GAN Training Acceleration Using Fréchet Descriptor-Based Coreset
Abstract
:1. Introduction
2. Related Works
2.1. Blob Identification
2.2. Coreset
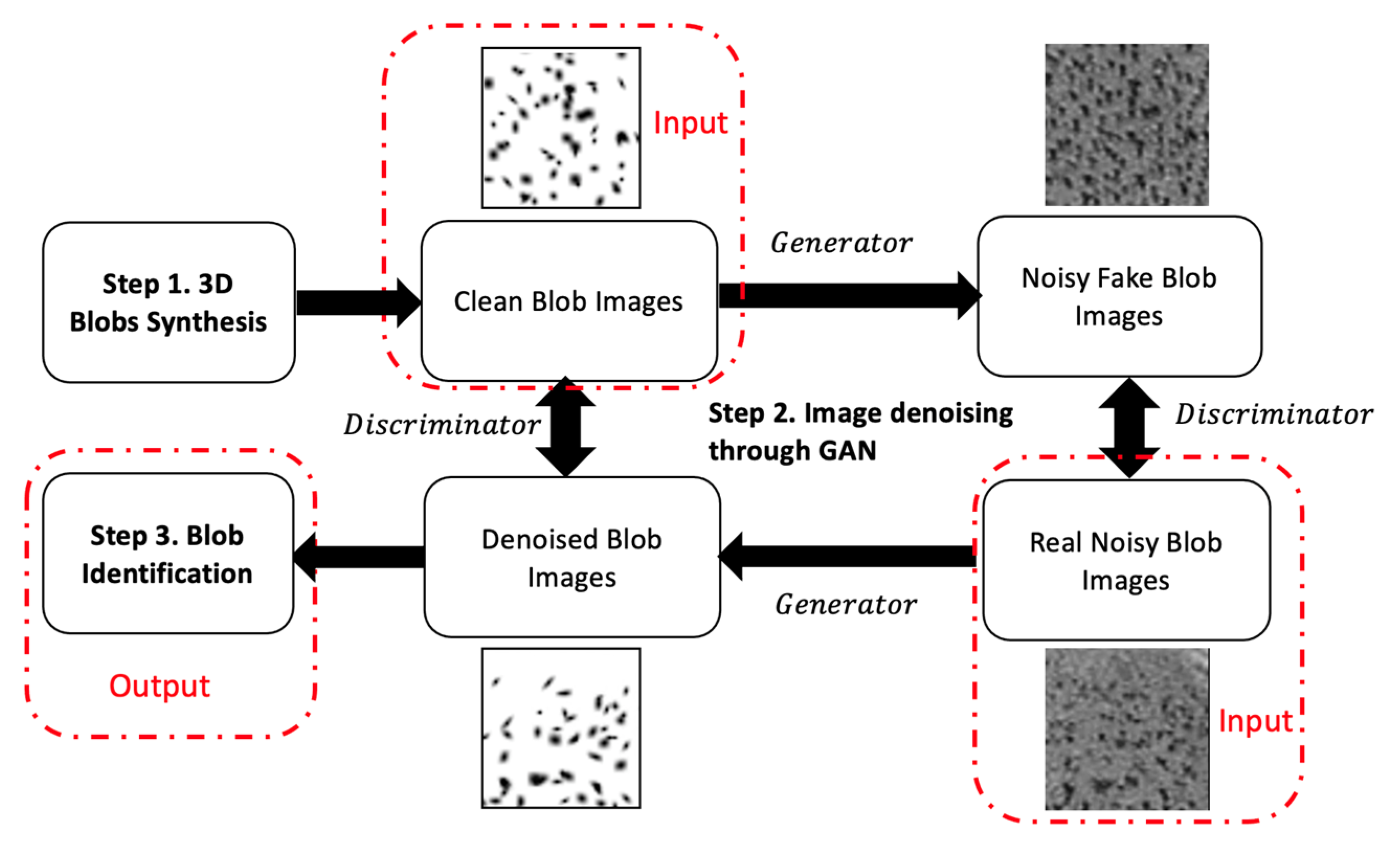
3. Methods
3.1. BlobGAN and Blob Descriptors
3.2. Fréchet Descriptor Distance
3.3. Dataset Sampling Based on Coreset
| Algorithm 1: Pseudocode for FDD-Coreset. |
| Input: target size 3D blob image datasets () with descriptors sets Output: Coreset () 1. Initialize Coreset 2. While 3. For 3D blob image from 4. Extract the blob descriptors sets of 5. For 3D blob image from 6. Extract the blob descriptors sets of 7. Calculate FDD distance between and : 8. Iteratively until find sample image = 9. Add in Coreset 10. End While 11. Return Coreset |
4. Experiments and Results
4.1. Training Dataset
4.2. Experiment I: Validation Experiments Using 3D Synthetic Image Data
- Peak Signal-to-Noise Ratio (PSNR) metric is to measure the performance of image denoising. Let the final 3D denoised blobs image be and the 3D blobs image without noises be , then PSNR is defined as follows:where is the possible maximum voxel values of .
