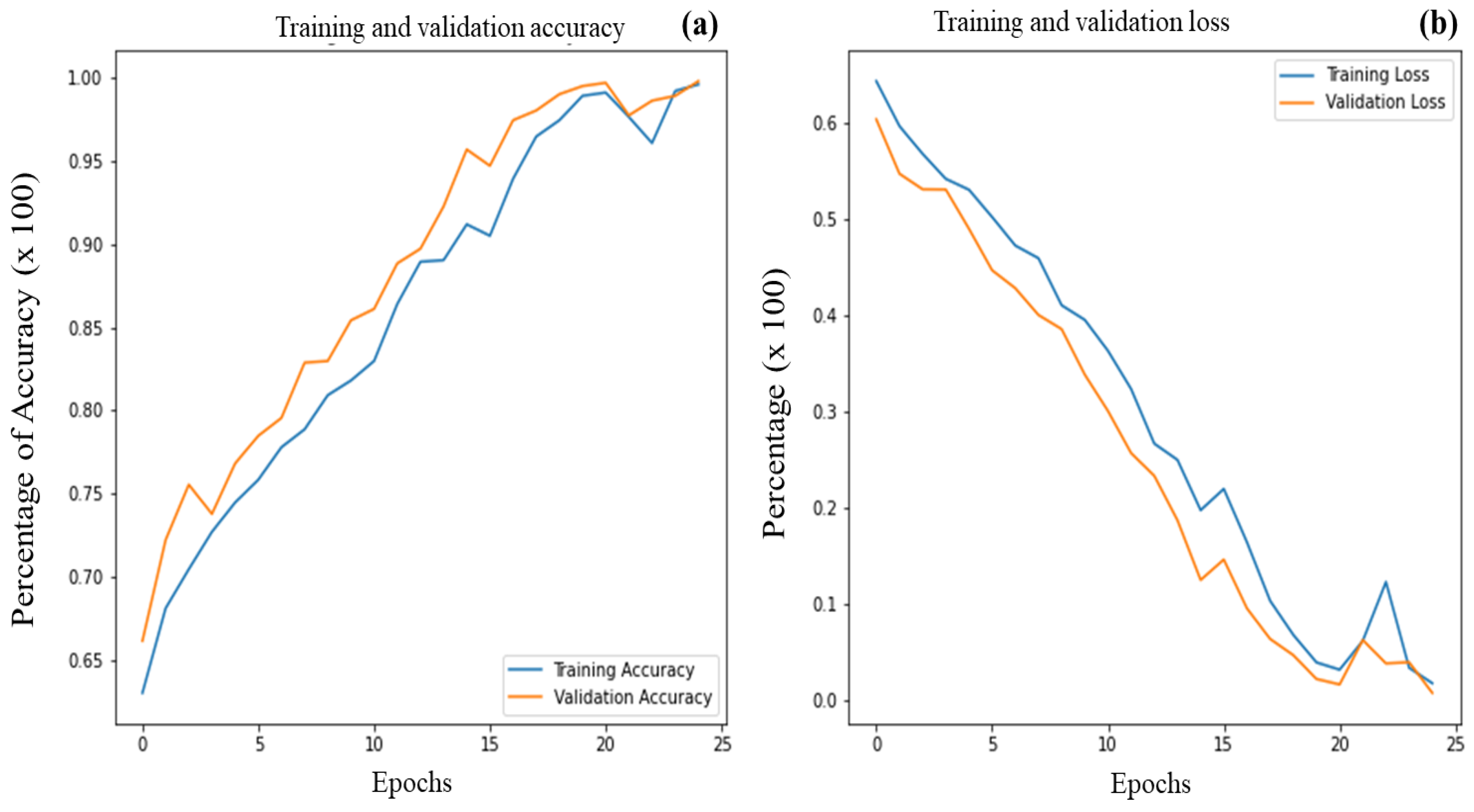
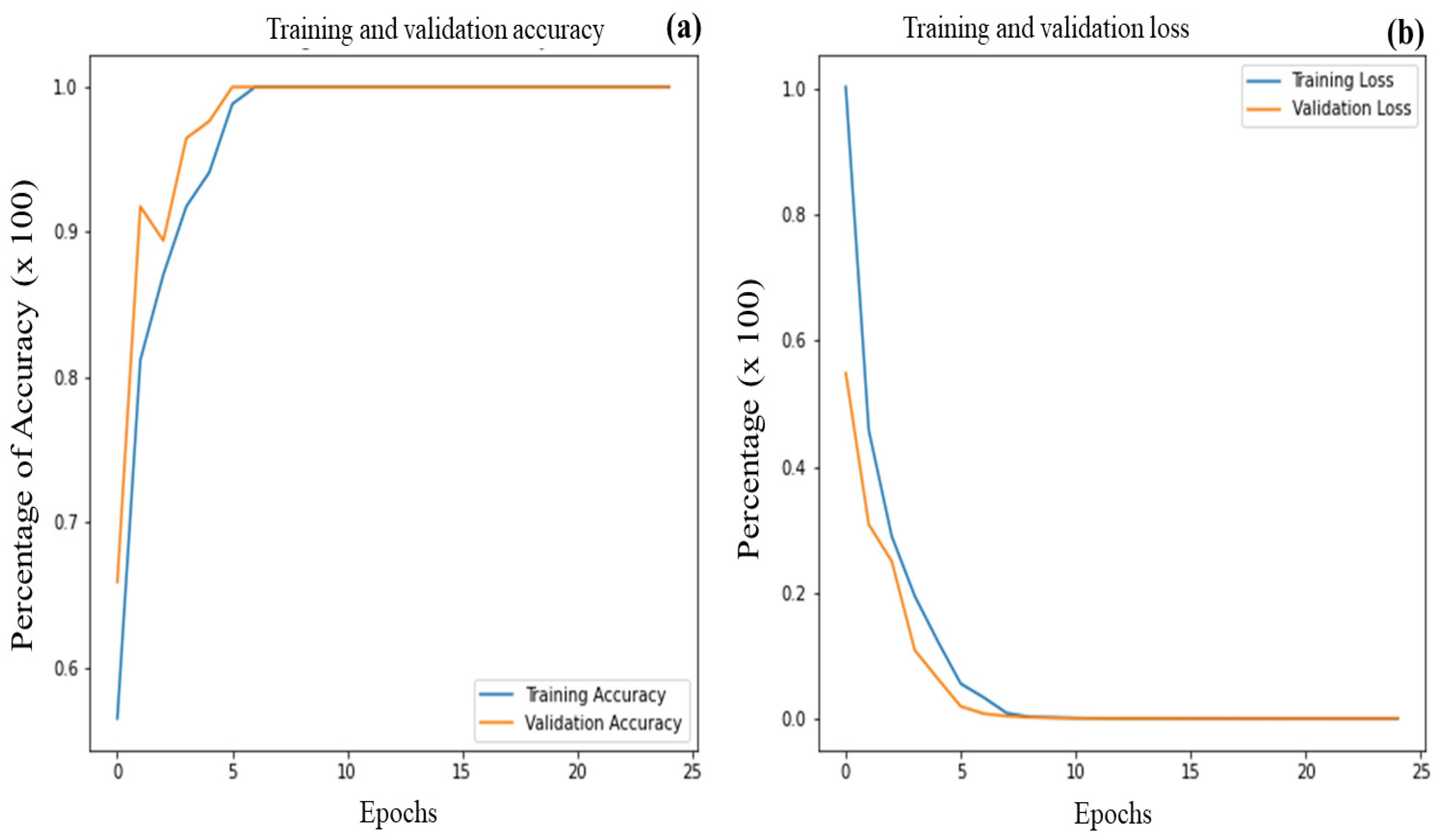
3.3. Identification of the Optimal Model
3.4. k-Fold Cross Validation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, A.; Christodorescu, R.; Agbariah, A.; Duda-Seiman, D.; Dahdal, D.; Man, D.; Kundnani, N.R.; Cretu, O.M.; Dragan, S. Cardiovascular Risk Prediction Parameters for Better Management in Rheumatic Diseases. Healthcare 2022, 10, 312. [Google Scholar] [CrossRef] [PubMed]
- El-Khafif, S.H.; El-Brawany, M.A. Artificial Neural Network-Based Automated ECG Signal Classifier. ISRN Biomed. Eng. 2013, 2013, 261917. [Google Scholar] [CrossRef]
- Mathews, S.M.; Kambhamettu, C.; Barner, K.E. A novel application of deep learning for single-lead ECG classification. Comput. Biol. Med. 2018, 99, 53–62. [Google Scholar] [CrossRef]
- Rubin, J.; Parvaneh, S.; Rahman, A.; Conroy, B.; Babaeizadeh, S. Densely connected convolutional networks for detection of atrial fibrillation from short single-lead ECG recordings. J. Electrocardiol. 2018, 51, S18–S21. [Google Scholar] [CrossRef]
- Fan, X.; Hu, Z.; Wang, R.; Yin, L.; Li, Y.; Cai, Y. A novel hybrid network of fusing rhythmic and morphological features for atrial fibrillation detection on mobile ECG signals. Neural Comput. Appl. 2020, 32, 8101–8113. [Google Scholar] [CrossRef]
- Chen, T.M.; Huang, C.H.; Shih, E.S.C.; Hu, Y.F.; Hwang, M.J. Detection and Classification of Cardiac Arrhythmias by a Challenge-Best Deep Learning Neural Network Model. iScience 2020, 23, 100886. [Google Scholar] [CrossRef]
- Liang, Y.; Yin, S.; Tang, Q.; Zheng, Z.; Elgendi, M.; Chen, Z. Deep Learning Algorithm Classifies Heartbeat Events Based on Electrocardiogram Signals. Front. Physiol. 2020, 11, 569050. [Google Scholar] [CrossRef]
- Ribeiro, A.H.; Ribeiro, M.H.; Paixão, G.M.M.; Oliveira, D.M.; Gomes, P.R.; Canazart, J.A.; Ferreira, M.P.S.; Andersson, C.R.; Macfarlane, P.W.; Wagner, M.; et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 2020, 11, 1760. [Google Scholar] [CrossRef]
- Sassi, R.; Bond, R.R.; Cairns, A.; Finlay, D.D.; Guldenring, D.; Libretti, G.; Isola, L.; Vaglio, M.; Poeta, R.; Campana, M.; et al. PDF–ECG in clinical practice: A model for long–term preservation of digital 12–lead ECG data. J. Electrocardiol. 2017, 50, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.H.; Hagiwara, Y.; Pang, W.; Lim, I.; Oh, S.L.; Adam, M.; Tan, R.S.; Chen, M.; Acharya, U.R. Application of stacked convolutional and long short-term memory network for accurate identification of CAD ECG signals. Comput. Biol. Med. 2018, 94, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep Learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Swapna, G.; Soman, K.P.; Vinayakumar, R. Automated detection of cardiac arrhythmia using deep learning techniques. Procedia Comput. Sci. 2018, 132, 1192–1201. [Google Scholar] [CrossRef]
