Abstract
Climate change has caused breeders to focus on varieties that are able to grow under unfavorable conditions, such as drought, high and low temperatures, salinity, and other stressors. In recent decades, progress in biotechnology and its related tools has provided opportunities to dissect and decipher the genetic basis of tolerance to various stress conditions. One such approach is the identification of genomic regions that are linked with specific or multiple characteristics. Cereal crops have a key role in supplying the energy required for human and animal populations. However, crop products are dramatically affected by various environmental stresses. Barley (Hordeum vulgare L.) is one of the oldest domesticated crops that is cultivated globally. Research has shown that, compared with other cereals, barley is well adapted to various harsh environmental conditions. There is ample literature regarding these responses to abiotic stressors, as well as the genomic regions associated with the various morpho-physiological and biochemical traits of stress tolerance. This review focuses on (i) identifying the tolerance mechanisms that are important for stable growth and development, and (ii) the applicability of QTL mapping and association analysis in identifying genomic regions linked with stress-tolerance traits, in order to help breeders in marker-assisted selection (MAS) to quickly screen tolerant germplasms in their breeding cycles. Overall, the information presented here will inform and assist future barley breeding programs.
1. Introduction
Barley (Hordeum vulgare L.) is one of the first domesticated crops in Old World agriculture [1]. This cereal was domesticated from its wild progenitor—H. spontaneum C. Koch––as a result of numerous selection processes by farmers under different environments in the Fertile Crescent region [2]. Both taxa are diploid (2n = 2x = 14) and mostly self-pollinated, without any impediments to their sustainable production [3]. Based on numerous reports and maps [4,5,6], the habitat of the wild progenitor H. spontaneum was distributed in a wide range of geographical areas from Israel and Jordan to South Turkey, Iraq, and Southwestern Iran. The natural habitats of this species have also been noted in Southwestern Asia, Egypt, Greece, Southern Tajikistan, and the Himalayas.
It has been reported that barley is the fourth largest global grain crop, after wheat, rice, and corn. Barley seeds have moderate amounts of protein, phosphorus, and calcium, as well as small amounts of B vitamins [7]. Furthermore, since the carbohydrate content in barley seeds is high, they are commonly used in breads, soups, stews, and other health products [8]. Barley also plays a key role as animal fodder, as barley silage is an important feed for animals, especially ruminants. Indeed, the high fiber and low protein content in silage makes it suitable for extensive cattle production [9]. Barley is categorized into two classes based on the number of rows of flowers in the spike: two-row barley, which has central florets that produce kernels and sterile lateral florets; and six-row barley, which has a spike with three spikelets notched on each side, each notch containing a small individual flower that develops a kernel [8]. Due to its higher sugar content, two-row barley is more commonly used for malt production, while the higher protein content of six-row barley makes it better suited for animal feed. However, Heuze et al. [10] reported that two-row and awnless varieties have higher nutritive quality as forage. The global average of seed production for this cereal was estimated to be ~3.50 ton h−1 in 2019 [11], and the average harvested area is an estimated 504,000 hectares worldwide. According to the Food and Agriculture Organization (FAO), countries such as Kuwait, Belgium, Ireland, Chile, The Netherland, Switzerland, France, the United Kingdom (UK), New Zealand, Germany, and Denmark are ranked as the top 10 countries in terms of barley seed production. Additionally, in 2020, Russia, Australia, Kazakhstan, Turkey, Canada, Spain, Ukraine, Iran, France, and Germany had extensive harvested areas compared with other countries.
2. Quantitative Trait Loci (QTL) and Its Role in Breeding Programs
Quantitative trait loci (QTL) mapping was initially proposed by Sax [12], who found that bean seed size (characteristic) was linked to seed coat color (a simple, monogenic trait). Thoday [13] extended this idea by highlighting that, if segregation of simple inherited monogenes could be used to identify connected QTLs, it should be possible to map and describe all QTLs involved in complicated characteristics. Traits exhibiting quantitative variation were previously explored using statistical analysis of suitable experimental populations based on means, variances, and covariances of relatives, without knowing the number and location of the underlying genes [14]. Such studies examined the distribution of phenotypes in populations, and associations between related individuals or lines. Investigations of morphological and agronomic aspects of maize [15] indicated that certain molecular markers explained a considerable part of the phenotypic variation of quantitative variables.
Plant breeding involves creating or assembling populations or germplasm collections with high genetic diversity, in order to identify individuals with robust or beneficial traits and use those individuals to develop improved cultivars [16]. The majority of important plant breeding qualities (e.g., height, yield, drought tolerance, etc.) are quantitative, also known as continuous, multifactorial, or complex traits. Quantitative traits are based on the combined activity of many genes and their interactions with the environment, and may vary across individuals over a given range to form a continuous distribution of phenotypes [17]. Since Nillson-Ehle [18] and East [19] proposed the multiple-factor theory, it has been considered that the genetic variation of a quantitative trait is controlled by the collective effects of various genes—QTLs [20,21]. As a result, many QTLs control the expression of a single phenotypic feature. Quantitative features increase difficultly for breeders, since performance only represents a portion of an individual’s hereditary values. Hence, since multiple genes or QTLs govern quantitative characteristics, plants with the same phenotype might have distinct alleles at each of these genes or QTLs. Conversely, plants with identical QTL genotypes may have distinct phenotypes when grown in different settings, and the impact of one QTL can be influenced by the plant’s allelic composition at other QTLs. As a result, the genotype cannot be inferred from the phenotype, necessitating the creation of specialized genetic stocks and their cultivation in carefully regulated conditions.
The two main aims of QTL mapping in plants are: to (i) improve our understanding of quantitative trait inheritance and genetic architecture within and between species, and (ii) discover markers that may be utilized as indirect selection tools in breeding [22]. Extensive QTL mapping studies in most commercially significant crops were conducted to identify molecular markers for marker-aided selection [21,23,24]. It is known that a phenotypic trait can be controlled by many independent loci of little effect, or a few genes of large effect. Moreover, beneficial QTL alleles are known to be inherited from parents who have beneficial QTL alleles [25]. Figure 1 shows the general flow of information from collecting phenotypic and genotypic data to QTL and association analyses.
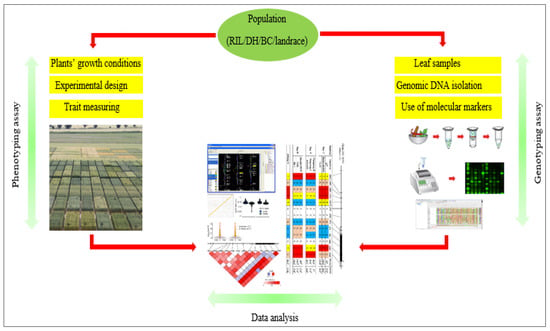
Figure 1.
The flow of information from different steps for QTL or association analysis from collecting data to statistical analysis.
3. Aims of the Review
In recent years, climate change has increased the intensity of abiotic stressors. Consequently, the effects of environmental stresses on cereal production have become increasingly important [26]. Among cereals, barley is adaptable to a wide range of climates. It has been shown that this crop can grow and mature in a shorter time compared with other small cereals. Hence, it has a greater tolerance to various abiotic stresses, especially heat and drought. In this review, we highlight QTL and association analyses related to abiotic stresses in barley, such as drought, salinity, temperature, water logging, and toxicity. This summary will help breeders to identify the important genomic regions linked to tolerance of these stresses in future research and breeding programs.
4. Response of Barley to Abiotic Stresses
Many crops’ yields can be adversely affected by abiotic stressors, particularly in species with limited resilience to unfavorable circumstances such as high and low temperatures, hydric limitation, floods, toxicity from mineral elements, excess salts, and nutritional deficiencies [27]. Generally, stress is a considerable deviation from the ideal growing conditions, causing alterations in all functioning levels that are initially reversible but later become permanent [28]. In order to prevent losses caused by environmental stressors, it is necessary to understand how plants respond to them, and to find a common mechanism of tolerance for a wide range of stress situations [29].
Plant breeders require access to novel genetic variants to develop varieties that can adapt to changing biotic and abiotic stressors, which then leads to new foods that provide nutritional and health advantages that customers desire. As global food consumption continues to grow, compounded by the consequences of climate change, the demand for successful crop breeding has increased. The few genes available to the top crop species has required the use of closely related families of domesticated plant species as a gene supply, with significant improvements in farm output [30]. Barley has been naturally hybridised long back from the wild parent H. spontaneum, and evolved from there. Wild relatives of domesticated barley have an excellent genetic resource for improving tolerance to abiotic stress conditions, such as heating, salt, and drought [31]. These genetic resources are easily crossed with domesticated crops and are widely available for breeding [32,33]. Thus, it fulfills the need for the supply of genes that improve abiotic and biotic stress tolerance in domestically managed barley [34]. With the recent discovery that current cultivars retain only 15–40% of alleles in the gene pools of this crop, the emphasis on using barley genetic resources to enhance tolerance has reemerged [35]. In the following sections, we briefly discuss each abiotic stress, and provide an overview of some relevant genomic regions linked with various agronomic and physiological features in natural and cultivated populations.
4.1. Salinity
An important environmental factor that affects the development of barley plants is salinity, which affects about 800 million hectares throughout the world [36]. Salinity causes significant losses in agricultural productivity, particularly in arid and semi-arid countries, where around 25% of irrigated land is salinized (FAO [37]). Several cations, such as sodium, calcium, magnesium, and anions, including chloride, sulfate, bicarbonate, and carbonate, make up the solute salts that effectively contribute to salinizing the soil [38]. It has been shown that soil salinity can reduce agricultural production by up to 50% in a wide range of economically important crops, including maize, rice, and barley, which are all susceptible to salt stress. Specifically, plants’ cellular metabolism is affected by salinity as a result of osmotic stress [39]. The high concentration of solutes in the soil decreases the hydric and osmotic potential, causing soil to retain more water [40], as well as ionic stress, which can induce ionic toxicity and nutritional imbalance from the high levels of Na+ and Cl− [39,41]. Compared with wheat, barley has a higher tolerance to salt, and a greater capacity to compartmentalize Na+ into vacuoles and thus maintain a high K+/Na+ ratio [42]. Although there are hints of more resistant cultivars, barley is nevertheless considered vulnerable to soil salinity, since it exhibits a gradual decline in growth as the concentration of salts in the environment increases [43].
The reduction in dry mass may represent the metabolic energy expenditure associated with adaptation to salinity and a reduction in carbon gain, which may be explained by the diversion of energy for the functional maintenance of tissues [44]. Since barley is the most salt-tolerant cereal [45], it can be regarded as a plant model species that is beneficial for understanding the complicated processes of salinity tolerance and resource use, which can then be applied towards increasing salinity tolerance of other cereal crops [46]. Although barley is known as a salt-tolerant crop, its growth and production are affected by ionic and osmotic potential in saline soils [47]. Moreover, it is worth noting that tolerance to salinity stress is a complex quantitative trait that is controlled by the interaction between physiological mechanisms and pathways of many genes [48]. Similar to other field crops, this stress negatively affects almost all aspects, i.e., phenology, morphology, physiology, and biochemistry processes, which ultimately reduces its economic value [49,50,51,52]. Figure 2 illustrates the most important effects of this stress on various aspects of plant growth and development.
