In this section, in the following, after a description of the features of the original dataset and the details of the obtained annotated corpus, the performances of the DNN trained using the self-made corpus will be discussed, measured in terms of precision, recall and F1-score and considering also the contribution of the closed-domain WE models.
4.1. B-NER Annotated Corpora
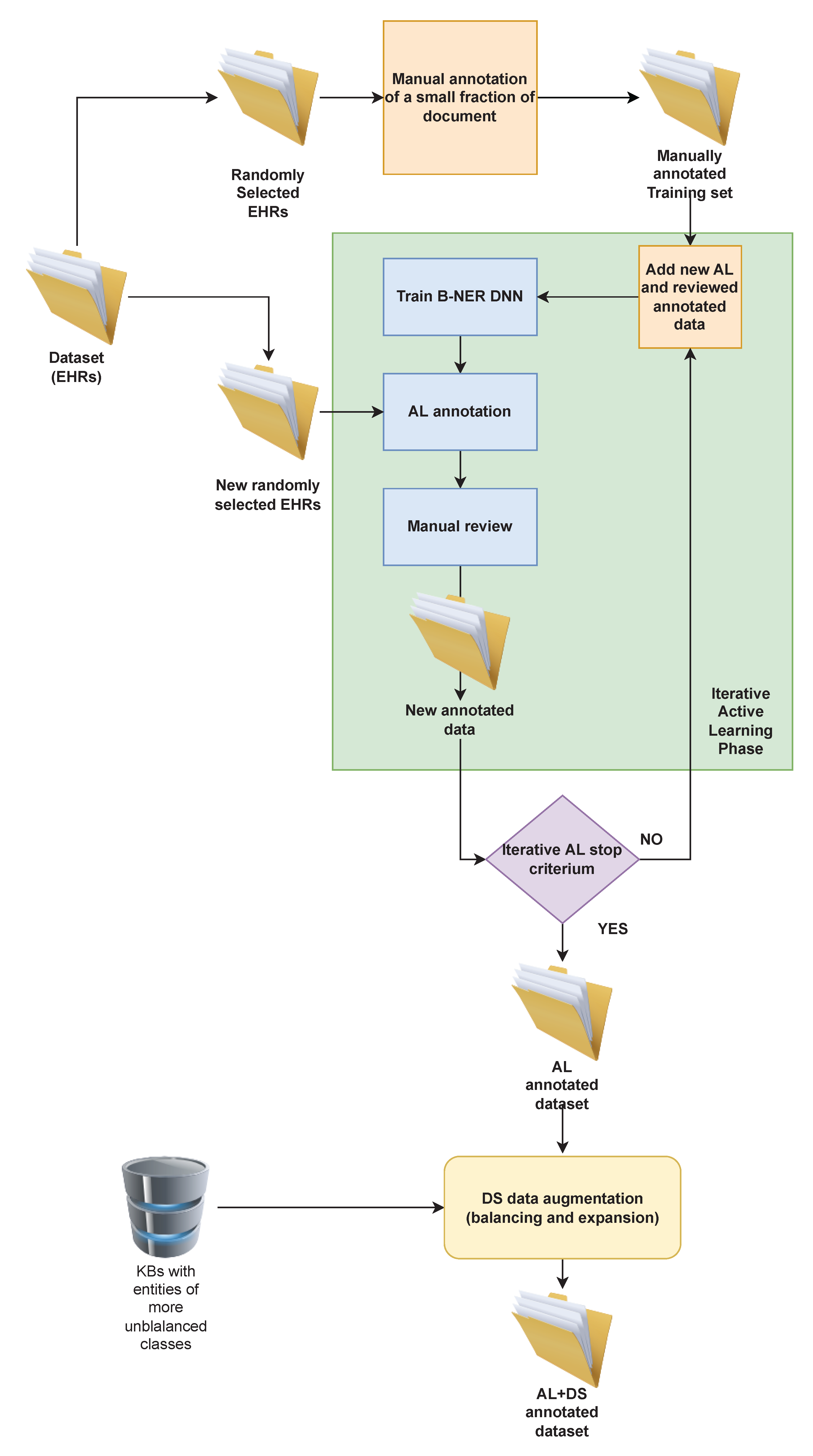
The original unannotated dataset is formed by the narrative parts of NL text extracted from a set of 1011 anonymized EHRs in the Italian language, which has a total word count of 1,657,970. In detail, the dataset contains EHRs acquired from the eHealth systems of some different hospitals in Italy. As mentioned above, the EHRs had been previously anonymized and they are related to patients admitted to different departments of the hospitals. The content of these documents is relatively homogeneous, containing the clinical diary of the patients, where the causes of the admission to the hospitals, the diseases, the prognosis, the follow-ups, the exams, the procedures and the prescriptions are described. Some sample sentences extracted from two different EHRs (translated into English) are reported below.
