1. Introduction
Metaheuristics (MH) are computational algorithms inspired by biological phenomena that solve a great diversity of complex scientific and engineering problems, particularly optimization. Some key features of metaheuristics are: (1) robustness, (2) self-organization, (3) adaptation and (4) decentralized control [
1]. Two of the most recognized branches in MH, which group several approaches, are Evolutionary Computation (EC) and Swarm Intelligence (SI) algorithms [
2]. Even though different metaphors inspire them, all have a set of parameters that the user must tune in a manual, adaptive, or self-adaptive manner to help the algorithm’s exploration/exploitation balance [
3]. However, in several MH, the use of a global best candidate solution to direct the search process toward the real optimum is common.
For instance, Harmony Search [
4,
5,
6] utilizes several individuals from the harmony memory (HM) to improvise a new harmony, and this individual is incorporated into the HM only if it is better than the worst individual in the whole population. Differential Evolution [
7] uses in most canonical variants [
8] a weighted difference of at least two parents to generate a new individual, which interchanges information with the actual individual and becomes the trial solution. The selection step compares the trial individual’s fitness against both the actual individual’s fitness and the best individual found so far. Another metaheuristic approach is Particle Swarm Optimization [
9], which considers the local best and the global best to make a velocity vector added to the actual individual to generate a new individual, whose fitness is compared against the local best and the global best and, if necessary, replaces the global best. The canonical Fireworks algorithm [
10,
11], whose population is composed of fireworks, modifies the individuals by consecutively applying the explosion, the Gaussian mutation, and the mapping operators. After such alterations, the new candidate solution’s fitness is compared against the best individual so far, and the individual with better fitness value is kept in memory. Based on some plants’ reproduction strategy, the authors in [
12] proposed the Flower Pollination Algorithm (FPA). This metaheuristic uses the abiotic, biotic, and switch operators to either locally or globally modify the actual candidate solution. As in the previous algorithms, FPA keeps tracking the best global individual found at each iteration. Another algorithm is the Gravitational Search algorithm [
13], in which the movements of the candidate solutions (called objects) consider their masses and accelerations to actualize their velocities and positions. As in other metaheuristics, the algorithm updates the global worst and the global best at each iteration. Other examples of metaheuristics that actualize the best solution found at each iteration are the Firefly Algorithm [
14,
15], Cuckoo Search [
16], Artificial Bee Colony optimization [
17,
18] and Chicken Swarm optimization [
19].
In order to solve high-dimensional problems and evaluate a large amount of data, it is necessary to improve the efficiency and execution times of evolutionary algorithms. Many authors consider parallel and distributed approaches of metaheuristic algorithms as alternatives to overcome the difficulties derived from the increase in data complexity. A recent review of parallel and distributed approaches for evolutionary algorithms is presented in [
20]. In addition, much research reports metaheuristics translated to parallel languages, such as CUDA (Compute Unified Device Architecture) [
21], which runs on parallel general-purpose graphic processing units (GPGPU). For example, a survey of GPU parallelization strategies for metaheuristics is conducted in [
22]. The authors of [
23] propose two approaches to parallelize the Gravitational Search algorithm using CUDA: one that modifies the whole population with a single kernel and another that utilizes multiple kernels to operate the complete population. In the multiple kernel version, the least parallelizable is the update mass kernel. At a given moment, all threads could be trying to write the global best to the same memory location.
With respect to DE, several proposals have been made to parallelize this algorithm, such as the one presented in [
24], where opposition-based learning and Differential Evolution are programmed with CUDA to optimize 19 benchmark functions. The authors cannot perform the fittest selection in parallel, and they propose a reduction approach to find the best individual at each iteration. A similar approach is found in [
25], where only four functions are tested up to 100 dimensions. In this case, the authors propose the use of an adaptive scaling factor and an adaptive crossover probability; even though the authors utilize the DE/rand/1/bin version, they also keep track of the best individual so far. To represent DE variants, a general pattern DE/
x/y/z is used, where
x describes the base vector selection type,
y represents the number of differentials and
z is the type of crossover operator, and if there is an asterisk symbol (*) it means that any valid operator can be used for that process. Thus, for DE/rand/1/bin, “rand” indicates that the individuals are randomly chosen, “1” specifies that only one vector difference is used to form the mutated population and “bin” means that a binomial crossover is applied.