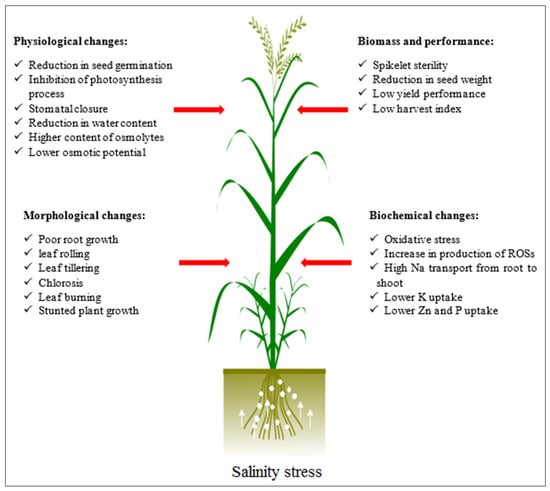
Figure 2.
Effect of high salt concentration on various aspects of plant growth and developmental stages.
As a consequence of domestication, selection, and breeding cycles [53], a wider genetic diversity (GD) is needed to reach the reproductive objectives, in order to compensate for the genetic degradation and loss of many alleles—not only in barley, but also in other crop species. The intrinsic ecological flexibility and stability of wild barley in highly varied settings may indicate that this is an excellent genetic resource for increased environmental and biotic stress tolerance in farmed barley [54]. To improve barley salinity tolerance in connection with climate change and genetic deterioration, it is essential to use GD available in the H. spontaneum wild subspecies [55]. When compared with wild emmer (T. turgidum ssp. dicoccoides), wild barley produces higher yields in salt stressed conditions, perhaps due to reduced sodium contents in the shoots [54]. For instance, Ebrahim et al. [55] investigated 47 wild barley accessions and 6 tolerant check cultivars under salinity stress conditions (300 mM NaCl). They found that salinity increased malondialdehyde (MAA) and proline, as well as Na+ and Ca+ in both the roots and the leaves. Moreover, they showed that H. spontaneum genotypes have been less affected by salt than check cultivars (H. vulgare L.). The concentration of Na+ was negatively correlated with the plant lifespan, while the leaf ratio of K+/Na+ was strongly linked with the membrane stability index. Mechanisms of ion exclusion (Na+) are probable in wild barley, resulting in homeostasis of K+/Na+, reduced root and Na+ flare-up, and ultimately a higher survival rate.
It has been shown that a greater level of salt tolerance is associated with an efficient mechanism for selective uptake of K+ over Na+ [56]. The reduction in K+ concentration occurs due to the presence of excessive Na+ in the growth medium. In other words, a high Na+ concentration is known to have an adversary effect on K+ uptake in plants [57,58]. Under salinity stress, this antagonism between the absorption of Na+ and K+ usually occurs at the root surface [59]. Salinity stress affected leaf growth of barley through changes in plasma membrane potential, osmolality, and transpiration [60]. Another consequence of high salinity on growth and development is its negative effect on stomata behavior and photosynthetic activities. Reduction in gas exchange due to photosynthetic processes could result from (i) toxic Na+ and Cl−, that reduce stomatal conductance and photosynthetic electron transport; (ii) reduced absorption and metabolism of carbon; and (iii) oxidative damage in the photosynthesis II apparatus (PSII) [61]. Moreover, salinity stress significantly increases the accumulation of reactive oxygen species (ROS) in plant cells. In turn, ROSs adversely affect chlorophyll membranes and ultimately decrease photosynthesis [62]. In a comparative study between wheat and barley, it was found that barley responded to salinity better than wheat, due to its high photosynthesis rate, chlorophyll content in their leaves, the capacity to alleviate the negative effects of ROS accumulation in cells, and other physiological mechanisms [63]. One of the main differences between barley and other crops is its germination potential and ability to establish seedling under severe salinity conditions. Understanding the genetic basis of seed germination under salt stress may improve barley plant growth, output, and even productivity. Thabet et al. [64] investigated salt stress tolerance in 121 spring barley accessions from across the globe by exposing them to 200 mM NaCl during seed germination and seedling growth. As expected, seed germination parameters and seedling-related traits were drastically lowered when salt stress was induced. Using 9K Single nucleotide polymorphisms (SNPs), a genome-wide association scan (GWAS) discovered numerous intriguing genomic areas (over 1500 potential genes), including 80 significant SNPs linked to the attributes investigated. Many genes encoding potassium channels, such as AKT2, were found on chromosome 1H. In addition, squamosa promoter-binding-like protein 6 at chromosome 5H was revealed to be associated with salinity stress tolerance during seedling developmental phase. The results showed that these genes may have a role in salt tolerance during the early phases of development, implying that early selection might increase salinity tolerance. Functional markers related to tolerance loci might aid in the development of barley cultivars that are more resistant to salt stress.
At the seedling and germination stages, genotypic diversity and separate quantitative trait loci (QTLs) were found to be linked with salt tolerance. This was an important attempt to bridge the genetic gap by focusing on early seedling survival characteristics and seedling vigor indicators in barley following germination under salt stress [65]. Identifying the genetic loci and alleles in existing germplasm is required for breeding efforts aimed at increasing salt tolerance during germination. Using 103 doubled haploid (DH) lines of the CM72/Gairdner population, Mwando et al. [65] applied QTL mapping to explore seedling vigor characteristics; 85 barley germplasms were used to validate the phenotypic response. They found that 150 mM NaCl stress considerably lowered all the observed behavioral characteristics. Several characters, such as seedling vigor index, root length, and root dry weight, showed a lesser decrease in DH populations and diversity barley germplasms compared with those that were exposed to 150 mM NaCl. A total of 6 QTLs consisting of 13 markers on chromosome 1H (3 markers), 3H (8 markers), and 4H (2 markers) were found in the DH population [65]. The regions of high potential for candidate genes were 3 QTLs on 1H and 2 QTLs on 3H with significantly associated markers (BmAC0032, bPb-9418, bPb-4741, bPb-45046, bPb-9624, bPb-3623, bPb-5666, and bPb-6383). On chromosome 3H, a QTL (flanked by markers bPb-4576 and bPb-9624) was associated with multiple salinity survival characteristics. This finding serves as the basis for future studies in selection genetic analysis [65].
Mwando et al. [66] investigated the characteristics of germinating seeds under controlled and salt stress conditions in 350 barley accessions. They found that multiple genes at distinct loci affected salt tolerance during germination and seedling development. This discovery aligns with the findings of Maddur [67] and Mano et al. [68], who also revealed that salt tolerance in barley is governed by separate genetic processes during the germination and seedling stages. Mano and Takeda [69] linked the S gene (rachilla hair length) to a genetic component determining salt tolerance during germination. Short-haired rachilla was shown to be substantially more tolerant of salt than long-haired rachilla in the Steptoe/Morex DH lines. Ullrich et al. [70] also utilized the Steptoe/Morex DH lines to locate the QTL linked with seed dormancy on the long arm of chromosome 5H. In general, detecting significant salt tolerance genes in seedlings or using markers associated with salt tolerance genes may provide accurate and consistent approaches in plant breeding.
Recently, Karahara and Horie [71] have reviewed the advances in QTLs or genetic loci (and their causal genes, if applicable) on salinity tolerance, with a specific focus on the essential functions of roots, such as ion homeostasis mediated by membrane transporters and signaling molecules under salinity stress. Our review focusses on relevant literature for developing barriers against apoplastic ion flow, such as Na+, as well as understanding the roles and components of the barrier structures under salt stress. For example, a set of 149 DH lines produced from a hybrid between Clipper (salt sensitive) and Sahara 3771 (salt resistant) barley genotypes were tested under natural saline-stress and non-stress settings; 14 characteristics were measured to identify the QTLs related with agronomic and physiological variables [72]. The genetic linkage map, which consisted of 517 molecular markers spanning a total of 1502.4 cM, was used to conduct QTL analysis using the composite interval mapping approach. Several traits, including days to heading, relative water content, chlorophyll content, plant height, spike length, days to maturity, biomass, grain yield, harvest index, grain number per spike, 1000-seed weight, Na+ and K+ concentrations, and K+/Na+ ratio were found to have 40 and 38 QTLs in normal and salinity environments, respectively. Most of the QTLs discovered were on chromosome 2H. Individual QTLs explained between 3.3 and 68.6% of the phenotypic variance. Under both environments, a significant QTL around the Vrs1 gene on chromosome 2H was found to be associated with biomass, grain number per spike, 1000-seed weight, plant height, and grain yield [72]. This QTL might be valuable in barley breeding efforts for marker-assisted selection to improve salt tolerance. In addition, QTLs for days to heading, biomass, spike length, grain number per spike, 1000-seed weight, and K+ content were discovered, all of which may be considered potential QTLs for breeding purposes. In another study, a set of 408 diversity arrays technology (DArT) markers were genotyped on 206 barley accessions from across the globe, and salt stress tolerance was assessed using the salinity tolerance score, a reliable characteristic created in prior research. A general linkage model (GLM) and a mixed linkage model (MLM) based on population structure and kinship were used to perform genome wide association analysis (GWAS) for salinity tolerance. In total, 24 significant marker–trait correlations discovered. In all techniques, a QTL on 4H with the closest marker of bPb-9668 was consistently observed [73].
Salinity stress tolerance was investigated in a DH population derived from the salinity resistant genotype CM72 and the salinity sensitive variety Gairdner, during the germination, seedling emergence, and first leaf full expansion development phases [74]. Deionized water and two NaCl treatments (150 mM and 300 mM NaCl) were used in the germination investigation. Three treatments were used to test salt tolerance in seedlings: control, and 150 mM NaCl injected during seedling emergence and the first leaf full expansion development phases. In this study, 12 QTLs on chromosomes 2H–6H were discovered, each accounting for 10–25% of the phenotypic variance. Fresh weight and dry weight per plant in salt stress during the first leaf full expansion development stage, as well as dry weight per plant in salt stress during seedling emergence, were associated with a QTL situated at 176.5 cM on chromosome 3H. On chromosome 2H, a persistent QTL for germination at both concentrations of salinity stress (150 and 300 mM NaCl) was found, although it was far away from the QTL connected to seedling stage salinity stress tolerance. Sayed et al. [75] tested an advanced backcross mapping population of barley in freshly reclaimed soil under two salinity levels of groundwater aquifers. A significant QTL linked with grain-weight-related features and the salt tolerance index (STI), which may be utilized to improve salinity tolerance, were found across the whole barley genome. Furthermore, the simple-sequence repeats (SSR) markers bPb-3739 (4H, 96.3 cM), AF043094A (5H, 156 cM), bPb-8161 (7H, 2.22 cM), and bPb-5260 (7H, 115.6 cM) were the most significant genomic areas pertaining to vernalization, dwarfing, and dehydrin genes, all of which are linked to salinity tolerance. A comparison of the Morex and Barke barley genomes revealed 436,640 InDels. Zhou et al. [76] generated 1140 InDel markers spanning the barley genome at an average genetic distance of 1 cM. A high-resolution melting (HRM) technique was used to genotype 55 InDel markers. The 1140 InDel markers were combined with 383 SSRs, 3909 gene-based SNPs, and 1544 DArT markers into a unified barley genetic map. In total, 6976 molecular markers (SSRs, DArTs, SNPs, and InDels) were combined into a single barley genetic map [76]. Quick map creation and gene fine-mapping will be enabled by HRM genotyping of InDel markers.