Found sub-capital fracture and dislocation of left shoulder and contusion of right hip caused by accidental fall at home.
Tomorrow follow-up exams.
Patient admitted to cardiology from 9 February to 19 February due to episodes of arrhythmia, likely secondary to chronic renal failure.
Eight different named-entity classes are identified, as shown in
Table 1, following UMLS semantic types [
52] and considering at the same time possible real-world applications of the trained ML models [
2].
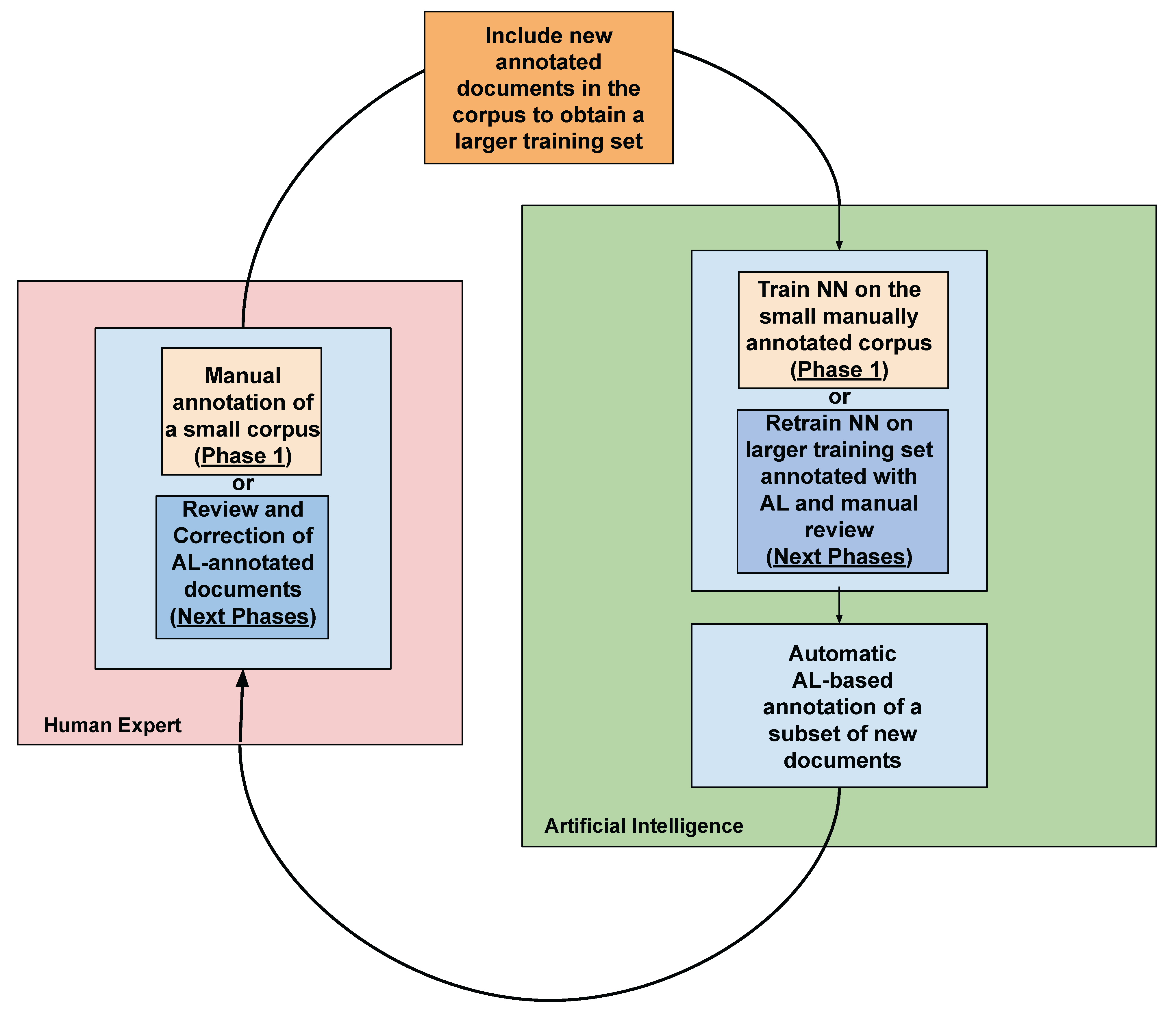
As explained in
Section 3.1, in the preliminary step of the annotation process a small set of documents formed by the text extracted from 25 randomly selected EHRs was manually annotated by two domain experts. The annotation procedure was conducted according to predefined guidelines, which describe general and specific annotation rules. The labelling process followed the
IOB notation [
53], i.e., each token belonging to an entity is labelled with the corresponding class adding the prefix
B (Begin) if it is the first token of the entity, the prefix
I for all subsequent tokens of the same multi-word entity and the tag
O (Outside) if the token does not belong to an entity. The result of the manual annotation is a small dataset, which counts 7421 tokens and 1963 named entities, as shown in the first row of
Table 2. The experts worked for approximately eight hours to produce this dataset, including the discussion about conflicts and disambiguation of the conflicting annotations. To the end of providing a stop criterion for the iterative AL phase (see
Section 3.1), a further test set, which counts 21,133 tokens, was also manually annotated.
This small dataset is used to train the DNN Bi-LSTM-CRF [
43] architecture. This DL model has been used to automatically annotate new documents randomly extracted from the whole dataset, starting the iterative AL phase. In each iteration, the human experts had to review the correctness of the annotations produced by the DL model, eventually correcting the wrong or the missing ones. They worked each step for approximately eight hours, but, in this case, they were able to annotate wider datasets, thanks to the reduced effort provided by the partial annotation of the data, as shown in
Table 2. The new data obtained in each iteration were added to the training set, producing a larger dataset, which was used to retrain the DL model. The same process was iterated and at each step the experts were able to speed up the annotation process, producing at the same time an increasing number of annotations thanks to the higher precision of the DL model trained on a larger and more complete dataset (see
Table 2 for the details). The iterative AL process was stopped after seven iterations (see