- Detection Error Rate (DER) is to measure the difference ratio between the number of detected blobs and the ground truth. DER can be calculated by Equation (10).where represents the # of ground truth blobs and represents the # of detected blobs.
- 3.
- Precision is to measure the fraction of retrieved blobs confirmed by the ground truth.
- 4.
- Recall is to measure the fraction of ground-truth data retrieved.
- 5.
- F-score is the overall performance of precision and recall.
- 6.
- Dice coefficient (Dice) is to measure the similarity between the segmented blob mask and the ground truth.where is the binary mask for segmentation result and is the binary mask for the ground truth.
- 7.
- Intersection over Union (IoU) is to measure the amount of overlap between the segmented blob mask and the ground truth.where is the binary mask for segmentation result and is the binary mask for the ground truth.
- 8.
- Blobness (PB) is to measure the likelihood of the objects with a blob shape. PB for each blob candidate from blobs set is calculated by Equation (17):where is the normalized 3D dark blobs image, represents the Hessian matrix and represents the principal minors of Hessian matrix.
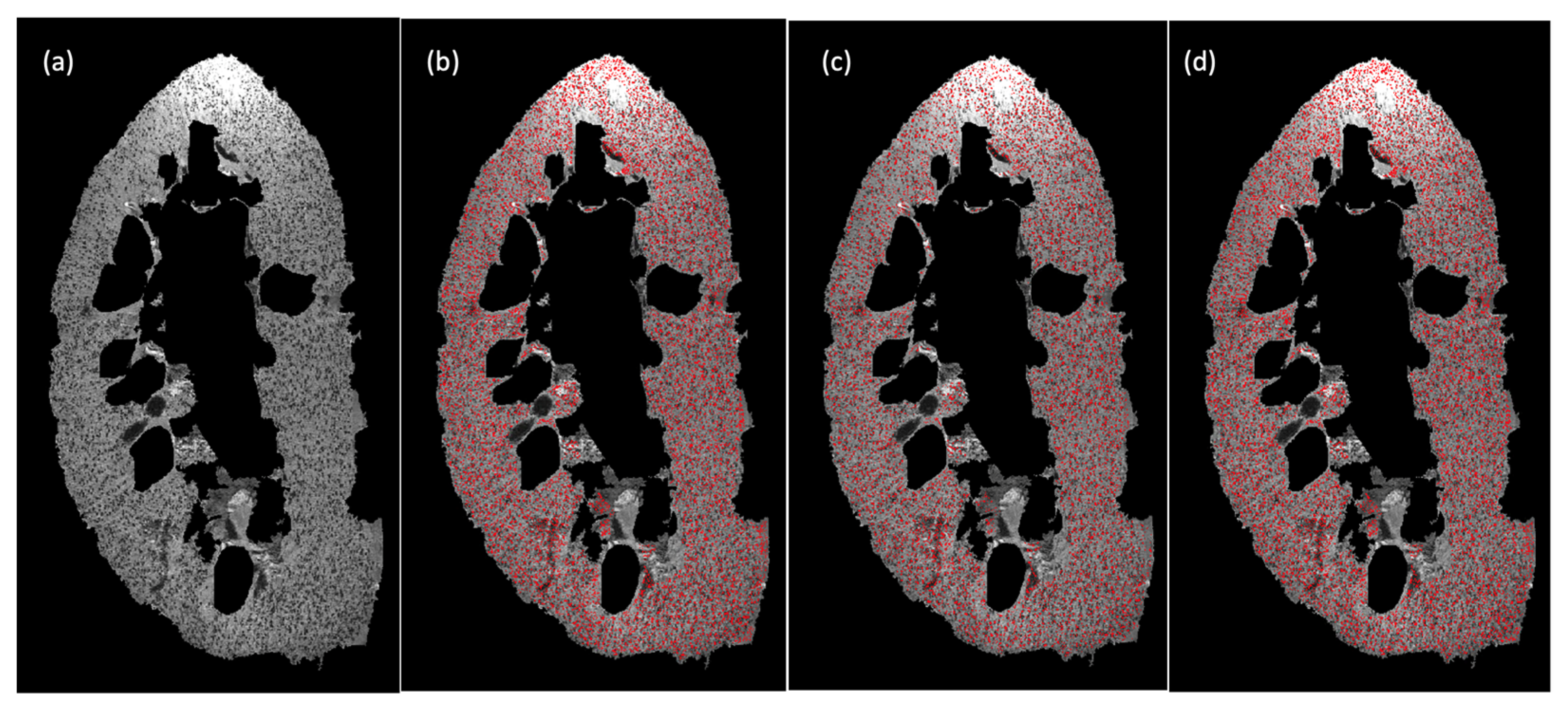
4.3. Experiment II: Validation Experiments Using 3D Human Kidney MR Images
4.4. Discussion: Clinical Translation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Han, C.; Hayashi, H.; Rundo, L.; Araki, R.; Shimoda, W.; Muramatsu, S.; Furukawa, Y.; Mauri, G.; Nakayama, H. GAN-based synthetic brain MR image generation. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 734–738. [Google Scholar] [CrossRef]
- Sandfort, V.; Yan, K.; Pickhardt, P.J.; Summers, R.M. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019, 9, 16884. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2242–2251. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Fang, Q.; Huang, Y.; Xu, K. Semi-supervised method for image texture classification of pituitary tumors via CycleGAN and optimized feature extraction. BMC Med. Inform. Decis. Mak. 2020, 20, 215. [Google Scholar] [CrossRef]
- Gu, J.; Yang, T.S.; Ye, J.C.; Yang, D.H. CycleGAN denoising of extreme low-dose cardiac CT using wavelet-assisted noise disentanglement. Med. Image Anal. 2021, 74, 102209. [Google Scholar] [CrossRef] [PubMed]
