Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence
Abstract
1. Introduction
- Transparent: open to the degree where humans can understand the decision-making mechanism.
- Justifiable: the decision can be supported or justified along each step.
- Informative: to provide reasoning and allow reasoning.
- Uncertainty yielding: does not follow hard-coded structure, but open to change.
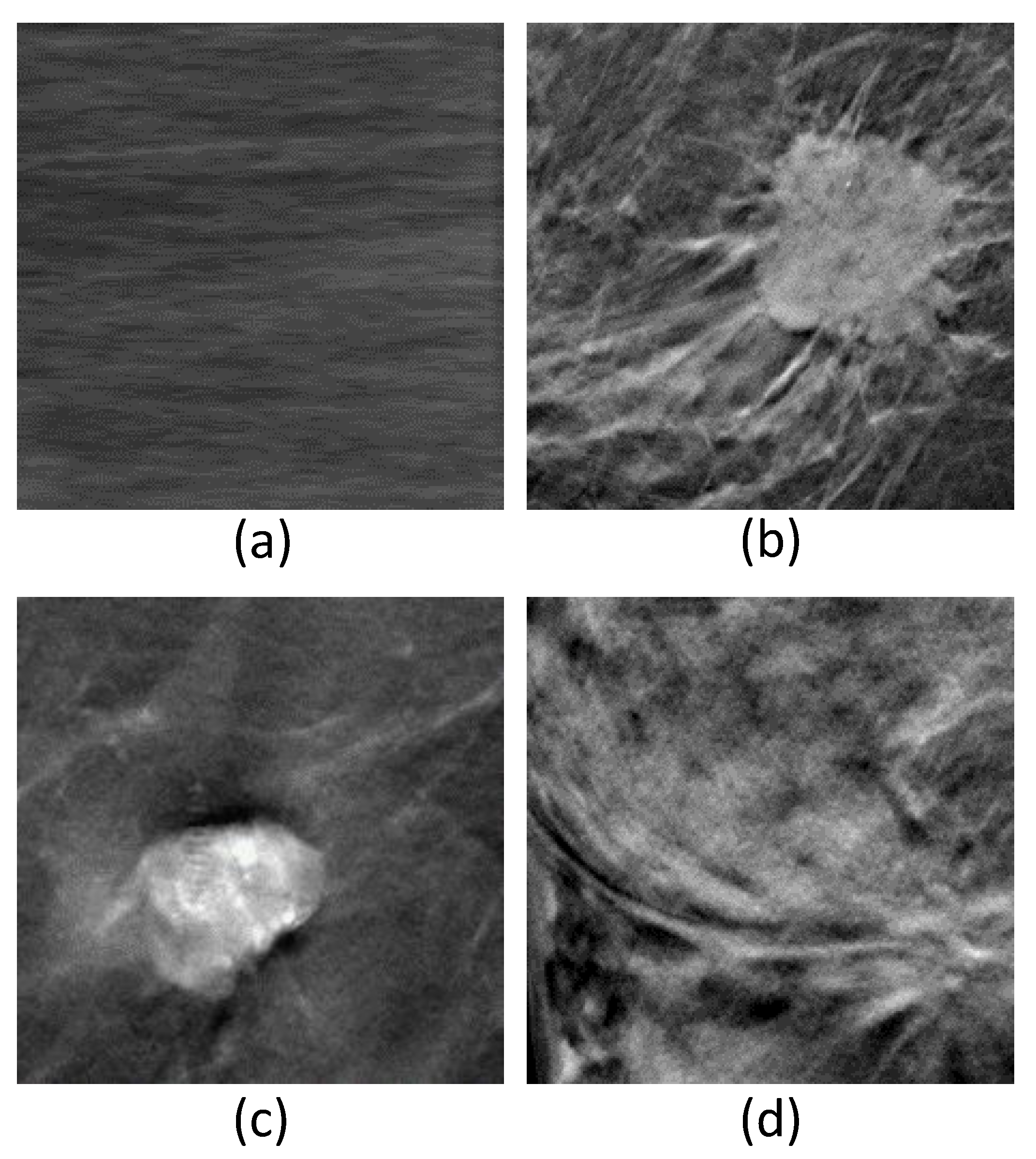
- Regular opacity (Oro) which includes the round, oval, and lobulated shapes;
- Irregular opacity (Ori);
- Architectural distortion shape (Ost).