Section 4.3) when no more notable performance improvements of the ML model were observed. At the end of the AL phase, a corpus counting 304,798 words and 60,669 entities was annotated.
The results shown in
Table 2 demonstrate that the proposed approach allows one to obtain a sensitive improvement of the time required for the annotation, with respect to a fully manual process. In the first preliminary step, a human expert was able to annotate a document collection formed by almost 8000 words in about 8 h, with a rate of 1000 words per hour. The dataset obtained through the AL phase counts 304,797 words: considering the same annotation rate of the preliminary step, the fully manual annotation of this dataset would have required about 300 h. The proposed AL process required seven steps where the experts reviewed and corrected the annotations of the new data obtained from the DNN for about 8 h for each step, with a total manual effort of 56 h. Moreover, the process required an average training time of the DNN equal to 1.5 h for each iteration (the training time increases with larger training sets) on the hardware used for the experimental assessment (see
Section 4.2). In summary, the proposed iterative AL phase required in total about 66 h, allowing one to obtain an annotated dataset in almost
of the time required by a fully manual annotation.
Table 3 shows the distribution of the classes in the dataset obtained at the end of the AL phase. We note that there are very few examples of DEP (Departments) and DRU (Drugs) classes. This skewed class distribution can limit the performances of the ML systems, in particular for these two specific classes (see next Table 7). Then, in order to mitigate the skewed class distribution, the annotated corpus was automatically augmented using DS with our proposed approach, exploiting knowledge sources related to the more imbalanced classes, such as a complete list of drugs and pharmaceutical substances extracted from the Pharmaceutical Reference Book officially maintained by the Agenzia Italiana del Farmaco (
https://farmaci.agenziafarmaco.gov.it/bancadatifarmaci/cerca-farmaco, accessed on 6 June 2022), the Italian government agency in charge for drug administration, and a list of medical departments was obtained from the main Italian medical centre (hospitals, clinical facilities, etc.) websites. These two KBs were used to expand the corpus, applying the data augmentation/oversampling, as described in
Section 3.2. The final resulting annotated corpus has a total word count equal to 1,699,028 and a total entity count equal to 424,776. In
Table 3, it is shown that the distribution of the samples after the DS augmentation clearly reduces the original skewness.