A survey of DE parallelized on GPGPU is found in [
26], tracking the research efforts to parallelize DE from computer networks since 2004, passing through the use of GPGPU with first (and therefore, with a lack of features) CUDA SDKs and ending with a brief exploration of DE and GPU-based frameworks. It is worth mentioning that, in most of the articles, the original operators are preserved and only the parallel implementation is performed using one kernel for each process. To have a general idea about the implementation of DE on GPUs,
Table 1 shows some proposals highlighting the implemented DE version, whether any operator was modified or not, the number and type of test functions and the maximum dimensionality. It can be observed that the idea of proposing modifications to differential evolution operators has not been widely investigated.
Some metaheuristics utilize two methods to update the global best (individual and fitness) by: (a) comparing it against the corresponding values of every individual in the population and (b) sorting the corresponding values of the entire population. In the first approach, parallelization implies that individuals will be trying to update the best fitness value in the actual iteration and the best individual to the same memory’s positions: this is the worst-case scenario. This could produce race conditions, which are undesirable because they could produce a loss of information. A better method is the sorting-based approach which can only be partially parallelized, in fact, this DE algorithm version is the one used for comparisons in this research.
This paper considers a modification, based on the stigmergy concept, to Differential Evolution (version best/1/bin, in this case the best individual solution of the DE population is selected as the target vector) to eliminate the global best utilization. For this purpose, the mutation and crossover operators are modified. Such adjustment, which we called Stigmergic Differential Evolution (SDE), has a similar performance to the original DE with a lower computational cost in some cases when programmed into GPGPU; mainly, it preserves a good convergence behavior as the dimensionality grows. Therefore, we consider that other metaheuristics that utilize the global best to guide the optimization process could benefit from the proposed methodology.
The remaining paper is as follows:
Section 2 explains the concept of stigmergy and explores some of its applications to solve engineering problems.
Section 3 gives an account of our proposal and the benchmark of optimization problems for the comparisons. In contrast,
Section 4 explains the experimental setup, shows the results and briefly discusses the results.
Section 5 concludes this work and gives some future venues around stigmergic metaheuristics.
2. Stigmergy and Some Stigmergic Applications
Some authors consider insect colonies as complex adaptive systems, with properties of: (1) spatial distribution; (2) decentralized control; (3) hierarchical organization of individuals; (4) adaptation to environmental changes, among others [
