Prediction of Overall Survival in Cervical Cancer Patients Using PET/CT Radiomic Features
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient and Treatment
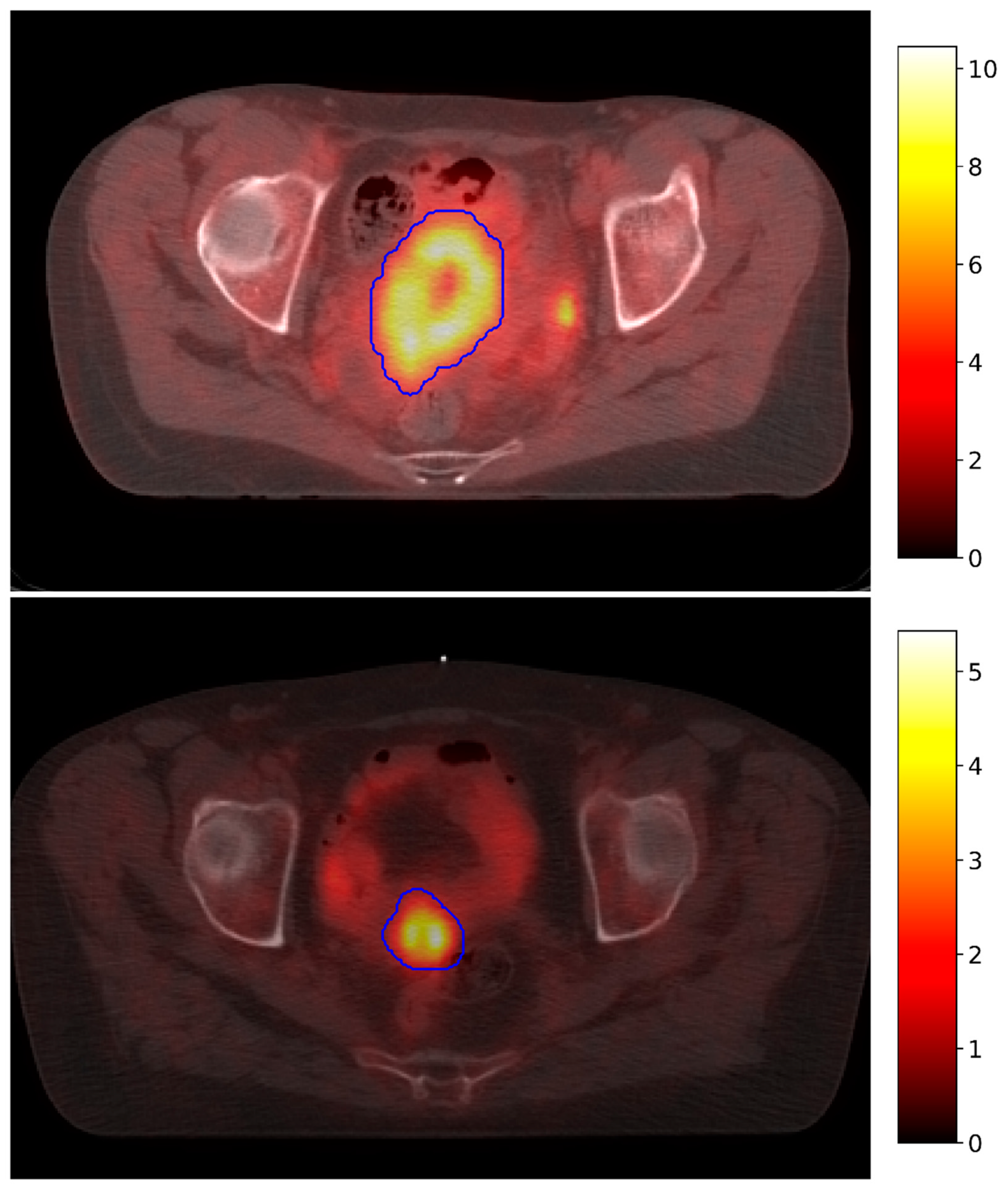
2.2. Data Acquisition
2.3. Database Description and Pre-Processing
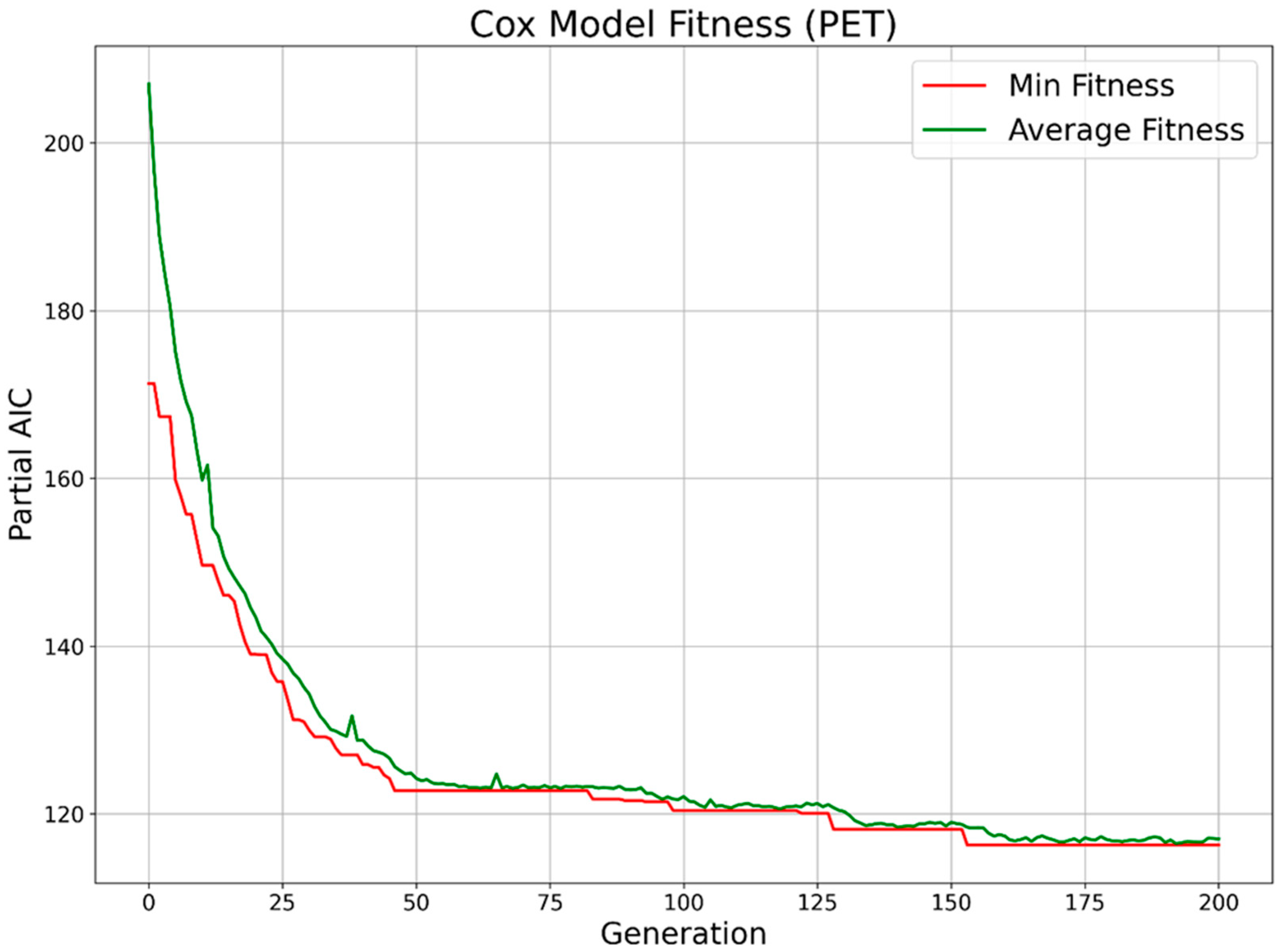
2.4. Feature Selection
2.5. Survival Prediction
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [PubMed]
- Kan, Y.; Dong, D.; Zhang, Y.; Jiang, W.; Zhao, N.; Han, L.; Fang, M.; Zang, Y.; Hu, C.; Tian, J.; et al. Radiomic signature as a predictive factor for lymph node metastasis in early-stage cervical cancer. J. Magn. Reson. Imaging 2018, 49, 304–310. [Google Scholar] [CrossRef]
- Mu, W.; Chen, Z.; Liang, Y.; Shen, W.; Yang, F.; Dai, R.; Wu, N.; Tian, J. Staging of cervical cancer based on tumor heterogeneity characterized by texture features on18F-FDG PET images. Phys. Med. Biol. 2015, 60, 5123–5139. [Google Scholar] [CrossRef]
- Lucia, F.; Visvikis, D.; Desseroit, M.-C.; Miranda, O.; Malhaire, J.-P.; Robin, P.; Pradier, O.; Hatt, M.; Schick, U. Prediction of outcome using pretreatment 18F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur. J. Pediatr. 2017, 45, 768. [Google Scholar] [CrossRef]
- Tsujikawa, T.; Rahman, T.; Yamamoto, M.; Yamada, S.; Tsuyoshi, H.; Kiyono, Y.; Kimura, H.; Yoshida, Y.; Okazawa, H. 18F-FDG PET radiomics approaches: Comparing and clustering features in cervical cancer. Ann. Nucl. Med. 2017, 31, 678–685. [Google Scholar] [CrossRef]