2. Related Studies
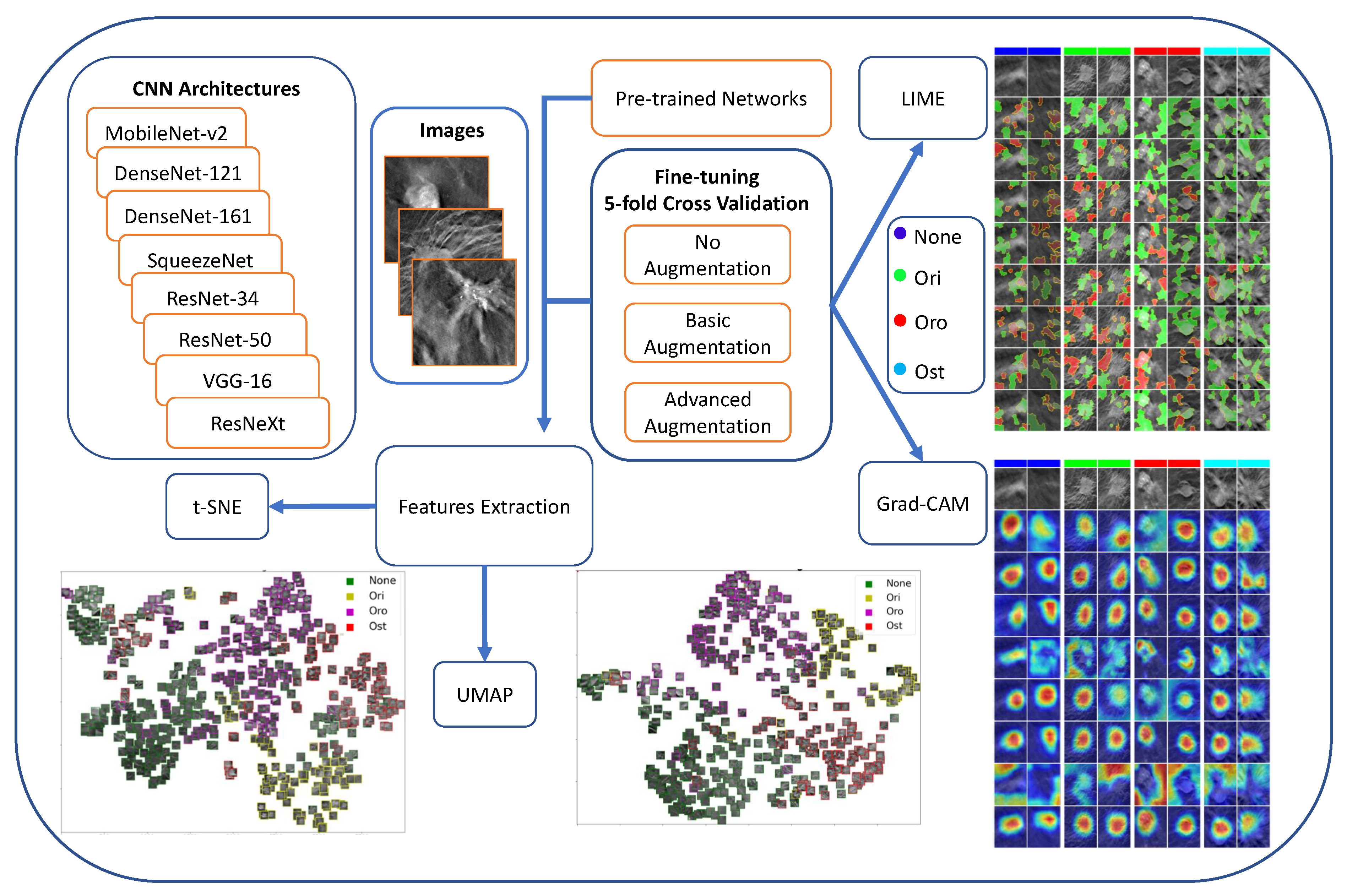
3. Materials and Methods
3.1. Dataset
3.2. CNN Models
- VGGThe VGG [50] comes in two famous versions, with 16 and 19 layers comprising 144 million parameters. This study considers the earlier VGG-16, which consists of several number of channels, 3 × 3 receptive fields, and a stride of 1. This model is composed of convolution layers, max pooling layers, fully connected layers with 5 blocks and each block with a max pooling layer, and extra convolutional layers contained in the last three blocks.
- ResNetThe deep neural networks suffer from the gradient vanish problem, which led to the development of residual network (ResNet) architecture. The ResNet takes care of the gradient vanishing problem and makes sure the performance remains satisfactory over the top and lower layers. ResNet comes with several variants where the number of layers is the distinguishing parameter among numerous architectures, however the underlying mechanism remains similar. This architecture utilizes skip connections between layers. The ResNet-34 and ResNet-50 [51] contain 34 and 50 layers and implement residual learning. This net is efficient to train and also improves the accuracy, which led us to utilize the two versions of network for multiclass classification purposes.
- ResNeXtThe ResNeXt, a counterpart of ResNet, is a specifically designed image classification network with very few tuneable parameters. It contains a series of blocks with a set of aggregations of similar topology with an additional dimension called cardinality. This cardinality, which creates major difference between its brother networks, competes with the depth and width of the network [52]. The simpler architecture based on VGG and ResNet with fewer parameters yields better accuracy on ImageNet classification dataset. The word NeXt in the name of the network refers to next dimension which surpasses ResNet-101, ResNet-152, ResNet-200, Inception-v3, and ResNet-v2 on the ImageNet dataset in accuracy.