17,
34]. For the second property, this means that there is no global control [
34], and therefore the interactions among individuals are distributed. To explain the behavior of insect colonies, researchers proposed the term stigmergy. Such a concept comes from entomology, and Pierre-Paul Grassé proposed it to explain the coordination and collaboration by the indirect communication [
35,
36] of social insects (particularly termites and ants) to complete complicated projects [
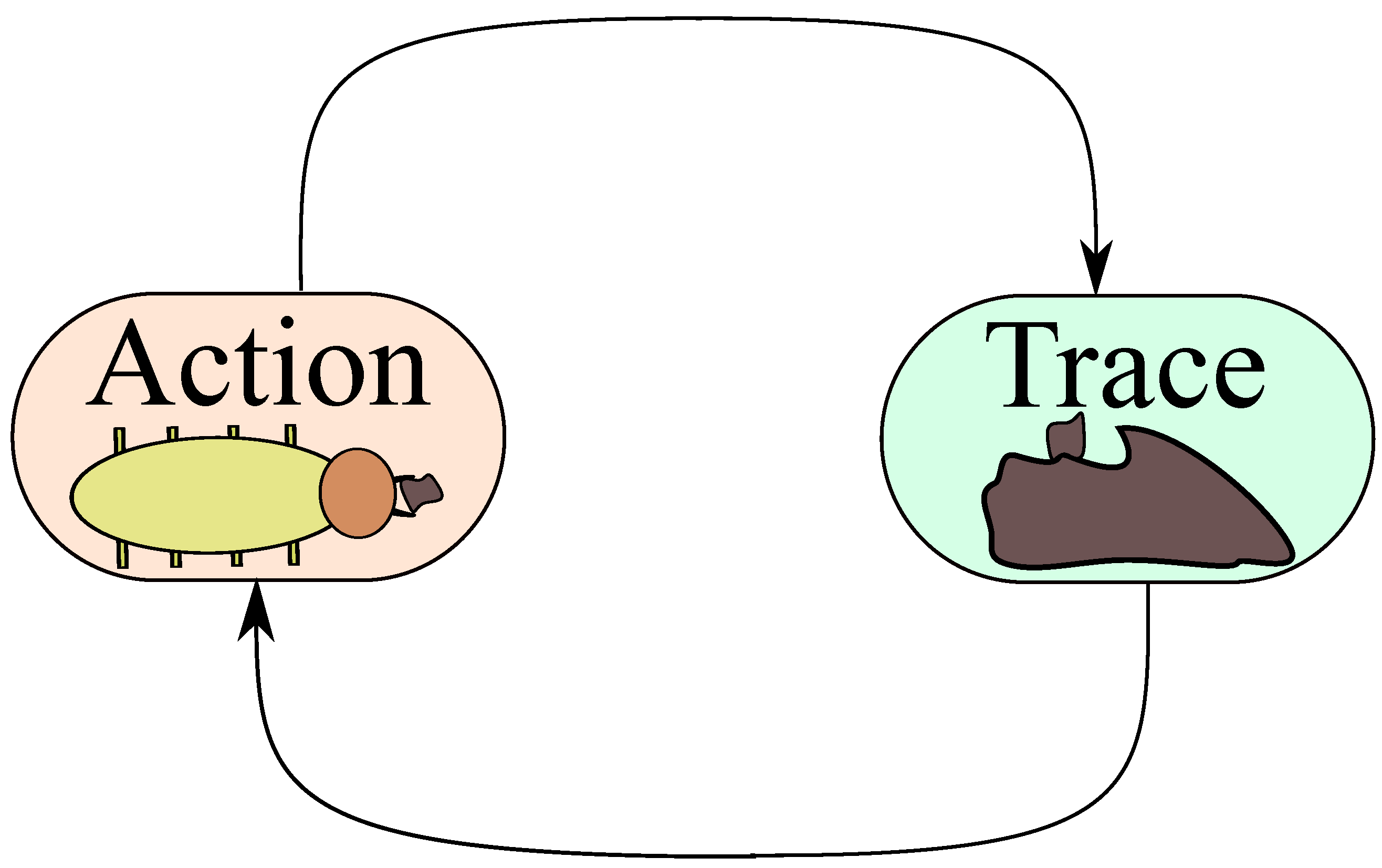
37]. In the original paper, Grassé considers that every individual in a colony that acts (ergon) on the medium leaves a trace (stigma), which at the same time stimulates another action from the same or another individual (
Figure 1). Actions and marks occur on the medium, and as mentioned, the latter only stimulate (but not force) the action of either the same or other individuals.
The concept of stigmergy has served as an inspiration to propose algorithms utilized to solve some problems in engineering [
38,
39,
40], and it also was used as an inspiration to propose one of the most known metaheuristic algorithms: Ant System (AS) [
41], the predecessor of Ant Colony Optimization (ACO) and its variants. In these approaches, it is the pheromone-based construction of trails that represents the stigmergic mechanism [
36,
42,
43].
ACO considers a population of artificial agents that keep some memory in the form of the pheromone of the best solutions visited so-far; those solutions are paths of a completely connected graph, constructed by selecting every vertex in the path in a probabilistic manner, being that vertices in the graph with the most artificial pheromones are the more likely to be selected. As the algorithm works in the graphs’ space to construct the solutions, it is natural that the first applications of this technique are in combinatorial optimization, such as the Traveling Salesman Problem or job-shop scheduling. The application of ACO to solve numerical optimization problems is not straightforward. Often, the proposals include hybridizations with other techniques, as the continuous ACO (CACO) [
44], continuous interacting ant colony (CIAC) [
45], or continuous ant colony system (CACS) [
42]; however, there are some research efforts using direct modifications to the original ACO. For instance, in his doctoral thesis, Korošec proposed two stigmergy-based modifications of the original ACO to solve numerical problems, including the Differential Ant-Stigmergy Algorithm [
36]. In the proposal, the authors present a fine-grained discretization of the continuous search space to construct a directed graph, to which they apply the operators Gaussian pheromone aggregation, pheromone evaporation and parameter precision to build a solution for the continuous optimization problem at hand. Korošec and colleagues later improved the original algorithm to tackle high-complexity problems or even dynamic problems [
43,
46] by using a pheromone operator based on the Cauchy distribution.