- Hiasa, Y.; Otake, Y.; Takao, M.; Matsuoka, T.; Takashima, K.; Carass, A.; Prince, J.L.; Sugano, N.; Sato, Y. Cross-Modality Image Synthesis from Unpaired Data Using CycleGAN. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Berlin, Germany, 2018; pp. 31–41. [Google Scholar] [CrossRef]
- Xu, Y. Novel Computational Algorithms for Imaging Biomarker Identification. Ph.D. Thesis, Arizona State University, Tempe, AZ, USA, 2022. [Google Scholar]
- Oulbacha, R.; Kadoury, S. MRI to CT Synthesis of the Lumbar Spine from a Pseudo-3D Cycle GAN. In Proceedings of the 2020 IEEE 17th Inter-national Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1784–1787. [Google Scholar] [CrossRef]
- Yang, S.; Kim, E.Y.; Ye, J.C. Continuous Conversion of CT Kernel Using Switchable CycleGAN With AdaIN. IEEE Trans. Med. Imaging 2021, 40, 3015–3029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, L.; Zheng, Y. Translating and Segmenting Multimodal Medical Volumes with Cycle- and Shape-Consistency Generative Adversarial Network. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 9242–9251. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Liu, J.; Liu, Y.; Fu, H.; Hu, Y.; Cheng, J.; Qi, H.; Wu, Y.; Zhang, J.; Zhao, Y. Structure and Illumination Constrained GAN for Medical Image Enhancement. IEEE Trans. Med. Imaging 2021, 40, 3955–3967. [Google Scholar] [CrossRef]
- Anoosheh, A.; Agustsson, E.; Timofte, R.; van Gool, L. ComboGAN: Unrestrained Scalability for Image Domain Translation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Salt Lake City, UT, USA, 18–22 June 2018; pp. 896–8967. [Google Scholar] [CrossRef] [Green Version]
- Nuha, F.U. Afiahayati Training dataset reduction on generative adversarial network. Procedia Comput. Sci. 2018, 144, 133–139. [Google Scholar] [CrossRef]
- DeVries, T.; Drozdzal, M.; Taylor, G.W. Instance Selection for GANs. Adv. Neural Inf. Process. Syst. 2020, 33, 13285–13296. [Google Scholar]
- Sinha, S.; Zhang, H.; Goyal, A.; Bengio, Y.; Larochelle, H.; Odena, A. Small-GAN: Speeding Up GAN Training Using Core-sets. In Proceedings of the 37th International Conference on Machine Learning, Virtual Event, 13–18 July 2020. [Google Scholar]
- Al-Kofahi, Y.; Lassoued, W.; Lee, W.; Roysam, B. Improved Automatic Detection and Segmentation of Cell Nuclei in Histopathology Images. IEEE Trans. Biomed. Eng. 2009, 57, 841–852. [Google Scholar] [CrossRef]