- Hong, S.; Zhou, Y.; Shang, J.; Xiao, C.; Sun, J. Opportunities and challenges of deep learning methods for electrocardiogram data: A systematic review. Comput. Biol. Med. 2020, 122, 103801. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.C.; Lee, C.C.; Yeng, C.H.; So, E.C.; Chen, Y.J. Attention mechanism-based convolutional long short-term memory neural networks to electrocardiogram-based blood pressure estimation. Appl. Sci. 2021, 11, 12019. [Google Scholar] [CrossRef]
- Ebrahimi, Z.; Loni, M.; Daneshtalab, M.; Gharehbaghi, A. A review on deep learning methods for ECG arrhythmia classification. Expert Syst. Appl. X 2020, 7, 100033. [Google Scholar] [CrossRef]
- Jiang, W.; Kong, S.G. Block-based neural networks for personalized ECG signal classification. IEEE Trans. Neural Netw. 2007, 18, 1750–1761. [Google Scholar] [CrossRef]
- Rahhal, M.M.A.; Bazi, Y.; Alhichri, H.; Alajlan, N.; Melgani, F.; Yager, R.R. Deep learning approach for active classification of electrocardiogram signals. Inf. Sci. 2016, 345, 340–354. [Google Scholar] [CrossRef]
- Wu, L.; Xie, X.; Wang, Y. ECG enhancement and r-peak detection based on window variability. Healthcare 2021, 9, 227. [Google Scholar] [CrossRef]
- Marston, H.R.; Hadley, R.; Banks, D.; Duro, M.D.C.M. Mobile self-monitoring ECG devices to diagnose arrhythmia that coincide with palpitations: A scoping review. Healthcare 2019, 7, 96. [Google Scholar] [CrossRef]
- Ullah, A.; Anwar, S.M.; Bilal, M.; Mehmood, R.M. Classification of arrhythmia by using deep learning with 2-D ECG spectral image representation. Remote Sens. 2020, 12, 1685. [Google Scholar] [CrossRef]
- Akula, R.; Mohamed, H. Automation algorithm to detect and quantify electrocardiogram waves and intervals. Procedia Comput. Sci. 2019, 151, 941–946. [Google Scholar] [CrossRef]
- Wang, L.H.; Yan, Z.H.; Yang, Y.T.; Chen, J.Y.; Yang, T.; Kuo, I.C.; Abu, P.A.R.; Huang, P.C.; Chen, C.A.; Chen, S.L. A classification and prediction hybrid model construction with the iqpso-svm algorithm for atrial fibrillation arrhythmia. Sensors 2021, 21, 5222. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, P.; Wang, S. Automated detection of atrial fibrillation in ECG signals based on wavelet packet transform and correlation function of random process. Biomed. Signal Process. Control 2020, 55, 101662. [Google Scholar] [CrossRef]