In a European winter barley strain, the dynamic response of seedling development to varied degrees of salt stress and PEG6000-mediated osmotic stress was examined with 4885 gene-based SNP markers. A final collection of 56 phenotypic features was subjected to genome-wide association mapping, leading to the discovery of 28 quantitative trait loci (QTL), 10 of which were associated with saline environments and 20 were involved in osmotic stress [77]. More than one growth condition revealed four loci on chromosomes 1H, 5H, and 6H. However, under salt stress, one colocalized QTL was engaged in both root and shoot development. Based on their physical proximity to the QTL peak markers, a collection of prospective candidate genes with putative pleiotropic effects on seedling development under various circumstances is offered. Mwando et al. [66] looked at seed-germination-related parameters in 350 barley accessions under control and salt stress conditions. Marker–trait associations (MTA) and the underlying candidate genes for salt tolerance during germination were discovered with the use of a genome-wide association study that included over 24,000 genetic markers. Across all chromosomes, they found 19 loci containing 52 significant salt-tolerance-associated markers, as well as four genes belonging to four families with functions underpinning the anticipated MTAs.
A population including 187 DH lines derived from a hybrid between TX9425 and Naso Nijo was tested for superoxide anion (O2−) and hydrogen peroxide (H2O2)—two primary ROS species produced during hypoxic stress—to discover the QTL for ROS tolerance in barley [78]. The results demonstrate that measuring ROS concentration after 48 h of hypoxia provides a rapid and accurate way to identify waterlogging-tolerant barley genotypes. Both superoxide (QSO.TxNn.2H) and hydrogen peroxide (QHP.TxNn.2H) were found to have the same QTL on chromosome 2H. This QTL was found in the same location as the QTLs for total waterlogging and salt tolerance described in earlier research, explaining 23% and 24% of the phenotypic variance for O2− and H2O2, respectively. Accordingly, ROS production was shown to be linked to both waterlogging and salt stress tolerance in the study. In a study conducted by Capo-chichi et al. [79], a recombinant inbred line (RIL) population including 185 individuals was subjected to QTL mapping analysis for 12 early seedling vigor (ESV)-related characteristics. A genomic map (1075.1 cM) was created using DArT markers, with an average adjacent-marker distance of 3.28 cM. In total, 46 significant QTLs for ESV-related characteristics were found. Across all chromosomes, 14 QTLs for biomass yield were discovered, 2 of which colocalized with QTLs for grain yield on 1H. The first and second leaf lengths, as well as the dry weight of the second leaf, biomass production, and grain yield, all demonstrated a high heritability (>30%). Meanwhile, grain and biomass yield showed a strong association, providing a clear picture of these qualities in the selection process. Capo-chichi et al. [79] showed that the DArT markers bPb-9280 and bPb-9108 on 1H were linked to a pleiotropic QTL related to the specific leaf area of the second leaf, biomass yield, and grain yield. These could be used to improve seed vigor through marker-assisted selection and facilitate future map-based cloning efforts.
Under control conditions, as well as exposure to 75 mM and 150 mM NaCl, 50 wild barley (Hordeum vulgare ssp. spontaneum, Hsp) introgression lines (ILs) were utilized to find QTL alleles that improved germination and seedling growth. Under salt stress, germination and seedling development features were diverse, and found to be highly heritable [80]. Furthermore, significant differences were found across treatments and for five salinity tolerance indicators. For the 10 phenotypes studied and the tolerance indices, a total of 90 and 35 significant QTL were found, respectively. Hsp introgression alleles were implicated in enhancing salt tolerance in 40 (43.9%) of the 90 QTL studied, including introgression lines S42IL-109 (2H), S42IL-116 (4H), S42IL-132 (6H), S42IL-133 (7H), S42IL-148 (6H), and S42IL-176 (6H) (5H) [75]. Seven unusual QTL alleles, including S42IL-127 (5H), 139 (7H), 125 (5H), 117 (4H), 118 (4H), 121 (4H), and 137 (4H), were effectively verified in wild barley ILs (7H).
Photosynthesis and transpiration are controlled by stomata, and these activities are crucial for plant responses to abiotic stressors such as salt. To find QTLs linked with stomatal and photosynthetic features relevant to salinity tolerance, Liu et al. [81] identified 22 significant QTLs in a population comprising 108 DH lines. These QTLs were found on all chromosomes except 5H, and account for 9.5–17.3% of the phenotypic variance. Under control conditions, QTLs for biomass, intercellular CO2 concentration, transpiration rate, and stomatal conductance colocalized. A QTL for biomass and one for transpiration rate under salinity stress were also found to be colocated. In addition, gas exchange and stomatal pore area were discovered to be linked. A QTL for salt tolerance was found among QTLs for grain yield and biomass on chromosome 3H. The potential genes for salt tolerance at this locus are suggested based on the draft barley genome. Table S1 shows additional details about the identified QTLs for salinity tolerance in barley [82,83,84,85,86,87].
4.2. Drought
The frequency and severity of hydric insufficiency are the most important variables in determining global output limits. Research has shown that this restriction is responsible for 60–70% of the variance in ultimate output. Since it is the most common source of yield losses throughout the world, developing more drought-resistant genotypes is critical for food security [88]. Indeed, drought tolerance is a major goal of breeding programs for semi-arid regions with rainfed conditions [89], as well as other areas with a history of poor water supply. To escape dryness, plants close their stomata, deepen their roots for water absorption, thicken their cell walls, and enhance the waxiness of their cuticles. These traits are genetically determined and vary between cultivars. Thus, the diversity among barley genotypes’ sensitivity to stress can lead to difference in mortality [90,91]. The productivity of barley is determined by genetic potential and environmental factors that interfere with plant function, particularly photosynthesis. The photosynthetic process is dependent on the capture of photosynthetically active radiation and its conversion to chemical energy. This process is impacted by a variety of abiotic and biotic stressors that can diminish leaf area at different phases of the crop’s growth and development [92]. For example, inadequate soil drainage conditions can affect barley yield. Plant tolerance to water scarcity can take several forms, depending on the species, length and intensity of the drought, developmental stage of the plant, organ, and most importantly, its genotype. In a study of the effect of water limitation in barley accessions, Oukarroum et al. [93] found that drought stress had a distinct influence on the cereal’s growth characteristics. Similarly, Cantero et al. [94] confirmed that the quantity of water in the plant decreased throughout the cycle under dry semi-arid environments, and that barley with genotypes for higher productive capacity retained reduced stomatal resistance and showed increased water-retaining potential. Drought stress has a substantial impact on grain production and all yield components, according to the findings of Mahmood et al. [95]. The average rate of grain production loss across years was 8.8–16.3%, while under extreme drought stress, the decrease is expected to reach 49–87% [96]. Figure 3 summarizes some important effects of drought stress on plant growth, as well as some considerable responses of plants to this stress.
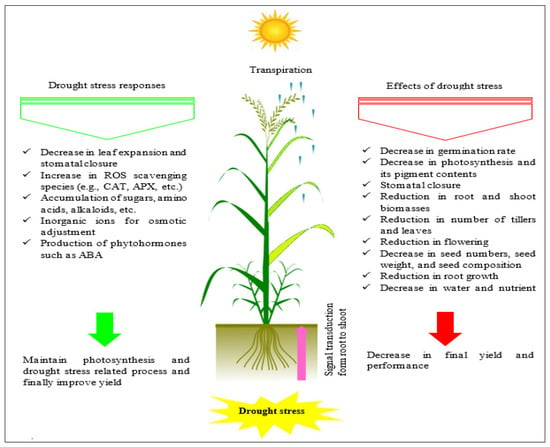
Figure 3.
Schematic of the information flow of drought stress on various mechanisms of plant growth and developmental phases, and responses of plants to this environmental stress.
Wild barley is a major source of genetic variation, and exploitation of this germplasm has been suggested as a means to increase drought tolerance. Since the root is the first organ to detect and react to water shortages [48], the capacity of plants to endure water shortages is largely determined by their extensive root systems [97]. Strong root system architecture (direction, depth, and branching) results in more contact between roots and soil, which improves water and nutrient absorption and has a direct, positive effect on yield performance [98,99]. Seminal and nodal roots emerge from the embryo and the base of the main stem, respectively, to form a fibrous root system for barley [97]. Nutrient-absorbing root hairs form on the lateral roots (secondary and tertiary roots) that arise from the main roots. The root is the most important organ for stress adaption, anchoring, and resource acquisition. Therefore, studying root system differences and selecting traits for drought tolerance could help to improve future global food security. Siddiqui et al. [100] focused on root system properties and how they interact with shoot architectural elements to cope with water shortages and preserve yield. They also used a microsynteny system to collect root-related drought-sensitive QTL/genes in cereal crops, including interspecies linkages, to simplify comparative genomic analysis. Finally, they offer a strategy for rapidly selecting root features required to generate robust crop types.
Barley has a tremendous genetic variety, owing to its extensive geographical distribution and climatic range. Only recently has this regional landrace and wild accessions diversity been studied using functional genomics [101]. Over the past two decades, many investigations identified QTLs linked with yield and its related features and physiological factors in scarcely cultivated crops under drought stress [33,102,103]. Table S2 lists some of the identified QTLs in different barley populations [33,75,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121]. Marok et al. [101] evaluated the collected data to identify potential genes involved in signaling and protective mechanisms that confer drought tolerance in barley germplasm lines. With this knowledge, wild and landrace barley populations may be improved for drought tolerance by enhancing transcription factor, heat shock or late embryogenesis protein (HSP) allelic variations, proline biosynthesis, or redox regulation [101]. With the capacity of bioinformatics tools and advances in sequencing and mass spectrometry, large-scale trials of genotypes may be planned to evaluate their physiological and biochemical responses to water deprivation while determining the genetic basis of their differences. Using such methods can help breeders find potential loci to improve drought tolerance in barley.