- Beeman, S.C.; Zhang, M.; Gubhaju, L.; Wu, T.; Bertram, J.F.; Frakes, D.H.; Cherry, B.R.; Bennett, K.M. Measuring glomerular number and size in perfused kidneys using MRI. Am. J. Physiol. Physiol. 2011, 300, F1454–F1457. [Google Scholar] [CrossRef]
- Beeman, S.C.; Cullen-McEwen, L.; Puelles, V.; Zhang, M.; Wu, T.; Baldelomar, E.J.; Dowling, J.; Charlton, J.R.; Forbes, M.S.; Ng, A.; et al. MRI-based glomerular morphology and pathology in whole human kidneys. Am. J. Physiol. Physiol. 2014, 306, F1381–F1390. [Google Scholar] [CrossRef]
- Kong, H.; Akakin, H.C.; Sarma, S.E. A Generalized Laplacian of Gaussian Filter for Blob Detection and Its Applications. IEEE Trans. Cybern. 2013, 43, 1719–1733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, T.; Bennett, K.M. Small Blob Identification in Medical Images Using Regional Features From Optimum Scale. IEEE Trans. Biomed. Eng. 2014, 62, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, T.; Beeman, S.C.; Cullen-McEwen, L.; Bertram, J.F.; Charlton, J.R.; Baldelomar, E.; Bennett, K.M. Efficient Small Blob Detection Based on Local Convexity, Intensity and Shape Information. IEEE Trans. Med. Imaging 2016, 35, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gao, F.; Wu, T.; Bennett, K.M.; Charlton, J.R.; Sarkar, S. U-Net with optimal thresholding for small blob detection in medical images. In Proceedings of the 2019 IEEE 15th International Conference on Automation Science and Engineering (CASE), Vancouver, BC, Canada, 22–26 August 2019; pp. 1761–1767. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, T.; Gao, F.; Charlton, J.R.; Bennett, K.M. Improved small blob detection in 3D images using jointly constrained deep learning and Hessian analysis. Sci. Rep. 2020, 10, 326. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Wu, T.; Charlton, J.R.; Gao, F.; Bennett, K.M. Small Blob Detector Using Bi-Threshold Constrained Adaptive Scales. IEEE Trans. Biomed. Eng. 2020, 68, 2654–2665. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Jiang, S.; Vishnoi, N. Coresets for clustering with fairness constraints. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; pp. 13285–13296. [Google Scholar]
- Sener, O.; Savarese, S. Active Learning for Convolutional Neural Networks: A Core-Set Approach. arXiv 2017, arXiv:1708.00489. [Google Scholar]
- Ulyanov, D.; Vedaldi, A.; Lempitsky, V. Instance Normalization: The Missing Ingredient for Fast Stylization. arXiv 2016, arXiv:1607.08022. [Google Scholar]