- DenseNetThe DenseNet [53], or in other cases, dense convolutional network, is a type of CNN designed to guarantee the maximum information flow between all layers in the network. The layers are subjected to align the feature map size and connect among each other, forming a dense network. The DenseNet works on feed-forward principle. Each layer in the network receives the input from the preceding layer, grabs the additional input, and hands it over to the following layer along with the feature map. All the layers follow a similar analogy. Differently from ResNets, in which the features are not combined through summation before they are passed into a layer, the feature combination is performed by the concatenation of these ones.DenseNet comes with several variants where the number of layers is the distinguishing parameter among numerous architectures, however the underlying mechanism remains similar. The DenseNet-121 and DenseNet-161 contain 121 and 161 layers and follow the feed-forward method. This net is efficient to train and also improves the accuracy, which led us to utilize the two versions of network for multiclass classification purposes.
- SqueezeNetThe SqueezeNet is another popular CNN model particularly known for its smaller size. The major motivations and reasons that caused this network to be smaller include the following: (a) during the procedure of training, the communication over the servers is shortened, (b) the minimum requirement of bandwidth for exporting a model from cloud to any other device is also cut, and (c) the smaller a model is, the less hardware and memory it requires to run.The SqueezeNet architecture is also simple; it contains 8 fire modules sandwiched between two convolutional layers. The sandwiched fire modules also contain a squeeze convolution layer with numerous filters of varying sizes. Each fire module comprises several filters that increase with respect to the network progression, being fewer in the start and more in the end. The SqueezeNet also utilizes the max pooling operation at several levels, including first and last layers.The SqueezeNet appears to achieve comparable accuracy to AlexNet on the ImageNet dataset with fifty-times-reduced number of parameters. It also offers scalability that implies that the size of SqueezeNet model can also be compressed to as low as 0.5 MB.
- MobileNet-v2The MobileNet-v2 [54], a depthwise separable convolutional network aimed at downsizing the model, is an architecture based on inverted residual connections. These residual connections appear between bottleneck layers. The total number of residual bottleneck layers in MobileNet-v2 count to 19 which follow the fully convolution layer comprising 32 filters. The network brings several benefits, including the time and memory savings with higher accuracy of results. The output of the model speaks to the validity of the architecture.
3.3. Explainable AI Methods
3.3.1. Perceptive XAI
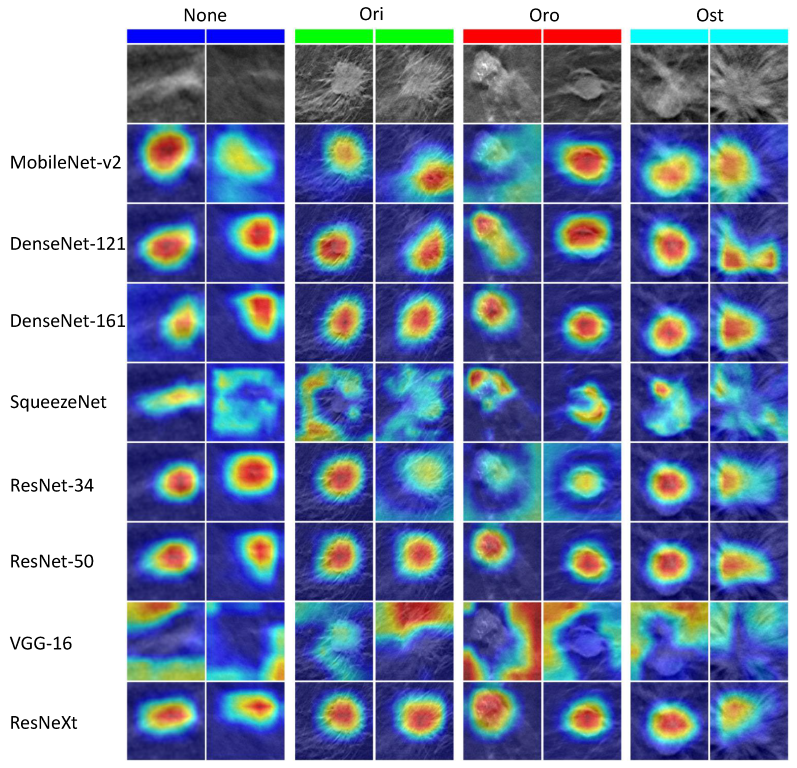
- Grad-CAMAccording to Das et al. [55], Grad-CAM can be classified as a back-propagation-based method, meaning that the algorithm makes several forward-passes (one or more) through the neural network and generates attributions during the back-propagation stage using partial derivatives of the activations. Contrary to the CAM, which requires a particular pattern of network under analysis, the Grad-CAM is the generalization that can be applied without any modifications in the DL model [56].The Grad-CAM produces a heatmap of the class activation in response to the input image and a class. In other words, for a particular provided class, the Grad-CAM produces approximate and comprehensible representations of the network’s decision-making mechanism in the form of a heatmap that translates to the feature importance. Specifically, in the last layers of a CNN, neurons look for semantic information associated with a specific class. In this layer, Grad-CAM uses the gradient flowing into it to assign a weight to each neuron according to its contribution to the decision in the classification task. The computed information is translated into a jet color scheme to depict the saliency zones, where the red color represents the higher intensity, i.e., pixels on which the network is focusing more for performing the classification, while the blue color represents the lower intensity of the focus.