Individual investigations have only found QTLs for water-soluble carbohydrates (WSC) [122]. Utilizing a comparative genomic approach, many QTLs related to drought tolerance in wheat were shown to colocalize with QTLs discovered in barley under drought conditions [123]. QTLs linked with drought tolerance, such as QDT and TxFr.2H, have been identified. Fan et al. [102] discovered the TxFr.5H gene on chromosome 5H, as well as QTLs related to proline content, such as QPCD.TxFr.3H and QPC-S.TxFr.3H, and QTLs for moisture content in barley drought-tolerant plants [102], which might be used for MAS in barley drought tolerance breeding programs. Drought tolerance without restricting yield potential is a major problem, and QTLs for stress-related characteristics that coincide with QTLs for productive potential should be prioritized when developing new cultivars [124]. Approximately 50 research studies have documented the complex genetic basis of drought tolerance and related characteristics in cereal crops using QTL approaches. In total, around 800 QTLs/MTAs (MTA) have been reported in these investigations. The QTLs are equally scattered on almost all chromosomes, most of which are physiologically characterized QTLs/MTAs (about 429), with agricultural (318) and root-architecture-related features (23) for cereal cultivations. Major QTLs (with 20% phenotypic variance) and stable QTLs (detected in more than 50% of settings) were among those detected [125].
Under drought conditions, stay-green (SG) is a very important feature. Gous et al. [126] used a DH population to discover QTL related to SG expression. Their study phenotyped the spikes (S), flag leaf (FL), and first leaf beneath the flag leaf (FL-1) visually, and by using single-photon avalanche diode measurements. The plant’s green leaf area was assessed visually by comparing the S and FL green areas. Composite interval mapping identified 10 SG QTL on chromosomes 3H, 4H, 5H, 6H, and 7H, 6 of which were linked to heat stress and 4 to water stress. Although these QTL were not colocated with previously described barley stress-response QTL, some SG QTL were mapped near previously described chromosomal locations. For example, two heat-stress QTL mapped to bPb-5529 on 5H, next to QTL for root length and root-to-shoot ratio. The identification of SG QTL in barley cultivated under heat and water stress conditions allows for high-throughput screening for these characteristics. In a study of 72 DH lines and a sensitive variety, Fan et al. [102] found two QTL for drought tolerance (leaf wilting) and one for salinity tolerance (plant survival). In three independent experiments, the QTL on 2H for drought tolerance explained 42% of the phenotypic variation. The drought tolerance QTL on 5H was less affected by agronomic traits and can be used in breeding programs. The draft barley genome sequence indicated that a candidate gene for this QTL was located on 5H. To investigate the relationships between DArTseq and SNP markers, as well as key physiological parameters associated with plant responses to drought, Wojcik-Jagła et al. [104] employed 109 spring barley genotypes with high or poor drought tolerance. Drought tolerance in four-leaf seedlings was phenotyped under controlled settings (growth chambers). There were 58 novel connections discovered, including 34 new markers (16 DArTseq and 18 SNP markers). Three of the markers yielded findings that were similar to those reported in a previous conventional biparental QTL mapping analysis. The greatest number of significant MTAs were found in the areas around markers on linkage group 2H. Additionally, five markers associated with photosystem II photosynthetic activity (PSII) were discovered on chromosome 4H. The sequences next to DArT markers on linkage group 6H had the fewest number of connections. In both biparental QTL and association studies, a chromosome 3H region associated with water usage efficiency and net photosynthetic rate may be especially useful, since these metrics correlate with plants’ capacity to stay highly productive under water deficiency stress. Abed and Belzile [105] used a GWAS analysis to identify SNP–trait relationships and potentially elucidate the complex genetic architecture of seven essential spring barley attributes. They revealed significant connections, some of which were shared with the single-SNP strategy. Their findings explained higher phenotypic variation (R2), which better suited the genetic model [Bayesian information criterion (BIC)]. The cumulative R2 and BIC of the single-SNP and haplotype-based techniques were comparable to the multi-SNP approach, with the haplotype-based approach improving. Despite low overlap, each strategy revealed previously verified QTLs, indicating that each approach may identify a distinct group of QTLs. A combined single-locus and multilocus GWAS technique may increase the ability to find significant QTLs and offer a more comprehensive picture of the genetic architecture of complex characteristics in barley.
Another study used a 9 K SNP chip to genotype 183 spring barley gene bank accessions from 23 countries to discover MTAs and candidate genes influencing agronomic and seed-associated attributes during post-anthesis drought stress [106]. The results of GWAS identified 97 significant MTAs related to kernel number and kernel weight per spike. Mora et al. (2016) discovered SNP markers related to 15 complex traits in a breeding population of barley assessed under differing water availability conditions in central Chile’s Mediterranean climate. Given the significant segregation distortion of markers, a QTL mapping mixed model was utilized to account for the genetic relatedness between genotypes. In rain-fed environments, 57 QTL were found, accounting for 5–22% of the phenotypic variance. In full irrigation, 84 SNPs were related with various characteristics, explaining 5–35% of the phenotypic variance. Most QTLs were found on chromosomes 2H and 3H; 12 of the 15 characteristics assessed had environment-specific genetic regions. Most QTL–trait relationships were environment- and trait-specific, although several were significant and persistent. Full irrigation revealed a significant genomic area on chromosome 1H that accounted for between 27% (SNP2711-234) and 35% (SNP1923-265) of the behavioral variance. In both rain-fed and irrigated conditions, the locus 1923-265 was discovered for grain yield, accounting for 9% and 18%, respectively. In spring barley, the drought-adaptive QTLs were located around dehydrin genes (Dhn4, Dhn7, Dhn8, and Dhn9) and aquaporins (e.g., HvPIP1;5, HvPIP2;7, and HvTIP2;1)—genes that are implicated in dehydration tolerance [107]. The relevance of QTL hotspots for seedling growth is shown by the colocalization of well-known key genes for barley development and flowering time. The role of HvPpd-H1 in the formation of the root system was discovered using association analysis [108]. Root QTL colocalization with HERK2, HvARF04, HvEXPB1, PIN5, PIN7, PME5, and WOX5 suggest further explorations of the functions of these genes in barley. As a result, the genes HvHOX2, HsfA2b, HvHAK2, and Dhn9, which are known to be implicated in the abiotic stress response, were found within stress-specific QTL areas and will be validated in the future [108].
Pham et al. [109] evaluated a nested association mapping population called HEB-25, which consists of 1420 BC1S3 lines developed by crossing 25 different wild barley accessions to the elite barley cultivar “Barke” under both the control and the drought-stressed conditions. Best linear unbiased estimators (BLUEs) for each trait were developed and utilized in the GWAS analysis. Many of the 14 quantitative trait loci (QTL) discovered had known inflorescence and developmental genes as colocalizers. The yield of cereal crops is determined by kernel weight (KW) and kernel number per unit area. To find loci regulating KW, Zhou et al. [127] employed two barley RIL populations, including Baudin/AWCS276 (BA) and Morex/AWCS276 (MA) with a common parent. A linkage map was provided for BA. In this population, many large effect QTLs influencing KW were discovered. The genotyping by sequence (GBS) data of 201 RILs from this group was used to create a linkage map with 5273 makers forming 1454 clusters. Each cluster was represented by a single marker, and the linkage map created with these markers has a total length of 1022.4 cM, with an average distance between loci of around 0.7 cM. Three of the large-effect loci influencing KW found in the BA population (found on 2HL, 6HL, and 7HL) were also found in the MA population under different environmental conditions. Moualeu-Ngangue et al. [110] measured morphological and physiological responses to drought stress in a group of 209 spring barley genotypes, and used a GWAS study to determine QTL and allele contributions for each of the variables studied. Almost all characteristics showed substantial water deficiency effects, and 37 phenotypic variables showed significant genotype × treatment (GT) interactions. During the vegetative stage experiment, GWAS found 77 significant loci related to 16 phenotypic variables, and a total of 85 significant loci were associated with 13 phenotypic traits during the generative stage trial [110]. For 17 phenotypic features, more than 110 loci for GT interaction were discovered, accounting for more than half of the genetic variation in several instances. Among the spring barley accessions, the genetic regulation of the nodal–root architectural and anatomical response to water deficiency was examined by Oyiga et al. [128]. The architectural and anatomical nodal–root response to water deprivation was linked to 11 QTL intervals in a GWAS analysis. Three and four QTL intervals, respectively, exhibited substantial impacts across seasons, and on root architectural and anatomical features. A total of 44 epistatically interacting SNP sites were discovered during a genome-wide epistasis analysis. Further research revealed that these QTL intervals include significant candidate genes, such as ZIFL2, MATE, and PPIB, whose activities have been linked to plant root adaptation responses to water shortage.
Moreover, a set of polymorphic SSR markers were used to discover MTAs 48 barley accessions. The MLM discovered a total of 12 significant MTAs. Importantly, the marker ABC252, on chromosome 2H, was shown to be associated with days to heading and 1000-kernal weight (KW), while the marker Bmag273 on chromosome 7H was found to be associated with spike length [111]. In multienvironmental experiments in Egypt, Sayed et al. [75] analyzed a mapping population of 298 doubled haploid lines. The results of composite interval mapping identified 35 QTLs. A significant QTL bPb-9110 (140.3 cM on 3H) was discovered to be related with the heading date. Furthermore, marker bPb-1213 on 1H was also linked to the heading date, as well as grain yield. This research also showed that the presence of unusual alleles in the gene HvFT4 (2H, 66 cM) had a substantial influence on the heading date. This gene was shown to be 24.9 cM from the photoperiod Ppd-H1 gene, and both genes play an important role in controlling flowering time in barley. It has been reported that Tibetan wild barley has a high level of genetic diversity and stress tolerance. A GWAS analysis has been carried out in both hydroponic and pot trials to find QTLs for drought tolerance utilizing 777 DArT markers and morphological and physiological features in 166 Tibetan wild barley accessions [112]. Wide genotypic diversity was reported for various variables, and population structure and kinship analyses revealed three subpopulations among the investigated accessions. Drought-tolerance-related traits, such as ATPase activity, antioxidant enzyme activities, MDA content, soluble protein content, and potassium concentration, were linked with 91 DArT markers. Water usage efficiency (WUE) is an important feature in drought conditions, since it indicates biomass output under low water circumstances. Experiments on 156 barley genotypes were carried out in greenhouse pots to determine WUE and other water consumption parameters, such as dry weight, fresh weight, total leaf water, and leaf water content [129]. The observed data were utilized for GWAS analysis, resulting in the discovery of 14 MTAs corresponding to 4 quantitative trait loci (QTL). Four MTAs related to WUE were found on chromosome 4H [129].
4.3. Temperature
Inadequate temperature is another environmental issue that has a detrimental impact on crops, especially when combined with another stressor, such as water scarcity. Indeed, water scarcity and heat are two of the most significant abiotic stressors limiting crop growth and production across the world [91]. As shown in sorghum [130], wheat, and barley [131], the combined impacts of heat and drought on yield are more detrimental than the effects of each stress separately [132]. Thermal factors influence a wide range of plant life activities, from seed germination and seedling emergence via soil temperature to vegetative growth and development of the entire plant via soil and air temperatures [133]. High temperatures can cause changes in cell membrane function, affecting metabolic pathways such as respiration and photosynthesis. This environmental component can alter the physicochemical characteristics of thylakoids, impairing electron transport, causing ionic imbalance, and causing ROSs. High temperature stress can affect the amount of carbon molecules produced by photosynthesis in plants with the C3 cycle, such as barley, which exhibits apparent photorespiration. Lower assimilate production tends to limit their allocation in seeds, resulting in thinner seeds with fewer stores intended for embryo development restart. Furthermore, it causes a high incidence of seedlings with poor development, which in the field increases the chance of creating a dominant plant within the population [134]. Detailed information regarding the effects of both high- and low-temperature stresses on plants is presented in Figure 4.