- Baldelomar, E.J.; Charlton, J.R.; Beeman, S.C.; Bennett, K.M. Measuring rat kidney glomerular number and size in vivo with MRI. Am. J. Physiol. Physiol. 2017, 314, F399–F406. [Google Scholar] [CrossRef]
- Aronov, B.; Har-Peled, S.; Knauer, C.; Wang, Y.; Wenk, C. Fréchet Distance for Curves, Revisited. In Algorithms—ESA 2006; Azar, Y., Erlebach, T., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4168, pp. 52–63. [Google Scholar] [CrossRef] [Green Version]
- Preuer, K.; Renz, P.; Unterthiner, T.; Hochreiter, S.; Klambauer, G. Fréchet ChemNet Distance: A Metric for Generative Models for Molecules in Drug Discovery. J. Chem. Inf. Model. 2018, 58, 1736–1741. [Google Scholar] [CrossRef] [Green Version]
- Unterthiner, T.; Van, S.; Kurach, K.; Marinier, R.; Michalski, M.; Gelly, S. Towards Accurate Generative Models of Video: A New Metric & Challenges. arXiv 2018, arXiv:1812.01717. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium. arXiv 2017, arXiv:1706.08500. [Google Scholar]
- Wolf, G.W. Facility location: Concepts, models, algorithms and case studies. Series: Contributions to Management Science. Int. J. Geogr. Inf. Sci. 2011, 25, 331–333. [Google Scholar] [CrossRef]
- Bennett, K.M.; Zhou, H.; Sumner, J.P.; Dodd, S.J.; Bouraoud, N.; Doi, K.; Star, R.A.; Koretsky, A. MRI of the basement membrane using charged nanoparticles as contrast agents. Magn. Reson. Med. 2008, 60, 564–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldelomar, E.J.; Charlton, J.R.; Beeman, S.C.; Hann, B.D.; Cullen-McEwen, L.; Pearl, V.M.; Bertram, J.F.; Wu, T.; Zhang, M.; Bennett, K.M. Phenotyping by magnetic resonance imaging nondestructively measures glomerular number and volume distribution in mice with and without nephron reduction. Kidney Int. 2016, 89, 498–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Coreset Samples | IED-Coreset | FDD-Coreset |
|---|---|---|
| 1 | 2.66 | 0.01 |
| 2 | 3325.27 | 1.17 |
| 3 | 13,366.92 | 2.00 |
| 4 | 33,394.27 | 3.39 |
| 5 | 66,640.30 | 4.66 |
| 6 | 116,509.83 | 5.89 |
| 7 | 186,813.33 | 7.67 |
| 8 | 279,370.76 | 9.30 |
| 9 | 398,713.87 | 10.71 |
| 10 | 546,784.21 | 12.78 |
| Coreset Samples | Entire Dataset | IED-Corese | FDD-Coreset |
|---|---|---|---|
| 10 | 89,927.46 | 547,825.60 | 1032.19 |
| 20 | -- | 1995.25 | |
| 30 | -- | 2896.44 |
| Metrics | Entire Dataset | = 10) | = 10) | = 20) | = 30) |
|---|---|---|---|---|---|
| PSNR | 12.539 0.143 | 13.000 0.753 | 12.084 0.423 | 15.758 0.373 | 11.489 0.216 |