The final corpus was split into a training set and a test set, randomly selecting about 15% of the data for the test and the remaining data for the training. In this way, the entity classes, respectively, in the training set and the test set are distributed as shown in
Table 4. The test set was used to assess the performance of the DNN with the annotated corpus.
Finally, a further test set was also manually annotated by the domain experts, extracting documents from a different medical domain document collection, with the purpose of assessing the quality of the corpus obtained with the proposed methodology. The aforementioned document collection, named hereinafter
out-of-corpus, is formed by short medical notes and diagnoses from various medical departments and counts 15,728 words and 3816 entities. A common problem of the Bi-LSTM CRF B-NER architecture is that it often fails to generalise to out of vocabulary words, namely words that do not appear in the training set [
54]. Thus, we tested the DL model also on the out-of-corpus test set, which contains many named entities not present in the original dataset.
4.3. Performances
To verify the effectiveness of the annotated corpus, we evaluated the performance of the same DNN used in the AL phase, trained on the obtained corpus. As explained above, we also tested different WE models to represent the input of the DNN, whose details are reported below.
Firstly, we considered a word2vec model [
26] trained on a general domain Italian language corpus, hereinafter called
W2V ISTI, formed by a Wikipedia dump and a collection of 31,432 novels [
48]. This document collection is very large (242,261,172 sentences and 2,534,600,769 words), and its content is related to many knowledge fields. The training parameters used for this model are: skip-gram algorithm, vector size 300, window size 10 and negative samples 10.
Then, a more specific biomedical closed domain text corpus, hereinafter
BIO-Corpus, was used to train the embedding models. This corpus was created considering different biomedical sources, in detail: (i) a dump of a selection of Italian Wikipedia pages related to medicine, biology, healthcare and other similar domains, following the procedure and the tools described in [
45]; (ii) the text extracted from the package leaflets of all drugs available in Italy, downloading all pdf files from Agenzia Italiana del Farmaco (AIFA) and extracting the corresponding text exploiting Apache Tika (
https://tika.apache.org/, accessed on 6 June 2022) and some specific Python scripts; (iii) the text extracted from the Italian Medical Dictionary of the
Corriere della Sera (
https://www.corriere.it/salute/dizionario/, accessed on 6 June 2022) through a set of custom web scraping Python scripts; and (iv) the text extracted from other Italian biomedical documents freely available online, such as scientific papers, presentations, technical reports and other things, exploiting also in this case Tika pipelines and Python scripts. The BIO-corpus is made up of 2,160,704 sentences and 511,649,310 words and it was used as a training set for five different WE models: two word2vec (W2V) [
26] models and two FastText (FT) models [
27], considering in both cases skip-gram and cbow algorithms and setting the vector size equal to 300, the window size equals to 10, the negative samples equals to 10 and, in the case of FastText embeddings, the char n-gram size varying from 3 to 6, as well as one contextual embedding model based on ELMo [
28]. These latter models trained on the BIO-Corpus were called, respectively,
W2V cbow,
W2V skip,
FT cbow,
FT skip and
ELMo.
Finally, we also tested the obtained annotated corpus by fine-tuning the BERT model pretrained on a very large general domain corpus, previously described in
Section 3.1.2.
Table 5 shows the results obtained on the manually annotated test set (see
Section 3.1) in the preliminary step of the AL phase when the DNN has been trained with few manually annotated data. The results are in terms of F1-Score, precision and recall averaged over all classes. It is possible to observe that
ELMo embeddings trained on the BIO-Corpus and used to represent the input to the DNN obtain performances sensibly higher than the other cases, despite a training set with few samples. This can provide substantial help to the experts during the next steps of the AL phase, further reducing the effort in the correction of wrong predictions.
Thus, this model was selected for further steps of the AL phase for the annotation of the B-NER corpus, as well as the input layer of the DNN used to test the effectiveness of the annotated corpus. Moreover, the experiments also considered the fine tuning of the BERT model pretrained on a general domain document collection, that being the current reference model for NER tasks in the literature and because it obtained performances comparable to the ELMo case in the preliminary step of the proposed approach.