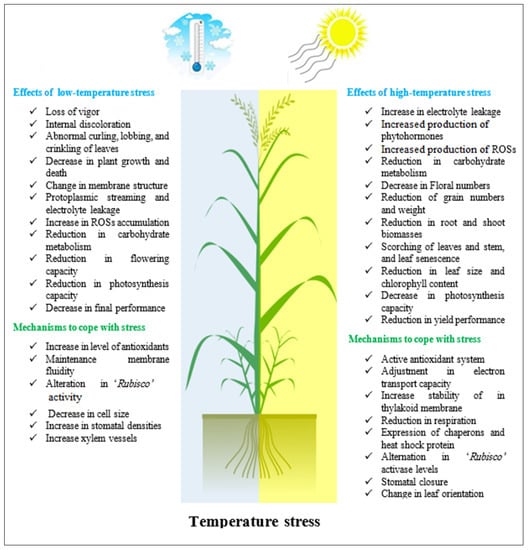
Figure 4.
The effects of high- and low-temperature stress on the growth and productivity of plants.
According to Brestic and Ivak [135], an increase in temperature to 37 °C may result in decreased stomatal conductance. Leaf photosynthesis, being the most heat-sensitive metabolic activity, has been shown to be disrupted by high temperatures [136]. Thermal stress on the plant photosynthetic system primarily affects the PSII and Rubisco’s carbon fixation [137,138]. Furthermore, chlorophyll fluorescence has been discovered to be a valid physiological measure for assessing damage to the photosynthetic machinery caused by heat stress [34]. The maximum quantum yield of PSII photochemistry (Fv/Fm) has long been used as a criterion for selecting heat-tolerant genotypes in plants. High temperature during the grain-filling stage, according to Gupta et al. [139], reduces photosynthetic ability, chlorophyll content, and grain production in wheat genotypes. Proline accumulation is an important adaptive response in crops exposed to high temperatures [140]. It is involved in cellular osmoregulation and protection of cellular structures against increased temperatures through cell water balance conservation and stabilization of biological membranes. The response of H. vulgare ssp. spontaneum genotypes to terminal heat stress was assessed by Bahrami et al. [141] using gas-exchange parameters, photosystem efficiency, proline buildup, cell membrane leakage, and grain yield characteristics. They reported that the adapted wild genotypes showed physiological mechanisms capable of retaining their production capacity and plasticity flow across time, which might be used to introduce heat tolerance by crossing with cultivated barley.
In this section, we highlight some important studies that reported the key QTLs and genes related to cold tolerance in barley (Table S3) [126,142,143,144,145,146,147,148,149,150]. The “Nure × Tremois” mapping population (NT) linkage map demonstrated the benefits of having numerous informative markers for linkage map development. Marker order and distance are preserved compared with published maps. The NT population is segregating for both vernalization need and low-temperature tolerance, while the “Dicktoo × Morex” population is segregating for photoperiod sensitivity and low-temperature tolerance [151]. These QTLs determined low-temperature tolerance, vernalization response (reaction to vernalization), and heading date on chromosome 5H. The location of cold-regulated genes (COR) relative to these QTLs, as well as the QTLs responsible for COR protein accumulation and the location of the representative CBF loci relative to these winter hardiness-related traits, were also determined [152]. The vernalization response QTL is found in the Triticeae region Vrn-1/Fr1, which is also the location of QTLs for all parameters of low-temperature tolerance [152]. Based on this research, COR14b and TMC-Ap3 are two distinct COR proteins that were found using composite interval mapping. The corresponding candidate gene is HvCBF4 or another member of the CBF locus grouped in this area. A CBF protein might interact with a CRT/DRE recognition site in the promoter of cor14b. In addition, Saade et al. [153] listed the important QTLs for low-temperature tolerance that have been discovered so far, such as FR-H1 and FR-H2. Although our understanding of barley’s reactions to abiotic stimuli is limited, the genetic diversity seen in farmed barley and its near wild relatives might be used to increase stress tolerance.
The results reported by Guerra et al. [142] illustrate the potential of allele mining on exome sequencing data obtained from over 400 barley accessions using case studies of frost tolerance and vernalization requirements. New deletions in VRN-first-H1’s intron were discovered and associated with a lower vernalization requirement, while the allelic diversity of HvCBF2a, HvCBF4b, and HvCBF14 was studied using a combination of SNP and read count analysis. HvCBF2 gene paralogs and copy number variations, as well as the HvCBF4b-HvCBF2a section, have been identified using this method. An MLM model revealed that HvCBF14, rather than HvCBF4b-HvCBF2a copy number variation, is more important in influencing frost tolerance in barley.
Low-temperature tolerance (LTT) and vernalization sensitivity are important features for barley seedling establishment, growth, and development. For facultative types, sensitivity to short-day photoperiod is a desirable trait. Munoz-Amatriain et al. [143] used a GWAS analysis with a panel of 882 barley accessions and a 9K SNP chip to map QTLs and identify candidate genes to obtain a better understanding of the genetics of these characteristics under cold stress conditions. For LTT, 15 loci were discovered, including five known and 10 new QTL/genes, which were evaluated as winter survival in 10 field tests and mapped using a GWAS meta-analysis. Furthermore, it was demonstrated that LTT is caused by the genes FR-H1, FR-H2, and FR-H3; candidate genes for FR-H3 were discovered. VRN-H1, VRN-H2, and PPD-H1 were the main factors of vernalization sensitivity.
4.4. Waterlogging
The bulk of grain-producing species’ growing fields have soils with a good draining status. However, flooding due to higher rainfall volumes may be one of the most common natural disasters in terms of adverse crop-growing circumstances. The sluggish diffusion of oxygen via the water may cause a reduction in oxygen availability for the roots, which may impair plant development in many vegetal species [91]. The lack of oxygen caused by floods may affect physiologic characteristics, such as carbon assimilation, macronutrient absorption, and root respiration metabolism [154]. Soil flooding causes a decrease in seed production and damages the chemical composition, physical, and physiological integrity of plants [155]. Changes in the radicular system and a decrease in photo-assimilated translocation, as well as a decrease in plant metabolic activity [156], result in a decrease in chlorophyll content, and can interfere with the efficiency and conversion of luminous radiation to chemical energy [157]. Yordanova and Popova [158] discovered a significant decrease in photosynthetic activity of the 1,5-bisphosphate carboxylase/oxygenase (RuBPCO) in barley plants that had been subjected to soil flooding.
As a consequence of waterlogging stress, oxidative stress may occur in cells and tissues. Increased production of ROSs is one of the plant’s responses to oxygen deprivation [51,159,160]. These ROSs are able to oxidize and break down biological components, such as lipids, proteins, carbohydrates, and nucleic acids, as well as affect enzymatic function [161]. ROS may be created by a variety of processes in plant roots under oxygen-limited circumstances, including plasma membrane (PM) NADPH, mitochondrial malfunction, and metal ion buildup [162]. Under O2 deprivation, increased iron and copper activity (together with other transition metals) in ionic and catalytically active chelated forms is commonly thought to be a key cause of the ROS burst through the conversion of H2O2 to very hazardous HO [162]. ROSs generated in oxygen-depleted environments also play an important part in plants’ developmental and adaptive responses to waterlogging stress as signaling molecules. ROS generation by PM NADPH oxidase and/or mitochondria seems to govern plant adaptation responses under oxygen-limited circumstances, according to a large body of evidence [78,163]. Unbalanced ROS production, on the other hand, may harm cellular components and cause them to malfunction. As a result of the detrimental effect of ROS overproduction in living tissues, the capacity of plants to generate antioxidant enzymes is often connected with vulnerability to environmental stressors, such as waterlogging [164]. Moreover, waterlogging causes anaerobic conditions in the root zone of plants. Climate change is increasing the occurrence of waterlogging, causing significant agricultural losses. Aerenchyma development, energy metabolism, and phytohormone signaling are all responses to waterlogging stress, as waterlogging affects biomass, photosynthesis, adventitious root growth, and aerenchyma formation [165] (Figure 5).
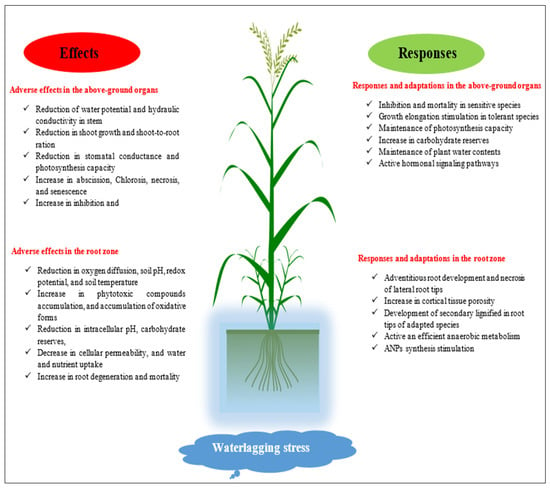
Figure 5.
Important effects of waterlogging stress, and plants’ responses to it.
Tong et al. [165] examined how waterlogging affects physiological and genetic pathways in four important grain crops. They also presented the state of knowledge on waterlogging tolerance mechanisms, genes, and QTL linked with waterlogging tolerance, as well as traditional and contemporary breeding approaches. Aerenchyma production under waterlogging stress is one of the most efficient strategies to overcome stress-induced hypoxia. Aerenchyma and radial O2 loss (ROL) barriers are examples of survival strategies of plants adapted to flooding stress. To protect plants against waterlogging stress, Jia et al. [166] studied the effect of phytohormones such as ethylene and ERF-VII transcription factors. Based on sequence similarity, ERF-VIIs from major crops and many other ERF-VIIs involved in plant flood tolerance were gathered and examined. Water supply impacts barley more than other crops, resulting in loss of output from 40 to 79% based on genotype, stages of growth, and duration of water supply [167]. One significant mechanism for water logging is the development of adventitious roots (ARs), as they help water and the absorption of nutrients under hypoxic stress [168]. Barley genotypes have significantly higher root porosity and faster development of aerenchyma in adventitious roots when exposed to water [169]. Luan et al. [170] reviewed seven different genotype barley physiological and morpho-anatomical responses to waterlogging stress. Slight reductions in plant height and SPAD (soil/plant development analysis), tillers, and shoot and root biomasses were observed in the tolerant varieties compared to sensitive varieties. The number of adventitious roots of tolerant genotypes was likewise much greater than in sensitive genotypes under water stress. Because of the greater ethylene content, lower ABA level, and less O2 buildup, the leaves of the waterlogging resistant cultivars exhibited larger intercellular gaps and better integrated chloroplast membrane structures [170].