| DER | 0.091 0.037 | 0.124 0.131 | 0.171 0.156 | 0.219 0.061 | 0.052 0.037 |
| Precision | 0.941 0.013 | 0.759 0.069 | 0.671 0.056 | 0.923 0.018 | 0.827 0.024 |
| Recall | 0.855 0.033 | 0.781 0.052 | 0.771 0.042 | 0.720 0.049 | 0.845 0.032 |
| F-score | 0.896 0.018 | 0.766 0.030 | 0.715 0.028 | 0.807 0.029 | 0.835 0.014 |
| Dice | 0.825 0.017 | 0.548 0.046 | 0.662 0.014 | 0.502 0.039 | 0.671 0.022 |
| IoU | 0.702 0.024 | 0.379 0.044 | 0.495 0.015 | 0.336 0.035 | 0.505 0.025 |
| (Ground Truth: 0.519) | 0.538 0.279 | 0.609 0.302 | 0.577 0.308 | 0.583 0.290 | 0.610 0.298 |
| Human Kidney | Entire Dataset | = 10) | = 10) |
|---|---|---|---|
| CF 1 | 91,325.20 | 547,800.21 | 1195.58 |
| CF 2 | 91,382.40 | 547,801.81 | 1180.38 |
| CF 3 | 91,515.60 | 547,772.61 | 1199.58 |
| Human Kidney | (Stereology) | (entiredataset) | Difference Ratio (%) | (IED-Coreset (k = 10)) | Difference Ratio (%) | (FDD-Coreset (k = 10)) | Difference Ratio (%) |
|---|---|---|---|---|---|---|---|
| CF 1 | 1.13 | 1.05 | 7.08 | 1.58 | 39.82 | 1.26 | 11.50 |
| CF 2 | 0.74 | 0.71 | 4.05 | 0.96 | 29.73 | 0.78 | 5.41 |
| CF 3 | 1.46 | 1.48 | 1.37 | 1.77 | 21.23 | 1.53 | 4.79 |
| Human Kidney | Mean aVglom (Stereology) | Mean aVglom (entire dataset) | Difference Ratio (%) | Mean aVglom (IED-Coreset (k = 10)) | Difference Ratio (%) | Mean aVglom (FDD-Coreset (k = 10)) | Difference Ratio (%) |
|---|---|---|---|---|---|---|---|
| CF 1 | 5.01 | 5.19 | 3.59 | 4.48 | 10.58 | 5.20 | 3.79 |
| CF 2 | 4.68 | 4.80 | 2.56 | 3.61 | 22.86 | 5.02 | 7.26 |
| CF 3 | 2.82 | 2.81 | 0.35 | 2.25 | 20.21 | 2.93 | 3.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Wu, T.; Charlton, J.R.; Bennett, K.M. GAN Training Acceleration Using Fréchet Descriptor-Based Coreset. Appl. Sci. 2022, 12, 7599. https://doi.org/10.3390/app12157599
Xu Y, Wu T, Charlton JR, Bennett KM. GAN Training Acceleration Using Fréchet Descriptor-Based Coreset. Applied Sciences. 2022; 12(15):7599. https://doi.org/10.3390/app12157599
Chicago/Turabian StyleXu, Yanzhe, Teresa Wu, Jennifer R. Charlton, and Kevin M. Bennett. 2022. "GAN Training Acceleration Using Fréchet Descriptor-Based Coreset" Applied Sciences 12, no. 15: 7599. https://doi.org/10.3390/app12157599
APA StyleXu, Y., Wu, T., Charlton, J. R., & Bennett, K. M. (2022). GAN Training Acceleration Using Fréchet Descriptor-Based Coreset. Applied Sciences, 12(15), 7599. https://doi.org/10.3390/app12157599