- Local Interpretable Model-Agnostic ExplanationsIn this study, the authors incorporated another well-known explanation technique based on model-agnostic phenomena known as LIME, that can be applied to any DL model. Specifically:
- -
- Local: states that LIME explains the behavior of the model by approximating its local behavior;
- -
- Interpretable: emphasizes the ability of the LIME to provide an output useful to understand the behavior of the model from a human point of view;
- -
- Model-Agnostic: means that LIME is not dependent on the model used; all models are treated as a black-box.
In our classification problem, the explanation of LIME remains simple. It takes the superpixels (a patch of pixels) of the original input image after generating a linear model, and generates several samples by exploiting the superpixels. The quick-shift algorithm is responsible for the computation of superpixels of an image. Thereafter, the perturbation images are generated and the final prediction is made.Afterwards, a heatmap appears over the image that highlights the important pixels, i.e., regions that contribute in classification. The positively contributing features are highlighted in green while the negatively contributing superpixels are colored in red. The LIME also allows to pick a threshold value to select the number of top contributing pixels, either positively or negatively.
3.3.2. Mathematically Explained XAI
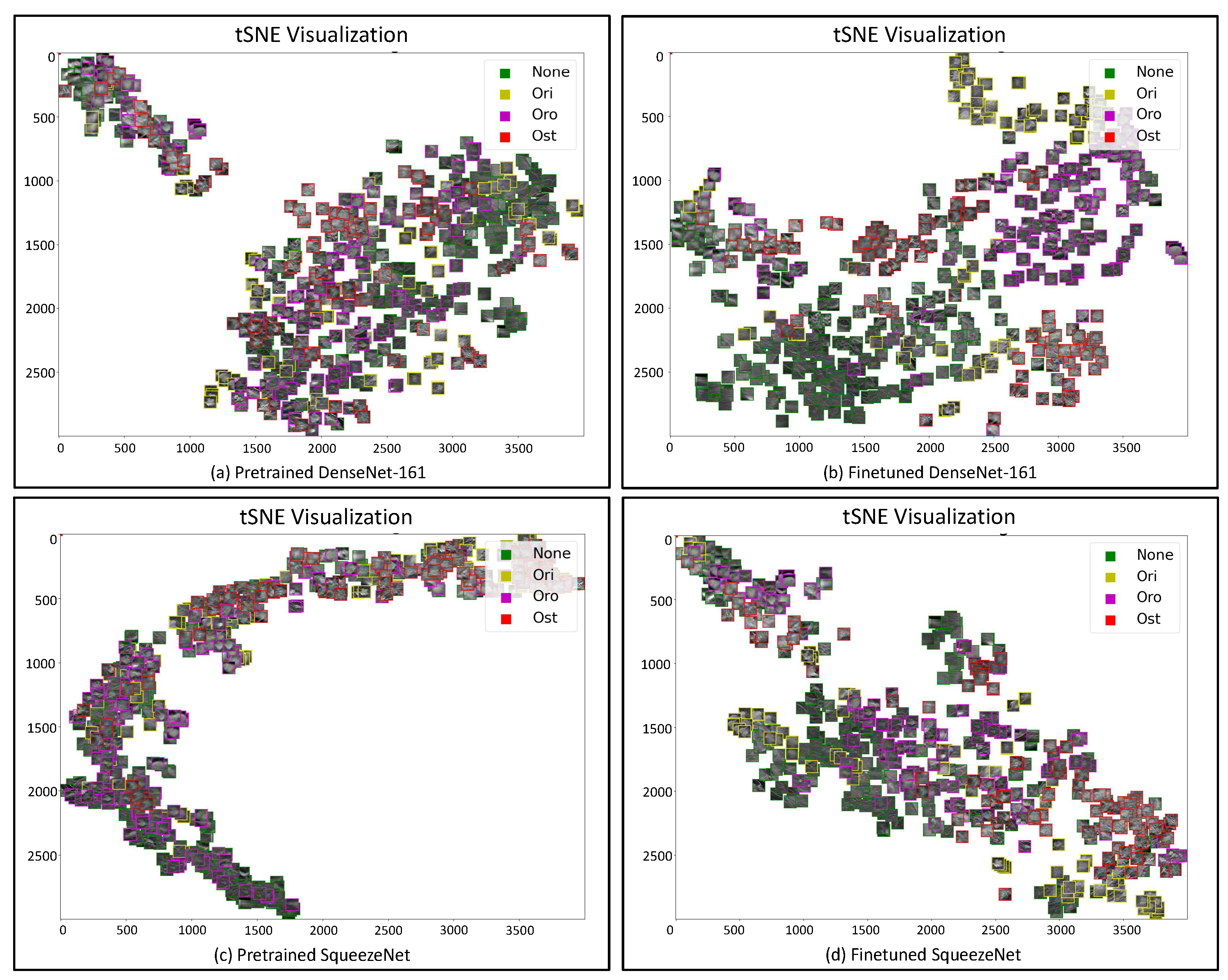
- T-Distributed Stochastic Neighbor Embedding (t-SNE)The t-SNE [57] is a variation of the SNE technique that makes the visualization of high-dimensional data possible by associating with each datum a location in lower dimensional space of two or three dimensions. It has been developed to face two issues that affect SNE technique:
- 1.