The impact of waterlogging timing and duration on yield is uncertain. To examine this, Liu et al. [171] used a variety of waterlogging treatments on barley genotypes with varying waterlogging tolerances. One experiment included four waterlogging treatments: ZS 12.5 for 1 or 2 months (WL1 and WL2), ZS 15 for 2 months (WL3), and 1 day before travelling for 15 days (WL4). Another experiment comprised only WL2 treatment. The reduction in the number of spikes per m2 and kernels per spike lowered the yield by 35% in W1 and by 52% in WL3. This was attributable to WL4’s influence on spikelet fertility and grain filling. The waterlogging susceptible cultivar showed the highest delays (1–8 ZS), whereas waterlogging resistant genotypes capable of aerenchyma production showed the least delays (0–4 ZS). Early phenological waterlogging prevented 23% yield loss in genotypes with aerenchyma development QTL. Waterlogging in late developmental stages lowered average final grain production by 70%.
Similar to studies on other abiotic stresses, there are many genetic studies on the discovery of genomic regions associated with waterlogging tolerance (Table S4) [78,172,173,174,175,176,177,178]. Waterlogging tolerance has been linked to a number of QTLs based on agronomic, physiological, and anatomical features. Several features such as plant height [179], grain yield [180], plant survival [181], leaf chlorosis [87], and plant biomass [182] under waterlogging stress have been used to conduct QTL analysis for waterlogging tolerance in barley. Since these QTLs have been found on all seven chromosomes, their practical use has been limited. Furthermore, the majority of this research has relied on quantitative traits, which might change across contexts. For example, a QTL discovered in one environment may not be detected in another [183]. Gill et al. [78] evaluated 187 barley DH lines under waterlogging and salinity stresses. They provided the first report of a large QTL mapping of waterlogging tolerance for both O2 and H2O2 based on ROS accumulation on chromosome 2H. The QTL for ROS was found in the same location as the QTLs for waterlogging and salinity tolerance. Using this characteristic as a physiological marker, the one QTL discovered permits precision mapping of the gene responsible for waterlogging and salinity tolerance. The presence of QTL for many abiotic stressors at this location on chromosome 2H suggests a particular mechanism for various stress tolerances, such as waterlogging and salt tolerance.
Li et al. [172] created a composite map using SSRs, RFLP, and DArT markers to discover QTLs linked with waterlogging tolerance in barley. Twenty QTLs for waterlogging tolerance were discovered in two barley DH populations. Several of these QTLs were verified by cross-season replication or population colocation. Qwt4-1, for example, contributed to both lowering barley leaf chlorosis and boosting plant biomass under waterlogging stress, whereas other QTLs regulated both leaf chlorosis and plant survival. Comparatively, improving waterlogging tolerance in barley is still in its infancy. Using the identified QTLs along with conventional field selection, waterlogging tolerance in barley can be improved. In addition, there may be a homoeologous link between QTLs influencing waterlogging tolerance in barley and in other crops [172]. Under waterlogging conditions, Mano et al. [184] used a panel of 110 individuals of an F2 population to investigate QTLs affecting adventitious root development (ARF) on the soil surface. ARF QTLs were found on chromosomes 1H, 3H, and 7H. These genomic regions were similar to those identified in a teosinte population (Zea mays ssp. huehuetenangensis). Broughton et al. [174] used a DH population to study the adventitious root porosity (gas-filled volume per unit root volume). On chromosome 4H, a single QTL for root porosity explained 35.7% of the phenotypic variance in aerated and oxygen-deficient environments, close to EBmac0701. When the population was screened in an earlier independent soil waterlogging experiment, this QTL contributed to waterlogging tolerance. Indeed, this QTL is syntenic with the Qaer1.02-3 QTL in maize and the Sub1A-1 gene in rice, both of which are involved in aerenchyma development.
The capacity of plants to retain extremely negative membrane potential (MP) is related to their tolerance to both salt and waterlogging stress. However, few efforts have been made to discover QTL underlying this feature. The plasma membrane potential of epidermal root cells from 150 DH lines was measured using noninvasive microelectrode ion flux measurements (MIFEs) technique by Gill et al. [185].
A significant MP QTL was found in hypoxia-exposed epidermal root cells. This QTL was found on 2H, among the QTLs for waterlogging and salinity tolerance. Furthermore, it has been revealed that MP has a key role in waterlogging and salinity tolerance. The fact that MP is regulated by a single main QTL shows the efficiency of single-cell phenotyping, and paves the way for fine mapping this QTL for better marker-aided selection [185].
In a comprehensive study, Zhang et al. [177] analyzed 632 QTL for drought, salinity, and waterlogging tolerance in barley. Only 195 significant QTL were utilized for meta-analysis to refine QTL locations for MAS. On the barley physical map, the meta-analysis highlighted significant QTL for drought, salinity, and waterlogging tolerance from multiple mapping populations. The meta-QTL (MQTL) locations were then utilized to identify candidate genes for drought, salinity, and waterlogging tolerance. For example, MQTL3H.4 and MQTL6H.2 regulate drought tolerance in barley. In addition, the salinity tolerance QTLs HvNax4 and HvNax3 were verified on MQTL1H.4 and MQTL7H.2, respectively. MQTL2H.1 and MQTL5H.3 were also targeted for enhancing salt tolerance in barley. MQTL4H.4 is the primary area that determines waterlogging tolerance in barley, including fine-mapped QTL for aerenchyma development. Hence, finding and refining MQTLs and candidate genes is critical for future MAS breeding success. Borrego-Benjumea et al. [176] genotyped 247 global spring barley genotypes using 35926 SNP markers. Biomass, spikes per plant, grains per plant, kernel weight per plant, height of plant, and chlorophyll content all showed significant phenotypic variation in the test barley panel. A GWAS for waterlogging tolerance revealed 17 genomic areas with 51 significant waterlogging tolerance-associated markers, accounting for 5.8–11.5% of the phenotypic variance; most QTL were located on chromosomes 1H, 2H, 4H, and 5H. Six new QTL were discovered, and eight candidate genes for abiotic stress responses were found near QTL for waterlogging tolerance.
4.5. Toxicity
Heavy metals are known to be naturally dispersed across the globe, but they are also introduced into the environment via human activities, such as mining, farming, sewage waste disposal, and industrial sludge [186,187]. This is a worldwide issue that must be addressed. All heavy metals are potentially hazardous, with varying degrees of toxicity depending on their biologic availability and the susceptibility of organisms exposed to the heavy metal. Lead (Pb) is an example of a heavy metal that is a well-known quickly accumulating contaminant in soils and sediments [188]. Pb is regarded as a serious toxin, and agricultural yield suffers significantly in Pb-contaminated soils. Chlorosis, decreased growth, blackening of the root system, disturbances in mineral nutrition and water balance, changes in hormone status, and effects on the structure and permeability of membranes are all toxic consequences of lead on plants [188]. Moreover, researchers are increasingly focusing on the combined impacts of metals on plants. For example, copper (Cu) was shown to have an antagonistic impact when combined with other divalent metals, such as Cd, Ni, and Pb. Many plants have been harmed in recent years as a result of increased contamination of the ground by heavy metals, prompting researchers to become interested in soil improvement, as well as the development and usage of resistant variants. The majority of key agricultural qualities, such as yield, grain quality, and tolerance, as well as tolerance to biotic and abiotic pressures in nature, are qualitatively complex traits. Lwalaba et al. [189] investigated the combined toxicity of cobalt (Co) and Cu on two barley genotypes with different degrees of Co tolerance. This toxicity reduced mineral uptake (Mn, Zn, and K), resulting in a significant reduction in plant growth and an increase in oxidative stress (ROS and MDA), while concurrently boosting the capacity to scavenge active oxygen species (AOS). This process is shown by enhanced glutathione (GSH), oxidized GSH (GSSG) and phytochelatin (PC) levels, as well as the related gene expression (HvPCS1 and HvGR1) [189]. Figure 6 shows how this stress alters plant growth.
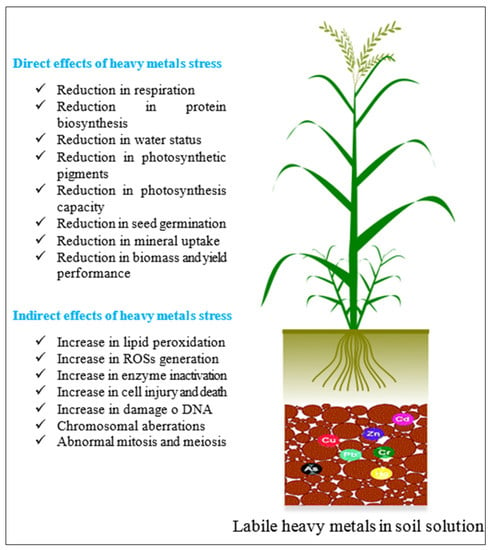
Figure 6.
Heavy metal stress and its effects on plants.
Barley production is also limited in major agricultural areas across the world due to soil acidity and aluminum toxicity. Aluminum (Al) toxicity prevents root cell division and elongation, lowering water and nutrient absorption as well as grain yield [190]. In Brazil, hazardous amounts of Al are found in more than half of the agricultural regions, and this feature limits barley production. As a result, some amount of genetic tolerance to hazardous levels of Al is critical for barley production in Brazil and other acidic soil locations. Cadmium (Cd) has been shown to decrease chlorophyll concentration, change nitrogen metabolism, and inhibit the photosynthetic system in plants [191]. Leaf chlorosis is the most visible indication of Cd poisoning in plants, and when the period of treatment and/or the Cd content increases, the shift from leaf chlorosis to necrosis occurs [192]. Extrusion across the plasma membrane, chelation in the cytoplasm, and vacuolar sequestration are only some of the strategies that plants have developed to adapt to Cd-contaminated environments [193]. The efflux pumping of Cd at the plasma membrane is mediated by transporters with heavy metal binding domains, such as pleiotropic drug tolerance-type ATP binding (ABC) protein (PDR9) and multi-antimicrobial extrusion protein (MatE) [194]. The tonoplast-based heavy-metal-transporting ATPase 3 (HMA3) aids in the sequestration of Cd into vacuoles [195]. Zn transporters such as Zrt (zinc-regulated transporter)/IRT-like protein 1 (ZIP) and the cation efflux family (cation efflux) proteins have also been shown to transport Cd ions [196,197].