A recently proposed stigmergic algorithm to solve numeric problems is the teaching–learning-based optimization [
47]. In the paper, students’ cooperation (ergon/action) through a board (stigma/mark) simulates the stigmergic process. Algorithmically, the authors create a ranking matrix (of the same size as the original population) for every individual, and they use a roulette wheel to select a guide student. Such guiding students serve as the mean to generate new solutions, which utilize as a standard deviation the weighted average of the remaining individuals. Therefore, the guiding student directs the complete exploration–exploitation process, diluting the stigmergic process as it happens in other metaheuristics.
In the next section, we explain our approach, the Stigmergic Differential Evolution, SDE. The algorithm eliminates the use of global best. Thus, the whole population controls the exploration–exploitation process in a coordinated but decentralized manner; moreover, to explore the efficiency of the proposal and its parallelization feasibility, we programmed it on GPGPU using the CUDA language.
5. Discussion
Stigmergic DE implements slight modifications to the original DE operators, so the computational time required to complete several iterations is practically the same, as suggested by the results in
Table 5 and
Table 6 and after applying the Wilcoxon test to such data.
Figure 4 shows a comparison between the execution time of DE and SDE for each test function, considering
and
. The comparison considers the difference in percentage between the execution times of each algorithm. In most cases (with the exception of the Brent and Griewank functions in these results), the proposal has a better time performance than its DE counterpart of about 17% in the best case. This behavior is similar in all these experiments; however, as already mentioned, SDE has a marginal advantage.
Moreover, both algorithms present similar problems in finding the global optimum in some functions. However, as can be seen from
Table 7 and
Table 8, in the case of the Rastrigin, Schwefel and Zacharov functions, the difference between the final fitness values obtained for SDE and DE is very large. SDE obtains fitness values lower than DE by an order of magnitude 2, and for the case of the Trid, Dixon and Price and Rosenbrock functions, DE performs better than SDE, but the difference is less significant. If the dimension increases (
Table 9 and
Table 10), only in the case of the Sum Powers function does DE have a better performance than SDE, but it is minimal, on the order of 0.0001. These results show the advantage of the proposal in high dimensions; empirically, this is due to the intrinsic parallelism of the stigmergy concept that was used as an inspiration to modify the original operators of a metaheuristic (DE, version best/1/bin in this case). The proposal avoids using the global best in the construction of new candidate solutions, thus simulating indirect communication among all individuals in the population.
In summary, under the same conditions for both metaheuristics, our proposal can find a similar result as DE (and better outcomes for some objective functions) but using fewer iterations on average (
Table 7,
Table 8,
Table 9 and
Table 10), especially when the problems are high-dimensional. Even if the iterations are insufficient to find the global optimum, SDE performs better than DE, so it can be a good alternative for solving multiple engineering optimization problems. As mentioned in [
59], applications can be in industrial engineering for the job-shop scheduling problem [
60,
61], in image processing to perform a multilevel thresholding process on a 2D histogram to segment images [
62,
63], in path planning to solve robotics challenges [
64], or to solve area coverage problems for wireless sensor networks (WSNs) [
65], to mention a few.
6. Conclusions
In this paper, we presented a modification to Differential Evolution that uses the global best to construct new candidate solutions. The adjustment considers stigmergy, a term taken from entomology to explain the indirect communication used by social insects to achieve complex projects. In the proposal, we eliminate the use of global best and instead we employ a local best. Like the real ants or termites, we utilize two neighbors of that local leader in the approach. The new candidate solution uses the leader’s information and two neighbors in conjunction with a crossover operation as in the original DE. The final step in the approach is trace updating, which could modify any of the three individuals and their fitness values, simulating distributed and indirect communication with all the individuals in the artificial colony. We called this changed metaheuristic Stigmergic Differential Evolution (SDE). Due to the elimination of the global best, SDE has features that make it more parallelizable than DE, therefore, both algorithms were programmed in the CUDA-C language.
We compared DE and SDE with 27 objective functions under the same circumstances. The results suggest that SDE achieves faster convergence than DE when the dimensionality increases and there are a larger number of individuals in the population, and it performs similarly to DE for 2D test functions, but DE converges in fewer iterations. It was demonstrated in [
66] that canonical DE is adversely altered by increments in dimensionality, which affects in a super-linear fashion the convergence to the global optimum of the metaheuristic. In that sense, even though SDE has average convergence properties (e.g.,
Table 5 and
Table 6), we claim that its main feature is the capability to maintain a good convergence behavior as dimensionality grows.
The proposed modification could apply to other metaheuristics that use a global best to construct a candidate solution. A possible future work would consider the stigmergy-based adaptation of the original operators in other metaheuristic algorithms and their comparison.