- Hao, P.; Gao, X.; Li, Z.; Zhang, J.; Wu, F.; Bai, C. Multi-branch fusion network for Myocardial infarction screening from 12-lead ECG images. Comput. Methods Programs Biomed. 2020, 184, 105286. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.H.; Nurmaini, S.; Partan, R.U. Automatic Classification of 15 Leads ECG Signal of Myorcadial Infarction Using Convolutional Neural Network. Appl. Sci. 2022, 12, 5603. [Google Scholar] [CrossRef]
- Babaoglu, I.; Findik, O.; Ülker, E. A comparison of feature selection models utilizing binary particle swarm optimization and genetic algorithm in determining coronary artery disease using support vector machine. Expert Syst. Appl. 2010, 37, 3177–3183. [Google Scholar] [CrossRef]
- Kumar, M.; Pachori, R.B.; Acharya, U.R. Characterization of coronary artery disease using flexible analytic wavelet transform applied on ECG signals. Biomed. Signal Process. Control 2017, 31, 301–308. [Google Scholar] [CrossRef]
- Kurt, I.; Ture, M.; Kurum, A.T. Comparing performances of logistic regression, classification and regression tree, and neural networks for predicting coronary artery disease. Expert Syst. Appl. 2008, 34, 366–374. [Google Scholar] [CrossRef]
- Kruger, G.H.; Latchamsetty, R.; Langhals, N.B.; Yokokawa, M.; Chugh, A.; Morady, F.; Oral, H.; Berenfeld, O. Bimodal classification algorithm for atrial fibrillation detection from m-health ECG recordings. Comput. Biol. Med. 2019, 104, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Polat, K.; Güneş, S. A new feature selection method on classification of medical datasets: Kernel F-score feature selection. Expert Syst. Appl. 2009, 36, 10367–10373. [Google Scholar] [CrossRef]
- Wen, J.; Xuan, S.; Li, Y.; Gao, Q.; Peng, Q. Image-segmentation algorithm based on wavelet and data-driven neutrosophic fuzzy clustering. Imaging Sci. J. 2019, 67, 63–75. [Google Scholar] [CrossRef]
- Badilini, F.; Erdem, T.; Zareba, W.; Moss, A.J. ECGScan: A method for conversion of paper electrocardiographic printouts to digital electrocardiographic files. J. Electrocardiol. 2005, 38, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Śmigiel, S.; Pałczyński, K.; Ledziński, D. Deep learning techniques in the classification of ecg signals using r-peak detection based on the ptb-xl dataset. Sensors 2021, 21, 8174. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, L.; Harless, C.; Member, S. Novel Tool for Complete Digitization of Paper Electrocardiography Data. IEEE J. Transl. Eng. Health Med. 2013, 1, 1800107. [Google Scholar] [CrossRef]