- The optimization of the cost function, by using a variation of SNE cost function (symmetrized) and using a Student’s t distribution for the computation of similarity between two datapoints in the lower-dimensional space.
- 2.
- The so-called “crowding problem”, by using a heavy-tailored distribution in low-dimensional space.
- Uniform Manifold Approximation and Projection (UMAP)The UMAP [58] is a nonlinear technique for the dimensionality reduction. It is based on three assumptions:
- 1.
- Data are uniformly distributed on an existing manifold;
- 2.
- Topological structure of the manifold should be preserved;
- 3.
- Manifold is locally connected.
The UMAP method can be divided into two main phases: learning a manifold structure in a high-dimensional space and finding the relative representation in the low-dimensional space. In the first phase, the initial step is to find the nearest neighbors for all datapoints, using the nearest-neighbor-descent algorithm. Then, UMAP constructs a graph by connecting the neighbors identified previously; it should be noticed that the data are uniformly distributed across the manifold, so the space between datapoints varies according to regions where data are denser or sparse. According to this assumption, it is possible to introduce the concept of ‘edge weights’: from each point, the distance with respect to the nearest neighbors is computed, so the edge weights between datapoints are computed, but there exists a problem of disagreeing edges.
3.4. Experimental Workflow
3.4.1. Data Augmentation Procedures
3.4.2. Training Procedures and Cross-Validation
3.5. Classification Performance Assessment
Explainable AI
4. Experimental Outcomes
4.1. Performance Module
4.1.1. Classification Results
4.1.2. Train and Validation Loss Trends
4.2. Area under the Curve and Number of Parameters Trade-Off
4.3. XAI Interpretation
4.3.1. t-SNE and UMAP
4.3.2. Class Activation Mapping
4.3.3. Local Interpretable Model-Agnostic Explanations
5. Discussion
5.1. Shape-Based Breast Cancer Classification
5.2. Explainable AI in Breast Cancer Classification
6. Limitations and Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, M.; Ayyoubzadeh, S.M.; Javanmard, Z.; Kalhori, S.R.N. A systematic review of decision aids for mammography screening: Focus on outcomes and characteristics. Int. J. Med. Inform. 2021, 149, 104406. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, Z. A review on image-based approaches for breast cancer detection, segmentation, and classification. Expert Syst. Appl. 2021, 182, 115204. [Google Scholar] [CrossRef]
- Kulkarni, S.; Freitas, V.; Muradali, D. Digital breast tomosynthesis: Potential benefits in routine clinical practice. Can. Assoc. Radiol. J. 2022, 73, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ma, J. Association between imaging characteristics and different molecular subtypes of breast cancer. Acad. Radiol. 2017, 24, 426–434. [Google Scholar] [CrossRef]
- Cai, S.Q.; Yan, J.X.; Chen, Q.S.; Huang, M.L.; Cai, D.L. Significance and application of digital breast tomosynthesis for the BI-RADS classification of breast cancer. Asian Pac. J. Cancer Prev. 2015, 16, 4109–4114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sickles, E.; D’Orsi, C.; Bassett, L. ACR BI-RADS® Mammography. In ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Lee, S.H.; Chang, J.M.; Shin, S.U.; Chu, A.J.; Yi, A.; Cho, N.; Moon, W.K. Imaging features of breast cancers on digital breast tomosynthesis according to molecular subtype: Association with breast cancer detection. Br. J. Radiol. 2017, 90, 20170470. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Yao, M.; Cai, D.; Yan, J.; Huang, M.; Yan, L.; Huang, H. Association between digital breast tomosynthesis and molecular subtypes of breast cancer. Oncol. Lett. 2019, 17, 2669–2676. [Google Scholar] [CrossRef]
- Hu, Z.; Tang, J.; Wang, Z.; Zhang, K.; Zhang, L.; Sun, Q. Deep learning for image-based cancer detection and diagnosis- A survey. Pattern Recognit. 2018, 83, 134–149. [Google Scholar] [CrossRef]
- Bevilacqua, V. Three-dimensional virtual colonoscopy for automatic polyps detection by artificial neural network approach: New tests on an enlarged cohort of polyps. Neurocomputing 2013, 116, 62–75. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Brunetti, A.; Trotta, G.F.; Dimauro, G.; Elez, K.; Alberotanza, V.; Scardapane, A. A novel approach for Hepatocellular Carcinoma detection and classification based on triphasic CT Protocol. In Proceedings of the 2017 IEEE congress on evolutionary computation (CEC), Donostia, Spain, 5–8 June 2017; pp. 1856–1863. [Google Scholar]