- Shen, W.-C.; Chen, S.-W.; Liang, J.-A.; Hsieh, T.-C.; Yen, K.-Y.; Kao, C.-H. 18Fluorodeoxyglucose Positron Emission Tomography for the Textural Features of Cervical Cancer Associated with Lymph Node Metastasis and Histological Type. Eur. J. Nucl. Med. Mol. Imaging 2017, 144, 646–1731. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Meta-Analysis Group, Medical Research Council Clinical Trials Unit. Reducing Uncertainties about the Effects of Chemoradiotherapy for Cervical Cancer: A Systematic Review and Meta-Analysis of Individual Patient Data From 18 Randomized Trials. J. Clin. Oncol. 2008, 26, 5802–5812. [Google Scholar] [CrossRef]
- Onal, C.; Reyhan, M.; Guler, O.C.; Yapar, A.F. Treatment outcomes of patients with cervical cancer with complete metabolic responses after definitive chemoradiotherapy. Eur. J. Pediatr. 2014, 41, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, P.; Sahdev, A. The role of18F-FDG PET CT in common gynaecological malignancies. Br. J. Radiol. 2017, 90, 20170283. [Google Scholar] [CrossRef] [PubMed]
- The Royal College of Radiologists; Royal College of Physicians and Surgeons of Glasgow; Royal College of Physicians of London; Royal College of Physicians of Edinburgh; British Nuclear Medicine Society; Administration of Radioactive Substances Advisory Committee. Evidence-based indications for the use of PET-CT in the United Kingdom 2016. Clin. Radiol. 2016, 71, e171–e188. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Zhu, H.; Xie, C.; Jin, X. Radiomics in cervical cancer: Current applications and future potential. Crit. Rev. Oncol./Hematol. 2020, 152, 102985. [Google Scholar] [CrossRef]
- Cox, D.R. Regression Models and Life-Tables. J. R. Stat. Soc. Ser. B Stat. Methodol. 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy; Jolesz, F.A., Ed.; Springer: New York, NY, USA, 2014; pp. 277–289. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Whitley, D. A genetic algorithm tutorial. Stat. Comput. 1994, 4, 65–85. [Google Scholar] [CrossRef]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle. In Selected Papers of Hirotugu Akaike; Parzen, E., Tanabe, K., Kitagawa, G., Eds.; Springer: New York, NY, USA, 1998; pp. 199–213. [Google Scholar] [CrossRef]