- Jambukia, S.H.; Dabhi, V.K.; Prajapati, H.B. Classification of ECG signals using machine learning techniques: A survey. In Proceedings of the 2015 International Conference on Advances in Computer Engineering and Applications, Ghaziabad, India, 19–20 March 2015; pp. 714–721. [Google Scholar] [CrossRef]
- Luukka, P.; Lampinen, J. A Classification Method Based on Principal Component Analysis and Differential Evolution Algorithm Applied for Prediction Diagnosis from Clinical EMR Heart Data Sets. In Computational Intelligence in Optimization; Springer: Berlin/Heidelberg, Germany, 2010; pp. 263–283. [Google Scholar] [CrossRef]
- Li, H.; Yuan, D.; Ma, X.; Cui, D.; Cao, L. Genetic algorithm for the optimization of features and neural networks in ECG signals classification. Sci. Rep. 2017, 7, 41011. [Google Scholar] [CrossRef] [PubMed]
- Garg, D.K. ECG Paper Records Digitization through Image Processing Techniques. Int. J. Comput. Appl. 2012, 48, 35–38. [Google Scholar]
- Yoo, H.; Han, S.; Chung, K. A frequency pattern mining model based on deep neural network for real-time classification of heart conditions. Healthcare 2020, 8, 234. [Google Scholar] [CrossRef]
- Sannino, G.; De Pietro, G. A deep learning approach for ECG-based heartbeat classification for arrhythmia detection. Futur. Gener. Comput. Syst. 2018, 86, 446–455. [Google Scholar] [CrossRef]
- Xu, S.S.; Mak, M.W.; Cheung, C.C. Towards End-to-End ECG Classification with Raw Signal Extraction and Deep Neural Networks. IEEE J. Biomed. Health Inform. 2019, 23, 1574–1584. [Google Scholar] [CrossRef]
- Maniruzzaman, M.; Rahman, M.J.; Ahammed, B.; Abedin, M.M. Classification and prediction of diabetes disease using machine learning paradigm. Health Inf. Sci. Syst. 2020, 8, 7. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Lih, O.S.; Hagiwara, Y.; Tan, J.H.; Adam, M. Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inf. Sci. 2017, 405, 81–90. [Google Scholar] [CrossRef]
- Yildirim, O.; Tan, R.S.; Acharya, U.R. An efficient compression of ECG signals using deep convolutional autoencoders. Cogn. Syst. Res. 2018, 52, 198–211. [Google Scholar] [CrossRef]
- Murat, F.; Yildirim, O.; Talo, M.; Baloglu, U.B.; Demir, Y.; Acharya, U.R. Application of deep learning techniques for heartbeats detection using ECG signals-analysis and review. Comput. Biol. Med. 2020, 120, 103726. [Google Scholar] [CrossRef] [PubMed]
- Rajeswari, K.; Vaithiyanathan, V.; Neelakantan, T.R. Feature Selection in Ischemic Heart Disease identification using feed forward neural networks. Procedia Eng. 2012, 41, 1818–1823. [Google Scholar] [CrossRef]