The AL-based iterative annotation stopped when no further improvements to the results were obtained. Seven iterations are considered empirically sufficient to produce in the AL phase an annotated corpus with 304,977 words and 60,669 entities.
Table 6 shows the performance improvements obtained in the test set at each step of the iterative AL procedure, using the ELMo model with BiLSTM CRF and the BERT model. As shown in
Table 6, increasing the size of the annotated corpus during the steps of the iterative AL phase improved the performances of both the ELMo and the BERT experiments. We also note that the ELMo model pretrained on the biomedical domain corpus performs slightly better when fewer data in the training set are available during the first iterations of the procedure, while, when larger training sets are obtained during the AL phases, the BERT model pretrained on a general domain corpus obtains slightly better results. In any case, both models obtain comparable performances, demonstrating that a simpler neural language model, such as ELMo, pretrained on the biomedical domain corpus obtains performances comparable with the ones produced by a more complex DNN, such as BERT, pretrained on a general domain corpus. Then, we focused the next phase of the experimental assessment only on the ELMo model, investigating the contribution of the DS data augmentation phase.
In
Table 7, the results of the ELMo experiment obtained in the last step of the AL phase are highlighted, showing the precision, recall and F1-Score obtained for each class of the dataset. Observing at the same time the left column of
Table 3, where the number of entities of each class are shown, and
Table 7, with the results obtained by the DNNs trained on the corpus obtained at the end of the AL phase, it is possible to note that the worst performances were obtained in the cases of the entities belonging to the more imbalanced classes, namely DRU (Drugs) and DEP (Departments), also limiting the average results.
We introduced the DS data augmentation phase in order to limit this issue. After the expansion and the balancing of the training set using the second part of the proposed approach, where new sentences are obtained leveraging DS with domain KBs containing lists of entities of two more imbalanced classes, the performance of the ELMo DNN trained on the training set obtained with both the AL and DS phases are sensibly improved, as shown in
Table 8. In this case, we reported only the results obtained by the best performing model, which was the Bi-LSTM CRF architecture with the ELMo embeddings. This behaviour is expected, due to overfitting issues of the BERT model trained on very large datasets [
55].
In particular, comparing the obtained results for DRU and DEP classes in
Table 8, where the DS augmentation for balancing and expansion were applied for the annotation of the training set after the AL, with the results achieved in the same class types shown in
Table 7, where only the AL is performed, it is possible to observe that the DS augmentation applied to the most unbalanced classes DEP and DRU provided a sensible performance boost. Moreover, we can also note an improvement in all the other classes thanks to the oversampling performed during the DS data augmentation.
To the end of further verifying the effectiveness of the final obtained annotated corpus, we also tested the DNN models on the out-of-corpus test set, previously described in
Section 4.1. This additional manually annotated test set was extracted from a different document collection, which contains many entities not present in the dataset used to build and annotate the training set.
Table 9 shows the results obtained by the ELMo BiLSTM CRF architecture trained on the final annotated corpus and tested on the out-of-corpus test set. It is worth noting that, despite a slight performance drop, the DNN model still performs at a good level, assessing the effectiveness of the obtained annotated training corpus.
In summary, these results demonstrate that the DS data augmentation phase is capable of further improving the quality of the dataset obtained from the previous iterative AL phase, mitigating the issues of the AL related to unbalanced classes and out-of-corpus named entities.
Finally, the next
Table 10 reports the metrics averaged on all classes obtained by each considered DNN model, namely the Bi-LSTM CRF with the various considered WE models as input layer and the fine-tuned BERT model, trained on the final annotated dataset (AL and DS) and tested on the out-of-corpus test set. The purpose of this last experiment is to evaluate the contribution of different neural language models on a corpus containing many named entities not present in the training set. The results in
Table 10 show that the WE model trained on a biomedical closed-domain document collection (W2V cbow, W2V skip, FT cbow and FT skip) provides sensible improvements with respect to the W2V ISTI model, trained on a general domain corpus. We also note that the WEs trained using the skipgram algorithm provide improved performance with respect to the cbow algorithm. The ELMo model produces the best performance, but the simpler W2V skip model also obtains good results, although it does not reach the performance obtained by more complex ELMo and BERT architectures. As in the previous case, the performances of the BERT model are limited by the overfitting issues, although we adopted a drop-out rate equal to
to limit them, following the literature [
55].