In general, multiple QTLs underlie these qualities, which are heavily impacted by the plants’ environment as well as the genes linked to heavy metal tolerance [188]. Table S5 includes a list of useful QTLs linked with heavy metal toxicity tolerance in barley [1,197,198,199,200,201,202]. Cadmium’s (Cd) high toxicity and easy absorption by plants has become a serious agricultural issue. Finding the gene(s) responsible for Cd accumulation/tolerance is critical for generating low-Cd agricultural cultivars. Derakhshani et al. [197] used the phenotypically polymorphic Oregon Wolfe Barley (OWB) mapping population to explore the genetic architecture and genetic control of Cd tolerance in barley. They identified one minor and one significant QTL on chromosomes 2H and 6H, respectively, after analyzing the Cd tolerance of 87 double haploid lines at the seedling stage. The significant QTL explained 47.24% of the phenotypic variation for chlorosis and 38.59% of the phenotypic variance for necrosis characteristics. Moreover, 16 Cd-tolerance-related candidate genes were discovered by combining the data from the identified QTLs with RNA-Seq analysis; 9 of these genes were metal ion transporters. Wang et al. [1] created a DH population from a cross between Suyinmai 2 (Cd-sensitive) and Weisuobuzhi (Cd-tolerant) to map QTLs. They measured chlorophyll content, developmental characteristics, metal concentration, and antioxidant enzyme activity in DH population lines and parents under control and Cd stress. A single QTL, qShCd7H, on chromosome 7H was connected to shoot Cd content, explaining 17% of the phenotypic variance. Gene silencing was employed to isolate, clone, and characterize the candidate gene. The identification of qShCd7H led to the discovery of a new gene, HvPAA1. A sequence comparison revealed that HvPAA1 has seven N-glycosylation domains.
The amount of vital nutrients and chemically equivalent hazardous analogues accumulating in cereal grains has a significant influence on crop nutritional quality and safety. Introgression lines (ILs) may be used to take advantage of naturally existing genetic variation in order to create superior varieties. A collection of 54 ILs of the wild ancestor H. vulgare ssp. spontaneum in the cultivated variety (cv. Scarlett) were studied using multi-element analysis on vegetative leaves, senesced flag leaves, and mature grains. Plants were grown on anthropogenically heavy-metal-contaminated soil acquired from an agricultural area, enabling the simultaneous identification of QTLs for the accumulation of critical nutrients and harmful trace elements in barley [202]. Reuscher et al. [202] found 25, 16, and 5 QTLs for the micronutrients Fe and Zn, and interfering toxin Cd, respectively. They linked QTL to candidate genes based on similarity to known metal homeostasis genes in Arabidopsis and rice by evaluating the gene content of the introgressions. Cu and Fe, rather than Cd, were remobilized preferentially from the flag leaf to growing grains, according to global comparative assessments. Grain micronutrient filling is a controlled and nutrient-specific mechanism that differs from vegetative micronutrient homoeostasis [202]. Wu et al. [198] also used genome-wide association mapping to look for Cd accumulation in different plant organs. They reported a high degree of genotypic heterogeneity in Cd content in all organs. Moreover, their results indicated a strong link between solution shoot Cd and soil culture, as well as between shoot Cd and grain Cd, but no link between root Cd and grain Cd. They also identified nine QTLs for root Cd, 21 for shoot Cd, 14 for root-to-shoot translocation, and 15 for grain Cd. On chromosome 5H, a shared QTL for shoot Cd and root-to-shoot translocation was discovered at 132.6 cM. On chromosomes 2H and 5H, two significant QTL for grain Cd were discovered. Hence, future cloning of potential genes for Cd accumulation and breeding low-Cd barley cultivars will be aided by the genetic diversity in Cd accumulation and key QTLs discovered in different studies.
Increased ion (manganese and iron) toxicity caused by changes in the soil redox potential under hypoxic conditions causes waterlogging stress, which disrupts plant metabolism. In a recent study, Huang et al. [199] discovered a link between tolerance to Mn2+ toxicity and waterlogging stress in barley, suggesting that enhancing Mn2+ toxicity tolerance might also improve waterlogging tolerance. Based on chlorophyll content and plant survival as selection criteria, a DH population was utilized to find QTL underlying tolerance to Mn2+ toxicity. At the seedling stage, this population had four significant QTL for plant survival under Mn2+ stress (QSur.yf.1H, QSur.yf.3H, QSur.yf.4H, and QSur.yf.6H). On chromosomes 3H and 6H, two significant QTL governing leaf chlorosis under Mn2+ stress were discovered near QSur.yf.3H and QSur.yf.6H. The main QTL, QSur.yf.3H, which was found at the marker Bmag0013, was responsible for 21% of the phenotypic variance. Soil elemental toxicity is also a result of floods. Excess water causes a gradual drop in soil redox potential, raising the concentration of Mn2+, which may be hazardous to plants if it exceeds a certain level. Using barley as a case study, this research intended to assess the proportional contribution of Mn2+ toxicity to waterlogging stress tolerance [175]. Twenty barley genotypes with varying levels of waterlogging stress tolerance were tested in the root rhizosphere for their capacity to deal with toxic (1 mM) quantities of Mn2+. Most waterlogging-tolerant genotypes (TX9425, Yerong, CPI-71284-48, and CM72) maintained chlorophyll content above 60% of the control value under Mn2+ toxicity, while sensitive genotypes (Franklin and Naso Nijo) exhibited 35% less chlorophyll than 35% of controls. Manganese content in leaves was not associated with visual Mn2+ poisoning symptoms, indicating that alternative Mn2+ tolerance mechanisms, such as avoidance (instead of tissue tolerance), may act in different tolerant genotypes [175]. The overall considerable (r = 0.60) connection between Mn2+ toxicity tolerance and waterlogging in barley showed that, at least in this species, plant breeding for waterlogging tolerance characteristics might be aided by targeting pathways involved in Mn2+ toxicity tolerance. Koochakpour et al. [200] used a spring barley population including 148 genotypes for whole genome association mapping under Mn stress. They used a mixed linear model (MLM with K + Q) to examine marker–phenotypic trait associations. In both non-stress and stress conditions, 39 significant MTAs were found. These markers were located on chromosomes 1H–5H. Many of the related markers were found in QTL-rich areas. Three important markers were found on chromosomes 3H and 5H, linked to MSL and awn length (AL), suggesting that these two traits are controlled by the same genetic regions. The findings showed that Mn tolerance was quantitatively inherited, and the identified QTLs might be useful for marker-assisted selection and gene discovery in barley.
Acid soil is a major issue in agricultural development across the globe. Toxic Al cations reduce root development and impairs yield in acidic soil. Although an acid soil tolerance gene has been discovered, it has not been exploited in malting barley breeding, since this gene has been connected to poor malting quality features [203]. Br2, a Brazilian malting barley variety, has been shown to be tolerant to acidic soil. Bian et al. [203] created a DH population from a hybrid between Br2 and the acid-sensitive Australian cultivar Hamelin. The DH population was genotyped using SSR, DArT, and gene-specific markers, and evaluated for acid soil tolerance in native acid soil and a hydroponic system with pH 4.2, pH 4.2 + Al, or pH 6.5. For all factors relevant to acid soil tolerance, a single QTL was discovered on chromosome 4H near the HvMATE gene. The HvMATE gene sequence alignment revealed 13 INDELs and 87 SNPs, each of which coded for a single amino acid variation between the 2 types. To identify the single nucleotide variation between Hamelin and Br2, a gene-specific marker was created. It was found that this marker cosegregated with aluminum tolerance, and accounted for 79% of the phenotypic variation in acid soil tolerance. In the study conducted by Navakoda et al. [201], a DH mapping population was used to investigate how Al tolerance is passed on in barley. A genetic map was built using genic markers and used to map QTL. Several QTLs for Al tolerance were discovered on chromosomes 2H, 3H, and 4H. In order to find probable candidate genes for Al tolerance, a sequence homology search was carried out to determine the likely function of the genes associated with the QTL. Some of these possibilities, in particular stress signal transduction, transcription regulatory factors, and cell metabolism, have been linked to stress/defense responses.
5. Conclusions
Finding the genomic areas associated with different attributes in barley under abiotic stress has helped to identify a region within the QTLs where the probable underlying candidate genes could be located. QTL mapping and association analyses are important genetic tools to identify and understand the relationships between stress tolerance and other agronomic and physiological characteristics, which may then be used in breeding programs for gene cloning and the development of stress-tolerant cultivars in barley. Among the various biotechnology tools, GWAS analysis has been efficiently utilized to discover novel genomic regions associated with different traits in crops, as it detects well the statistical correlation between phenotypic and genotypic data within natural populations. Overall, the information presented in this review improves the knowledge about the effects of various abiotic stresses on different aspects of growth and development in barley, as well as the genomic regions associated with stress tolerance and other agro-morphological and biochemical features in early and whole plant stages. Moreover, this review highlights the use of molecular markers in the MAS approach to barley breeding programs to improve tolerance to various abiotic stresses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app12105189/s1, Table S1: Reports on some of the identified genomic regions linked with various agronomic, physiological, biochemical, root system architecture, and quality features in barley under salinity stress. Table S2. Reports on the identified genomic regions linked with various agronomic, physiological, biochemical, root system architecture, and quality features in barley under water deficit stress. Table S3. List of studies that identified genomic regions linked with various agronomic, physiological, biochemical, root system architecture, and quality features in barley under low-temperature stress. Table S4. List of reports that identified genomic regions linked with various agronomic, physiological, biochemical, root system architecture, and quality features in barley under waterlogging stress. Table S5. List of reports that identified genomic regions linked with various agronomic, physiological, biochemical, root system architecture, and quality features in barley under heavy metal stress.
Author Contributions
All authors equally have contributed in this work. All authors have read and agreed to the published version of the manuscript.
Funding
Open access funding provided by University of Helsinki. Peter Poczai thanks the support of the Eötvös Research Fund.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, X.K.; Gong, X.; Cao, F.B.; Wang, Y.Z.; Zhang, G.P.; Wu, F.B. HvPAA1 encodes a P-Type ATPase, a novel gene for cadmium accumulation and tolerance in barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2019, 20, 1732. [Google Scholar] [CrossRef]
- Yan, S.; Sun, D.; Sun, G. Genetic divergence in domesticated and non-domesticated gene regions of barley chromosomes. PLoS ONE 2015, 10, e0121106. [Google Scholar] [CrossRef][Green Version]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World. In The Origin and Spread of Cultivated Plants in West Asia, Europe, and the Nile Valley; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Harlan, J.R.; Zohary, D. Distribution of wild wheats and barley. Science 1996, 153, 1074–1080. [Google Scholar] [CrossRef]
- Nevo, E. Origin, evolution, population genetics and resources for breeding of wild barley, Hordeum spontaneum, in the Fertile Crescent. In Barley: Genetics, Biochemistry, Molecular Biology and Biotechnology; Shewry, P.R., Ed.; International, The Alden Press: Oxford, UK, 1992; pp. 19–43. [Google Scholar]
- Bothmer, R.V.; Jacobsen, N.; Baden, C.; Jorgensen, R.B.; Linde-Laursen, I. An Ecogeographical Study of the Genus Hordeum; IPGRI: Rome, Italy, 1995; p. 129. [Google Scholar]
- Singh, L.; Park, R.F.; Dracatos, P.; Ziems, L.; Singh, D. Understanding the expression and interaction of Rph genes conferring seedling and adult plant resistance to Puccinia hordei in barley. Can. J. Plant Pathol. 2021, 43, 218–226. [Google Scholar] [CrossRef]
- Vaezi, B.; Pour-Aboughadareh, A.; Mohammadi, R.; Armion, M.; Mehraban, A.; Hossein-Pour, T.; Dorri, M. GGE biplot and AMMI analysis of barley yield performance in Iran. Cereal Res. Commun. 2017, 45, 500–511. [Google Scholar] [CrossRef]
- OECD. Consensus Document on Compositional Considerations for New Varieties of Barley (Hordeum vulgare L.): Key Food and Feed Nutrients and Anti-Nutrients; Report No: 12, Environment Directorate; OECD: Paris, France, 2004. [Google Scholar]
- Heuzé, V.; Tran, G.; Nozière, P.; Bastianelli, D.; Lebas, F. Cowpea (Vigna unguiculata) Forage. Feedipedia, a Programme by INRA, CIRAD, AFZ and FAO. Last Updated on October 20. 2015. Available online: https://www.feedipedia.org/node/233 (accessed on 14 February 2018).