- Al-Ani, M.; Rawi, A.A. A rule-based expert system for automated ecg diagnosis. Int. J. Adv. Eng. Technol. 2014, 6, 1480–1492. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Is object localization for free?—Weakly-supervised learning with convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 685–694. [Google Scholar] [CrossRef]
- Cireşan, D.C.; Meier, U.; Gambardella, L.M.; Schmidhuber, J. Convolutional neural network committees for handwritten character classification. In Proceedings of the 2011 International Conference on Document Analysis and Recognition, Beijing, China, 18–21 September 2011; Volume 10, pp. 1135–1139. [Google Scholar] [CrossRef]
- Yuan, Y.; Xun, G.; Suo, Q.; Jia, K.; Zhang, A. Wave2Vec: Deep representation learning for clinical temporal data. Neurocomputing 2019, 324, 31–42. [Google Scholar] [CrossRef]
- Jayaraman, S.; Swamy, P.; Damodaran, V.; Venkatesh, N. A Novel Technique for ECG Morphology Interpretation and Arrhythmia Detection Based on Time Series Signal Extracted from Scanned ECG Record. Adv. Electrocardiograms-Methods Anal. 2012, 10, 21785. [Google Scholar] [CrossRef][Green Version]
- Carolina Sparavigna, A. An Image Processing Approach Based on Gnu Image Manipulation Program Gimp to the Panoramic Radiography. Int. J. Sci. 2015, 1, 57–67. [Google Scholar] [CrossRef]
- Biswas, D. Novel Gray Scale Conversion Techniques Based on Pixel Dept. J. Glob. Res. Comput. Sci. 2011, 2, 79–82. [Google Scholar]
- Pratt, W.K. Morphological Image Processing. In Digital Image Processing, 3rd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2003; pp. 401–441. [Google Scholar] [CrossRef]
- Naam, J.; Suharinto, C.; Sumijan, S. Digitalisasi Grafik Elektrokardiogram dengan Teknik Pixel Indexing. Pros. SISFOTEK 2017, 1, 172–176. [Google Scholar]
- Tabassum, T.; Ahmad, M. Numerical data extraction from ECG paper recording using image processing technique. In Proceedings of the 2020 11th International Conference on Electrical and Computer Engineering (ICECE), Dhaka, Bangladesh, 17–19 December 2020; pp. 355–358. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A Real-Time QRS Detection Algorithm. IEEE Trans. Biomed. Eng. 1985, BME-32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Sedghamiz, H. Matlab Implementation of Pan Tompkins ECG QRS. 2014, Code Available at the File Exchange Site of MathWorks. 2014. Available online: https://fr.mathworks.com/matlabcentral/fileexchange/45840-complete-pan-tompkins-implementationecg-qrs-detector (accessed on 15 June 2022).