- Bevilacqua, V.; Altini, N.; Prencipe, B.; Brunetti, A.; Villani, L.; Sacco, A.; Morelli, C.; Ciaccia, M.; Scardapane, A. Lung Segmentation and Characterization in COVID-19 Patients for Assessing Pulmonary Thromboembolism: An Approach Based on Deep Learning and Radiomics. Electronics 2021, 10, 2475. [Google Scholar] [CrossRef]
- Chugh, G.; Kumar, S.; Singh, N. Survey on machine learning and deep learning applications in breast cancer diagnosis. Cogn. Comput. 2021, 13, 1451–1470. [Google Scholar] [CrossRef]
- Houssein, E.H.; Emam, M.M.; Ali, A.A.; Suganthan, P.N. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review. Expert Syst. Appl. 2021, 167, 114161. [Google Scholar] [CrossRef]
- Wu, J.; Hicks, C. Breast Cancer Type Classification Using Machine Learning. J. Pers. Med. 2021, 11, 61. [Google Scholar] [CrossRef]
- Khan, S.; Islam, N.; Jan, Z.; Din, I.U.; Rodrigues, J.J.C. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 2019, 125, 1–6. [Google Scholar] [CrossRef]
- Yadav, S.S.; Jadhav, S.M. Thermal infrared imaging based breast cancer diagnosis using machine learning techniques. Multimed. Tools Appl. 2022, 81, 13139–13157. [Google Scholar] [CrossRef]
- Ragab, D.A.; Attallah, O.; Sharkas, M.; Ren, J.; Marshall, S. A framework for breast cancer classification using multi-DCNNs. Comput. Biol. Med. 2021, 131, 104245. [Google Scholar] [CrossRef]
- Ghiasi, M.M.; Zendehboudi, S. Application of decision tree-based ensemble learning in the classification of breast cancer. Comput. Biol. Med. 2021, 128, 104089. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Satapathy, S.C.; Guttery, D.S.; Górriz, J.M.; Wang, S.H. Improved breast cancer classification through combining graph convolutional network and convolutional neural network. Inf. Process. Manag. 2021, 58, 102439. [Google Scholar] [CrossRef]
- Mokni, R.; Gargouri, N.; Damak, A.; Sellami, D.; Feki, W.; Mnif, Z. An automatic Computer-Aided Diagnosis system based on the Multimodal fusion of Breast Cancer (MF-CAD). Biomed. Signal Process. Control 2021, 69, 102914. [Google Scholar] [CrossRef]
- Shi, J.; Vakanski, A.; Xian, M.; Ding, J.; Ning, C. EMT-NET: Efficient multitask network for computer-aided diagnosis of breast cancer. arXiv 2022, arXiv:2201.04795. [Google Scholar]
- Shen, Y.; Wu, N.; Phang, J.; Park, J.; Liu, K.; Tyagi, S.; Heacock, L.; Kim, S.G.; Moy, L.; Cho, K.; et al. An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization. Med. Image Anal. 2021, 68, 101908. [Google Scholar] [CrossRef] [PubMed]
- Saffari, N.; Rashwan, H.A.; Abdel-Nasser, M.; Kumar Singh, V.; Arenas, M.; Mangina, E.; Herrera, B.; Puig, D. Fully Automated Breast Density Segmentation and Classification Using Deep Learning. Diagnostics 2020, 10, 988. [Google Scholar] [CrossRef]
- Shrivastava, N.; Bharti, J. Breast tumor detection and classification based on density. Multimed. Tools Appl. 2020, 79, 26467–26487. [Google Scholar] [CrossRef]
- Kopans, D. Mammography, Breast Imaging; JB Lippincott Company: Philadelphia, PA, USA, 1989; Volume 30, pp. 34–59. [Google Scholar]
- Kisilev, P.; Sason, E.; Barkan, E.; Hashoul, S. Medical image description using multi-task-loss CNN. In Deep Learning and Data Labeling for Medical Applications; Springer: Berlin/Heidelberg, Germany, 2016; pp. 121–129. [Google Scholar]
- Singh, V.K.; Rashwan, H.A.; Romani, S.; Akram, F.; Pandey, N.; Sarker, M.M.K.; Saleh, A.; Arenas, M.; Arquez, M.; Puig, D.; et al. Breast tumor segmentation and shape classification in mammograms using generative adversarial and convolutional neural network. Expert Syst. Appl. 2020, 139, 112855. [Google Scholar] [CrossRef]
- Kim, S.T.; Lee, H.; Kim, H.G.; Ro, Y.M. ICADx: Interpretable computer aided diagnosis of breast masses. In Proceedings of the Medical Imaging 2018: Computer-Aided Diagnosis, Houston, TX, USA, 10–15 February 2018; Volume 10575, p. 1057522. [Google Scholar]