- Harrell, F.E.; Califf, R.M.; Pryor, D.B.; Lee, K.L.; Rosati, R.A. Evaluating the Yield of Medical Tests. JAMA 1982, 247, 2543. [Google Scholar] [CrossRef]
- Hatt, M.; Majdoub, M.; Vallières, M.; Tixier, F.; Le Rest, C.C.; Groheux, D.; Hindié, E.; Martineau, A.; Pradier, O.; Hustinx, R.; et al. 18F-FDG PET Uptake Characterization Through Texture Analysis: Investigating the Complementary Nature of Heterogeneity and Functional Tumor Volume in a Multi–Cancer Site Patient Cohort. J. Nucl. Med. 2014, 56, 38–44. [Google Scholar] [CrossRef] [PubMed]
| Selection Method (CT Features) | Cross-Validated CI | Number of Features |
|---|---|---|
| Genetic Algorithm | 0.798 ± 0.105 | 9 |
| LASSO Regression | 0.696 ± 0.140 | 8 |
| Baseline | 0.435 ± 0.148 | 105 |
| Selection Method (PET Features) | Cross-Validated CI | Number of Features |
|---|---|---|
| Genetic Algorithm | 0.801 ± 0.127 | 9 |
| LASSO Regression | 0.700 ± 0.138 | 5 |
| Baseline | 0.509 ± 0.198 | 105 |
| Feature (PET) | Coef. | Std. Err. | Coef. Lower 95% | Coef. Upper 95% | p-Value |
|---|---|---|---|---|---|
| Max2DDiamSlice | 0.884 | 0.621 | −0.333 | 2.101 | 0.155 |
| SurfaceVolumeRatio | 1.270 | 0.480 | 0.329 | 2.212 | 0.008 |
| Max2DDiamRow | 1.052 | 0.598 | −0.119 | 2.223 | 0.078 |
| Correlation | −2.408 | 1.238 | −4.835 | 0.019 | 0.052 |
| Energy | 1.046 | 0.474 | 0.118 | 1.974 | 0.027 |
| Maximum | 1.560 | 0.861 | −0.128 | 3.248 | 0.070 |
| Minimum | 1.815 | 0.832 | 0.185 | 3.445 | 0.029 |