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; Tan, R.S. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, M.; Chen, T.; Sun, Z.; Ma, Y.; Yu, B. Recent advances in convolutional neural network acceleration. Neurocomputing 2019, 323, 37–51. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- The Theano Development Team; Al-Rfou, R.; Alain, G.; Almahairi, A.; Angermueller, C.; Bahdanau, D.; Ballas, N.; Bastien, F.; Bayer, J.; Belikov, A.; et al. Theano: A Python framework for fast computation of mathematical expressions. arXiv 2016, arXiv:1605.02688. [Google Scholar]
- Duraj, K.; Piaseczna, N.; Kostka, P.; Tkacz, E. Semantic Segmentation of 12-Lead ECG Using 1D Residual U-Net with Squeeze-Excitation Blocks. Appl. Sci. 2022, 12, 3332. [Google Scholar] [CrossRef]

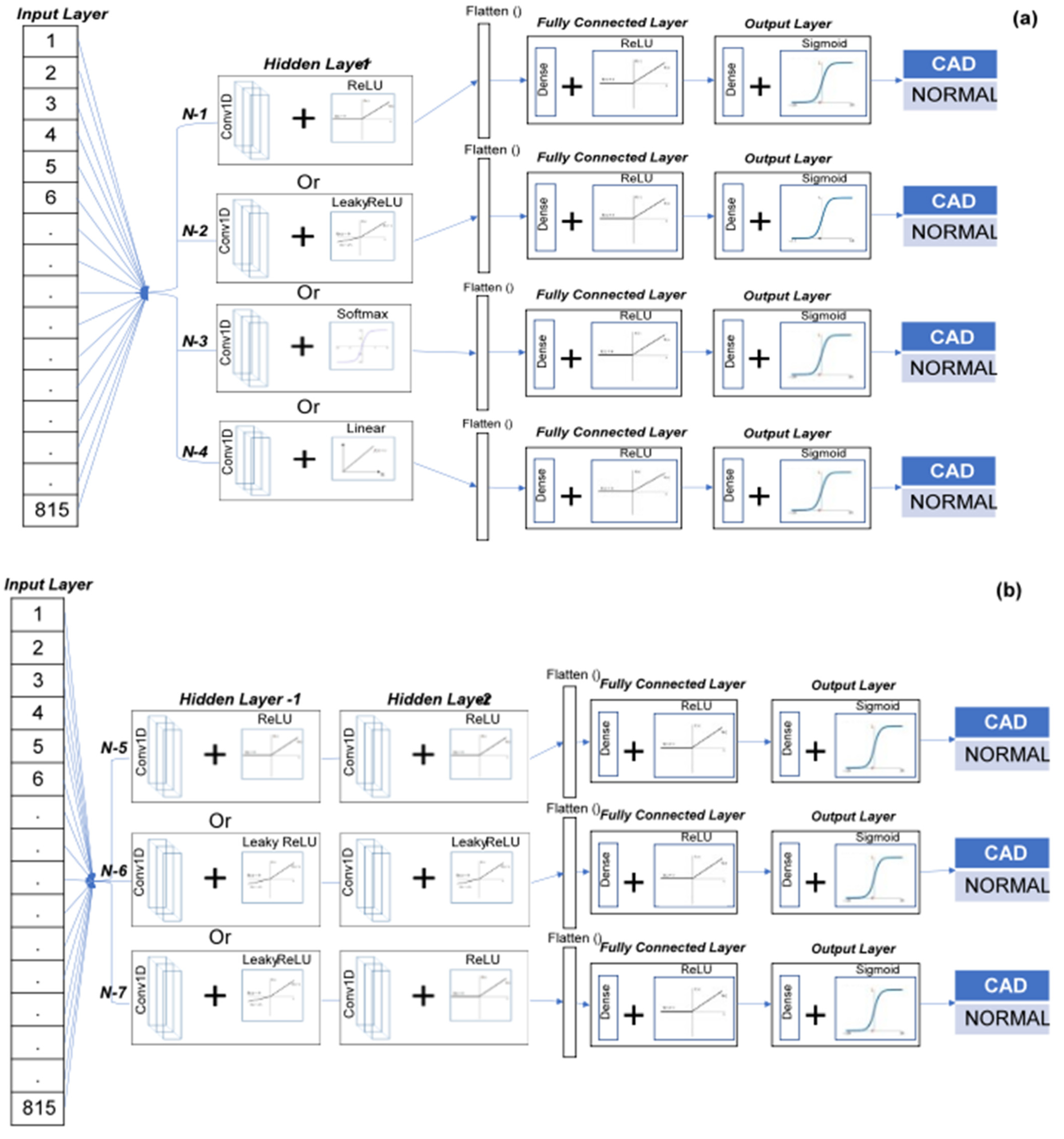



| Single-Layer CNN | Two-Layer CNN | Three-Layer CNN | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Network (N) | ||||||||||
| N-1 | N-2 | N-3 | N-4 | N-5 | N-6 | N-7 | N-8 | N-9 | N-10 | |
| Input | (815, 1) | (815, 1) | (815, 1) | (815, 1) | (815, 1) | (815, 1) | (815, 1) | (815, 1) | (815, 1) | (815, 1) |
| Hidden_1 Layer | Conv1D + ReLU (814, 64) | Conv1D + LeakyReLU (814, 64) | Conv1D + SoftMax (814, 64) | Conv1D + Linear (814, 64) | Conv1D + ReLU (814, 64) | Conv1D + LeakyReLU (814, 64) | Conv1D + LeakyReLU (814, 64) | Conv1D + ReLU (814, 64) | Conv1D + LeakyReLU (814, 64) | Conv1D + ReLU (814, 64) |
| Hidden_2 Layer | None | None | None | None | Conv1D + ReLU (813, 32) | Conv1D + LeakyReLU (813, 32) | Conv1D + ReLU (813, 32) | Conv1D + ReLU (813, 32) | Conv1D + ReLU (813, 32) | Conv1D + LeakyReLU (813, 32) |
| Hidden_3 Layer | None | None | None | None | None | None | None | Conv1D + LeakyReLU (812, 16) | Conv1D + ReLU (812, 16) | Conv1D + ReLU (812, 16) |
| Flatten | [52096] | [52096] | [52096] | [52096] | [26016] | [26016] | [26016] | [156624] | [156624] | [156624] |
| Fully-Connected | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) |
| Output | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) |
| Single-Layer CNN | Two-Layer CNN | Three-Layer CNN | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Network (N) | ||||||||||
| N-1 | N-2 | N-3 | N-4 | N-5 | N-6 | N-7 | N-8 | N-9 | N-10 | |