- Samek, W.; Montavon, G.; Vedaldi, A.; Hansen, L.K.; Müller, K.R. Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer Nature: Berlin/Heidelberg, Germany, 2019; Volume 11700. [Google Scholar]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Gulum, M.A.; Trombley, C.M.; Kantardzic, M. A Review of Explainable Deep Learning Cancer Detection Models in Medical Imaging. Appl. Sci. 2021, 11, 4573. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A survey on explainable artificial intelligence (xai): Toward medical xai. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4793–4813. [Google Scholar] [CrossRef]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable ai via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2022, 77, 29–52. [Google Scholar] [CrossRef]
- Suh, Y.J.; Jung, J.; Cho, B.J. Automated breast cancer detection in digital mammograms of various densities via deep learning. J. Pers. Med. 2020, 10, 211. [Google Scholar] [CrossRef]
- Ricciardi, R.; Mettivier, G.; Staffa, M.; Sarno, A.; Acampora, G.; Minelli, S.; Santoro, A.; Antignani, E.; Orientale, A.; Pilotti, I.; et al. A deep learning classifier for digital breast tomosynthesis. Phys. Medica 2021, 83, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Sickles, E.A.; D’Orsi, C.J.; Bassett, L.W.; Appleton, C.M.; Berg, W.A.; Burnside, E.S.; Mendelson, E.B.; Morris, E.A.; Creech, W.E.; Butler, P.F.; et al. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013; pp. 39–48. [Google Scholar]
- Bevilacqua, V.; Brunetti, A.; Guerriero, A.; Trotta, G.F.; Telegrafo, M.; Moschetta, M. A performance comparison between shallow and deeper neural networks supervised classification of tomosynthesis breast lesions images. Cogn. Syst. Res. 2019, 53, 3–19. [Google Scholar] [CrossRef]
- Skaane, P.; Bandos, A.I.; Niklason, L.T.; Sebuødegård, S.; Østerås, B.H.; Gullien, R.; Gur, D.; Hofvind, S. Digital mammography versus digital mammography plus tomosynthesis in breast cancer screening: The Oslo Tomosynthesis Screening Trial. Radiology 2019, 291, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qin, G.; He, Q.; Sun, L.; Zeng, H.; He, Z.; Chen, W.; Zhen, X.; Zhou, L. Digital breast tomosynthesis versus digital mammography: Integration of image modalities enhances deep learning-based breast mass classification. Eur. Radiol. 2020, 30, 778–788. [Google Scholar] [CrossRef]
- Mendel, K.; Li, H.; Sheth, D.; Giger, M. Transfer learning from convolutional neural networks for computer-aided diagnosis: A comparison of digital breast tomosynthesis and full-field digital mammography. Acad. Radiol. 2019, 26, 735–743. [Google Scholar] [CrossRef]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Richter, C.; Cha, K. Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. Phys. Med. Biol. 2018, 63, 095005. [Google Scholar] [CrossRef]
- Fotin, S.V.; Yin, Y.; Haldankar, H.; Hoffmeister, J.W.; Periaswamy, S. Detection of soft tissue densities from digital breast tomosynthesis: Comparison of conventional and deep learning approaches. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis. International Society for Optics and Photonics, San Diego, CA, SUA, 27 February–3 March 2016; Volume 9785, p. 97850X. [Google Scholar]
- Hamouda, S.; El-Ezz, R.; Wahed, M.E. Enhancement accuracy of breast tumor diagnosis in digital mammograms. J. Biomed. Sci. 2017, 6, 1–8. [Google Scholar] [CrossRef]
- Sakai, A.; Onishi, Y.; Matsui, M.; Adachi, H.; Teramoto, A.; Saito, K.; Fujita, H. A method for the automated classification of benign and malignant masses on digital breast tomosynthesis images using machine learning and radiomic features. Radiol. Phys. Technol. 2020, 13, 27–36. [Google Scholar] [CrossRef]
- Boumaraf, S.; Liu, X.; Ferkous, C.; Ma, X. A new computer-aided diagnosis system with modified genetic feature selection for bi-RADS classification of breast masses in mammograms. BioMed Res. Int. 2020, 2020, 7695207. [Google Scholar] [CrossRef]
- Masud, M.; Eldin Rashed, A.E.; Hossain, M.S. Convolutional neural network-based models for diagnosis of breast cancer. Neural Comput. Appl. 2020, 1–12. [Google Scholar] [CrossRef]
- Lou, M.; Wang, R.; Qi, Y.; Zhao, W.; Xu, C.; Meng, J.; Deng, X.; Ma, Y. MGBN: Convolutional neural networks for automated benign and malignant breast masses classification. Multimed. Tools Appl. 2021, 80, 26731–26750. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Das, A.; Rad, P. Opportunities and challenges in explainable artificial intelligence (xai): A survey. arXiv 2020, arXiv:2006.11371. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Van Der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Meta-AI. PyTorch Transforms. 2022. Available online: https://pytorch.org/vision/stable/transforms.html (accessed on 5 December 2021).