| 10Percentile | −3.731 | 1.360 | −6.397 | −1.065 | 0.006 |
| ZoneEntropy | −3.393 | 1.812 | −6.945 | 0.158 | 0.061 |
| Feature (CT) | Coef. | Std. Err. | Coef. lower 95% | Coef. Upper 95% | p-Value |
|---|---|---|---|---|---|
| SmallDepHighGLEmph | 2.579 | 0.781 | 1.048 | 4.109 | <0.005 |
| LargeDepLowGLEmph | −1.069 | 0.546 | −2.139 | 0.001 | 0.050 |
| JointAverage | −4.234 | 1.240 | −6.665 | −1.803 | <0.005 |
| DifferenceEntropy | −1.124 | 0.527 | −2.157 | −0.091 | 0.033 |
| Imc2 | −1.258 | 0.911 | −3.044 | 0.529 | 0.168 |
| Imc1 | −2.679 | 1.339 | −5.304 | −0.054 | 0.045 |
| Skewness | −1.458 | 0.603 | −2.640 | −0.276 | 0.016 |
| Energy | 1.054 | 0.295 | 0.476 | 1.632 | <0.005 |
| Contrast | −2.093 | 1.122 | −4.292 | 0.106 | 0.062 |
| Feature | Coef. | Std. Err. | Coef. Lower 95% | Coef. Upper 95% | p-Value |
|---|---|---|---|---|---|
| SmallDepHighGLEmph (CT) | 1.425 | 0.563 | 0.32 | 2.529 | 0.011 |
| JointAverage (CT) | −2.226 | 1.078 | −4.338 | −0.113 | 0.039 |
| Imc2 (CT) | −2.545 | 1.073 | −4.648 | −0.442 | 0.018 |
| Imc1 (CT) | −4.401 | 1.389 | −7.123 | −1.678 | <0.005 |
| Skewness (CT) | −1.114 | 0.754 | −2.592 | 0.365 | 0.140 |
| Energy (CT) | 0.676 | 0.339 | 0.011 | 1.341 | 0.046 |
| Contrast (CT) | −4.655 | 1.329 | −7.260 | −2.049 | <0.005 |
| Max2DDiamSlice (PET) | 1.650 | 0.680 | 0.317 | 2.983 | 0.015 |
| SurfaceVolumeRatio (PET) | 2.126 | 0.679 | 0.796 | 3.456 | <0.005 |
| Maximum (PET) | 1.336 | 0.699 | −0.034 | 2.705 | 0.056 |
| 10Percentile (PET) | −1.350 | 0.705 | −2.732 | 0.031 | 0.055 |
| Model (without Recurrence) | No. Concordant Pairs | No. Discordant Pairs | Concordance Index |
|---|---|---|---|
| Cox (CT) | 41 | 17 | 0.707 |
| Cox (PET) | 17 | 41 | 0.293 |
| Cox (PET + CT) | 32 | 26 | 0.552 |