| Input | (9792, 1) | (9792, 1) | (9792, 1) | (9792, 1) | (9792, 1) | (9792, 1) | (9792, 1) | (9792, 1) | (9792, 1) | (9792, 1) |
| Hidden_1 Layer | Conv1D + ReLU (9791, 64) | Conv1D + LeakyReLU (9791, 64) | Conv1D + SoftMax (9791, 64) | Conv1D + Linear (9791, 64) | Conv1D + ReLU (9791, 64) | Conv1D + LeakyReLU (9791, 64) | Conv1D + LeakyReLU (9791, 64) | Conv1D + ReLU (9791, 64) | Conv1D + LeakyReLU (9791, 64) | Conv1D + ReLU (9791, 64) |
| Hidden_2 Layer | None | None | None | None | Conv1D + ReLU (9790, 32) | Conv1D + LeakyReLU (9790, 32) | Conv1D + ReLU (9790, 32) | Conv1D + ReLU (9790, 32) | Conv1D + ReLU (9790, 32) | Conv1D + LeakyReLU (9790, 32) |
| Hidden_3 Layer | None | None | None | None | None | None | None | Conv1D + LeakyReLU (9789, 16) | Conv1D + ReLU (9789, 16) | Conv1D + ReLU (9789, 16) |
| Flatten | [626624] | [626624] | [626624] | [626624] | [313280] | [313280] | [313280] | [156624] | [156624] | [156624] |
| Fully-Connected | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) | Dense + ReLU (16) |
| Output | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) | Dense + Sigmoid (1) |
| Extracted Data vs. Original Data | Mean (Δ) | Median (Δ) | Correlation (Δ) |
|---|---|---|---|
| Mean | −0.0311 | 0.02102 | 0.98 |
| Standard Deviation | 0.02423 | 0.03541 | 0.9 |
| Minimum | −0.06378 | 0.06784 | 0.89 |
| Maximum | 0.02785 | 0.02516 | 0.97 |
| Single-Layer CNN | Two-Layer CNN | Three-Layer CNN | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Network (N) | ||||||||||
| N-1 | N-2 | N-3 | N-4 | N-5 | N-6 | N-7 | N-8 | N-9 | N-10 | |
| Precision | 72.50% | 84% | 32% | 97% | 97.50% | 98% | 99% | 100% | 96.50% | 99% |
| Recall | 74.50% | 81.50% | 50% | 95.50% | 98% | 98% | 99% | 99% | 98% | 99% |
| F1-Score | 72.50% | 82% | 39% | 96% | 97.50% | 97.50% | 98% | 100% | 97.50% | 99% |
| Accuracy | 73% | 84% | 64% | 96% | 98% | 98% | 99% | 100% | 97% | 100% |
| Validation Loss. | 0.4825 | 0.37 | 0.65 | 0.1269 | 0.0773 | 0.069 | 0.022 | 0.00075 | 0.0098 | 0.00412 |
| Single-Layer CNN | Two-Layer CNN | Three-Layer CNN | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Network (N) | ||||||||||
| N-1 | N-2 | N-3 | N-4 | N-5 | N-6 | N-7 | N-8 | N-9 | N-10 | |
| Precision | 29.50% | 98% | 29.50% | 97% | 29.50% | 97.00% | 98% | 99% | 99% | 99% |