| Transform | No Aug | Basic Aug | Adv Aug |
|---|---|---|---|
| RandomRotation90 | ✗ | ✓ | ✓ |
| RandomRotation180 | ✗ | ✓ | ✓ |
| RandomRotation270 | ✗ | ✓ | ✓ |
| RandomHorizontalFlip | ✗ | ✓ | ✓ |
| RandomVerticalFlip | ✗ | ✓ | ✓ |
| ColorJitter | ✗ | ✗ | ✓ |
| Normalization | ✓ | ✓ | ✓ |
| Architecture | Area under the Curve (AUC) | ||
|---|---|---|---|
| No Aug (None, Ori, Oro, Ost) | Basic Aug (None, Ori, Oro, Ost) | Adv Aug (None, Ori, Oro, Ost) | |
| MobileNet-v2 | 91.9 ± 1.1 | 92.4 ± 0.9 | 93.6 ± 1.2 |
| 97.4 ± 0.4 | 98.0 ± 0.6 | 97.6 ± 0.9 | |
| 95.2 ± 1.3 | 95.9 ± 1.1 | 96.3 ± 0.9 | |
| 95.8 ± 0.7 | 96.6 ± 0.5 | 96.5 ± 0.7 | |
| 95.1 | 95.7 | 96.0 | |
| DenseNet-121 | 90.1 ± 1.2 | 93.9 ± 1.9 | 94.5 ± 1.3 |
| 94.2 ± 1.4 | 98.5 ± 0.6 | 98.2 ± 0.8 | |
| 89.9 ± 1.7 | 95.5 ± 0.6 | 96.7 ± 0.8 | |
| 92.9 ± 1.8 | 97.1 ± 0.8 | 97.2 ± 1.2 | |
| 91.8 | 96.2 | 96.6 | |
| DenseNet-161 | 94.8 ± 0.9 | 95.8 ± 1.0 | 96.4 ± 0.5 |
| 97.6 ± 1.4 | 99.1 ± 0.7 | 99.4 ± 0.2 | |
| 95.8 ± 1.3 | 97.8 ± 1.0 | 98.7 ± 0.7 | |
| 97.0 ± 0.9 | 98.2 ± 0.3 | 98.0 ± 0.7 | |
| 96.3 | 97.7 | 98.2 | |
| SqueezeNet | 50.9 ± 3.0 | 56.6 ± 5.6 | 62.7 ± 8.1 |
| 85.9 ± 3.2 | 84.3 ± 1.4 | 86.4 ± 2.9 | |
| 68.9 ± 5.6 | 67.6 ± 3.8 | 71.7 ± 7.2 | |
| 83.8 ± 2.6 | 86.2 ± 3.7 | 87.6 ± 3.1 | |
| 72.4 | 73.7 | 77.1 | |
| ResNet-34 | 92.0 ± 0.8 | 94.5 ± 1.0 | 95.4 ± 0.6 |
| 96.2 ± 0.8 | 98.6 ± 0.5 | 98.9 ± 0.5 | |
| 94.7 ± 1.7 | 97.6 ± 0.4 | 97.4 ± 1.0 | |
| 96.1 ± 1.3 | 97.6 ± 0.7 | 97.7 ± 0.7 | |
| 94.8 | 97.1 | 97.3 | |
| ResNet-50 | 93.8 ± 1.1 | 95.3 ± 1.2 | 96.2 ± 0.6 |
| 98.0 ± 0.5 | 99.4 ± 0.3 | 99.3 ± 0.3 | |
| 95.8 ± 0.8 | 97.8 ± 0.6 | 97.9 ± 0.7 | |
| 97.0 ± 1.0 | 97.8 ± 0.9 | 98.5 ± 0.4 | |
| 96.1 | 97.6 | 98.0 | |
| VGG-16 | 90.6 ± 1.7 | 92.5 ± 1.4 | 93.6 ± 1.3 |
| 98.1 ± 0.6 | 98.9 ± 0.6 | 97.7 ± 0.7 | |
| 96.1 ± 0.7 | 96.7 ± 0.7 | 97.2 ± 0.7 | |
| 96.6 ± 0.4 | 97.7 ± 0.6 | 98.1 ± 0.6 | |
| 95.3 | 96.4 | 96.6 | |
| ResNeXt | 94.1 ± 1.0 | 96.1 ± 0.7 | 95.8 ± 0.7 |
| 97.7 ± 0.7 | 99.3 ± 0.2 | 99.0 ± 0.7 | |
| 96.0 ± 0.8 | 97.9 ± 0.7 | 98.2 ± 0.8 | |
| 97.1 ± 0.5 | 98.3 ± 0.3 | 98.2 ± 0.9 | |
| 96.2 | 97.9 | 97.8 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, S.M.; Buongiorno, D.; Altini, N.; Berloco, F.; Prencipe, B.; Moschetta, M.; Bevilacqua, V.; Brunetti, A. Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence. Appl. Sci. 2022, 12, 6230. https://doi.org/10.3390/app12126230
Hussain SM, Buongiorno D, Altini N, Berloco F, Prencipe B, Moschetta M, Bevilacqua V, Brunetti A. Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence. Applied Sciences. 2022; 12(12):6230. https://doi.org/10.3390/app12126230
Chicago/Turabian StyleHussain, Sardar Mehboob, Domenico Buongiorno, Nicola Altini, Francesco Berloco, Berardino Prencipe, Marco Moschetta, Vitoantonio Bevilacqua, and Antonio Brunetti. 2022. "Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence" Applied Sciences 12, no. 12: 6230. https://doi.org/10.3390/app12126230
APA StyleHussain, S. M., Buongiorno, D., Altini, N., Berloco, F., Prencipe, B., Moschetta, M., Bevilacqua, V., & Brunetti, A. (2022). Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence. Applied Sciences, 12(12), 6230. https://doi.org/10.3390/app12126230