| RSF (CT) | 16 | 42 | 0.276 |
| RSF (PET) | 30 | 28 | 0.517 |
| RSF (PET + CT) | 30 | 28 | 0.517 |
| Model (with Recurrence) | No. Concordant Pairs | No. Discordant Pairs | Concordance Index |
|---|---|---|---|
| Cox (CT) | 45 | 13 | 0.776 |
| Cox (PET) | 36 | 22 | 0.621 |
| Cox (PET + CT) | 45 | 13 | 0.776 |
| RSF (CT) | 26 | 32 | 0.448 |
| RSF (PET) | 30 | 28 | 0.517 |
| RSF (PET + CT) | 30 | 28 | 0.517 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carlini, G.; Curti, N.; Strolin, S.; Giampieri, E.; Sala, C.; Dall’Olio, D.; Merlotti, A.; Fanti, S.; Remondini, D.; Nanni, C.; et al. Prediction of Overall Survival in Cervical Cancer Patients Using PET/CT Radiomic Features. Appl. Sci. 2022, 12, 5946. https://doi.org/10.3390/app12125946
Carlini G, Curti N, Strolin S, Giampieri E, Sala C, Dall’Olio D, Merlotti A, Fanti S, Remondini D, Nanni C, et al. Prediction of Overall Survival in Cervical Cancer Patients Using PET/CT Radiomic Features. Applied Sciences. 2022; 12(12):5946. https://doi.org/10.3390/app12125946
Chicago/Turabian StyleCarlini, Gianluca, Nico Curti, Silvia Strolin, Enrico Giampieri, Claudia Sala, Daniele Dall’Olio, Alessandra Merlotti, Stefano Fanti, Daniel Remondini, Cristina Nanni, and et al. 2022. "Prediction of Overall Survival in Cervical Cancer Patients Using PET/CT Radiomic Features" Applied Sciences 12, no. 12: 5946. https://doi.org/10.3390/app12125946
APA StyleCarlini, G., Curti, N., Strolin, S., Giampieri, E., Sala, C., Dall’Olio, D., Merlotti, A., Fanti, S., Remondini, D., Nanni, C., Strigari, L., & Castellani, G. (2022). Prediction of Overall Survival in Cervical Cancer Patients Using PET/CT Radiomic Features. Applied Sciences, 12(12), 5946. https://doi.org/10.3390/app12125946