| Recall | 50.00% | 97.50% | 50.00% | 96.50% | 50.00% | 97.50% | 97% | 99% | 99% | 99% |
| F1-Score | 37.00% | 96.50% | 37.00% | 97% | 37.00% | 96.50% | 97% | 100% | 100% | 100% |
| Accuracy | 59% | 98% | 59% | 97% | 59% | 97% | 98.30% | 99% | 99% | 99% |
| Validation Loss | 0.68 | 0.054 | 0.6878 | 0.079 | 0.68 | 0.052 | 0.025 | 0.00022 | 0.00017 | 0.00028 |
| Single-Lead ECG Data | Twelve-Lead ECG Data | |||
|---|---|---|---|---|
| Accuracy (%) | Standard Deviation | Accuracy (%) | Standard Deviation | |
| Split-1 | 99.8 | 0.006 | 98.7 | 0.021 |
| Split-2 | 99.5 | 0.011 | 99.4 | 0.013 |
| Split-3 | 99.3 | 0.01 | 99.3 | 0.011 |
| Split-4 | 99.3 | 0.012 | 98.8 | 0.019 |
| Split-5 | 99.7 | 0.008 | 99.1 | 0.017 |
| Average | 99.5% | 0.009 | 99.1% | 0.016 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasudeva, S.T.; Rao, S.S.; Panambur, N.K.; Shettigar, A.K.; Mahabala, C.; Kamath, P.; Gowdru Chandrashekarappa, M.P.; Linul, E. Development of a Convolutional Neural Network Model to Predict Coronary Artery Disease Based on Single-Lead and Twelve-Lead ECG Signals. Appl. Sci. 2022, 12, 7711. https://doi.org/10.3390/app12157711
Vasudeva ST, Rao SS, Panambur NK, Shettigar AK, Mahabala C, Kamath P, Gowdru Chandrashekarappa MP, Linul E. Development of a Convolutional Neural Network Model to Predict Coronary Artery Disease Based on Single-Lead and Twelve-Lead ECG Signals. Applied Sciences. 2022; 12(15):7711. https://doi.org/10.3390/app12157711
Chicago/Turabian StyleVasudeva, Shrivathsa Thokur, Shrikantha Sasihithlu Rao, Navin Karanth Panambur, Arun Kumar Shettigar, Chakrapani Mahabala, Padmanabh Kamath, Manjunath Patel Gowdru Chandrashekarappa, and Emanoil Linul. 2022. "Development of a Convolutional Neural Network Model to Predict Coronary Artery Disease Based on Single-Lead and Twelve-Lead ECG Signals" Applied Sciences 12, no. 15: 7711. https://doi.org/10.3390/app12157711
APA StyleVasudeva, S. T., Rao, S. S., Panambur, N. K., Shettigar, A. K., Mahabala, C., Kamath, P., Gowdru Chandrashekarappa, M. P., & Linul, E. (2022). Development of a Convolutional Neural Network Model to Predict Coronary Artery Disease Based on Single-Lead and Twelve-Lead ECG Signals. Applied Sciences, 12(15), 7711. https://doi.org/10.3390/app12157711