- FAO. Special Report: 2021 FAO Crop and Food Supply Assessment Mission to the Syrian Arab Republic—December 2021; FAO: Rome, Italy, 2021. [Google Scholar]
- Sax, K. The association of size differences with seed-coat pattern and pigmentation in Phaseolus vulgaris. Genetics 1923, 8, 552–560. [Google Scholar] [CrossRef]
- Thoday, J.M. Location of polygenes. Nature 1961, 191, 368–370. [Google Scholar] [CrossRef]
- Kearsey, M.J.; Farquhar, A.G.L. QTL analysis; where are we now? Heredity 1998, 80, 137–142. [Google Scholar] [CrossRef]
- Stuber, C.W.; Lincoln, S.E.; Wolff, D.W.; Helentjaris, T.; Lander, E.S. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 1992, 132, 823–839. [Google Scholar] [CrossRef]
- Moose, S.P.; Mumm, R.H. Molecular plant breeding as the foundation for 21st Century crop improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef]
- Sham, P.; Bader, J.S.; Craig, I.; Odonovan, M.; Owen, M. DNA pooling: A tool for large-scale association studies. Nat. Rev. Genet. 2002, 3, 862–871. [Google Scholar] [CrossRef]
- Nillson-Ehle, H. Kreuzunguntersuchungen an Hafer und Weizen. Acta Univ. Lund. 1909, 5, 1–122. [Google Scholar]
- East, E.M. Studies on size inheritance in nicotiana. Genetics 1916, 1, 164–176. [Google Scholar] [CrossRef]
- Xu, Y. Quantitative trait loci: Separating, pyramiding, and cloning. Plant Breed. Rev. 1997, 15, 85–139. [Google Scholar]
- Xu, Y. Molecular Plant Breeding; CABI: Wallingford, UK, 2010; p. 736. [Google Scholar]
- Bernardo, R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 2008, 48, 1649–1664. [Google Scholar] [CrossRef]
- Xu, S. Mapping quantitative trait loci using multiple families of line crosses. Genetics 1998, 148, 517–524. [Google Scholar] [CrossRef]
- Collard, B.; Mace, E.; Mcphail, M.; Wenzl, P.; Cakir, M.; Fox, G.; Poulsen, D.; Jordan, D. How accurate are the marker orders in crop linkage maps generated from large marker datasets? Crop Pasture Sci. 2009, 60, 362–372. [Google Scholar] [CrossRef]
- Bradshaw, H.D. Molecular genetics of populus. In Biology of Populus and Its Implications for Management and Conservation; Part 1; Stettler, R.F., Bradshaw, H.D., Heilman, P.E., Hinckley, T.M., Eds.; NRC Research Press: Ottawa, ON, Canada, 1996; pp. 183–199. [Google Scholar]
- Pour-Aboughadareh, A.; Yousefian, M.; Moradkhani, H.; Poczai, P.; Siddique, K.H.M. STABILITYSOFT: A new online program to calculate parametric and non-parametric stability statistics for crop traits. Appl. Plant Sci. 2019, 7, e1211. [Google Scholar] [CrossRef]
- Szareski, V.J.; Carvalho, I.R.; Rosa, T.C.; Dellagostin, S.M.; de Pelegrin, A.J.; Barbosa, M.H.; Pegoraro, C. Wild Species: An Alternative for Rice Breeding under Abiotic Stress Conditions. Am. J. Plant Sci. 2018, 9, 1093–1104. [Google Scholar] [CrossRef]
- Larcher, W. Ecofisiologia Vegetal; Rima Artes: São Carlos, Brazil, 2004. [Google Scholar]
- Shanker, A.; Venkateswarlu, B. Abiotic Stress Response in Plants: Physiological, Biochemical and Genetic Perspectives; InTech Open: Rijeka, Croatia, 2011; p. 360. [Google Scholar]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef]
- Paterson, A.H.; Lin, Y.R.; Li, Z.; Schertz, K.F.; Doebley, J.F.; Pinson, S.R.; Liu, S.C.; Stansel, J.W.; Irvine, J.E. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 1995, 269, 1714–1718. [Google Scholar] [CrossRef] [PubMed]
- Arzani, A.; Ashraf, A. Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit. Rev. Plant Sci. 2016, 35, 146–189. [Google Scholar] [CrossRef]
- Honsdorf, N.; March, T.J.; Berger, B.; Tester, M.; Pillen, K. High-throughput phenotyping to detect drought tolerance QTL in wild barley lines introgression. PLoS ONE 2014, 9, e97047. [Google Scholar] [CrossRef] [PubMed]
- Jedmowski, C.; Ashoub, A.; Momtaz, O.; Brüggemann, W. Impact of drought, heat, and their combination on chlorophyll fluorescence and yield of wild barley (Hordeum spontaneum). J. Bot. 2015, 2015, 120868. [Google Scholar] [CrossRef]
- Kilian, B.; Ozkan, H.; von Kohl, J.; Haeseler, A.; Barale, F.; Deusch, O.; Salamini, F. Haplotype structure at seven barley genes: Relevance to gene pool bottlenecks, phylogeny of ear type and site of barley domestication. Mol. Genet. Genom. 2006, 276, 230–241. [Google Scholar] [CrossRef]
- Zhang, H.; Irving, L.J.; McGill, C.; Matthew, C.; Zhou, D.; Kemp, P. The effects of salinity and osmotic stress on barley germination rate: Sodium as an osmotic regulator. Ann. Bot. 2010, 106, 1027–1035. [Google Scholar] [CrossRef]
- FAO. Food and Agriculture Organization of the United Nations. In Global Network on Integrated Soil Management for Sustainable Use of Salt-Affected Soils; FAO: Rome, Italy, 2000. [Google Scholar]
- Toppa, E.V.B.; Brambilla, W.P. O melhoramento de plantas e a salinidade dos solos. Rev. Verde Agroecol. Desenvolv. Sustent. 2011, 6, 21–25. [Google Scholar]
- Fatemi, F.; Hashemi-Petroudi, S.H.; Nematzadeh, G.; Askari, H.; Abdollahi, M.R. Exploiting differential gene expression to discover ionic and osmotic-associated transcripts in the halophyte grass Aeluropus littoralis. Biol. Proced. Online 2019, 21, 14. [Google Scholar] [CrossRef]
- Gheyi, H.R. Problemas de salinidade na agricultura irrigada. In Agricultura, Sustentabilidade e o Semiárido; Oliveira, J.R.C., Assis-Junior, T.S., Romero, R.N., Silva, R.E., Eds.; Sociedade Brasileira de Ciências do Solo: Fortaleza, Brazil, 2000; pp. 329–346. [Google Scholar]
- Willadino, L.; Camara, T.R. Tolerância das plantas à salinidade: Aspectos fisiológicos e bioquímicos. Encicl. Biosfera. 2010, 6, 11. [Google Scholar]
- James, R.; Munns, R.; Caemmerer, S.; Trejo, C.; Miller, C.; Condon, T.A. Photosynthetic capacity is related to the cellular and sub-cellular partitioning of Na+, K+ and Cl− in salt-affected barley and durum wheat. Plant Cell Environ. 2006, 29, 2185–2197. [Google Scholar] [CrossRef]
- Loomis, R.S.; Coonor, D.J. Crop Ecology: Productivity and Management in Agricultural Systems; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Richardson, S.G.; McCree, K.J. Carbon balance and water relations of sorghum exposed to salt and water stress. Plant Physiol. 1985, 79, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Long, N.; Dolstra, V.O.; Malosetti, M.; Kilian, B.; Graner, A.; Visser, R.G.F.; Linden, D. Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2013, 126, 2335–2351. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.L.; Ribot, S.A.; Dolstra, O.; Niks, R.E.; Visser, R.E.; Linden, C. Identification of quantitative trait loci for ion homeostasis and salt tolerance in barley (Hordeum vulgare L.). Mol. Breed. 2013, 31, 137–152. [Google Scholar] [CrossRef]
- El Sabagh, A.; Hossain, A.; Islam, M.S.; Barutcular, C.; Hussain, S.; Hasanuzzaman, M.; Akram, T.; Mubben, M.; Nasim, W.; Fahad, S.; et al. Drought and salinity stresses in barley: Consequences and mitigation strategies. Austral. J. Crop Sci. 2019, 13, 810. [Google Scholar] [CrossRef]
- Ahmadi, J.; Pour-Aboughadareh, A.; Fabriki-Ourang, S.; Mehrabi, A.A.; Siddique, K.H.M. Screening wild progenitors of wheat for salinity stress at early stages of plant growth: Insight into potential sources of variability for salinity adaptation in wheat. Crop Pasture Sci. 2018, 69, 649–658. [Google Scholar] [CrossRef]
- Ahmed, S. Effect of Soil Salinity on the Yield and Yield Components of Mungbean. Pak. J. Bot. 2009, 41, 263–268. [Google Scholar]
- Abd El-Monem, A.A.; El-Habbasha, S.F.; Hozayn, M. Mitigation salinity stress effects on barley (Hordeum vulgare L.) growth, yield and some physiological aspects by Hemin. J. Appl. Sci. Res. 2013, 9, 2411–2421. [Google Scholar]
- Pour-Aboughadareh, A.; Mehrvar, M.R.; Sanjani, S.; Amini, A.; Nikkhah-Chamanabad, H.; Asadi, A. Effects of salinity stress on seedling biomass, physiochemical properties, and grain yield in different breeding wheat genotypes. Acta Physiol. Plant. 2021, 43, 98–122. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Sanjani, S.; Nikkhah-Chamanabad, H.; Mehrvar, M.R.; Asadi, A.; Amini, A. Identification of salt tolerant barley genotypes using multiple-traits index and yield performance at the early growth and maturity stages. Bull. Natl. Res. Cent. 2021, 45, 117. [Google Scholar] [CrossRef]
- Ma, X.; Li, C.; Wang, A.; Duan, R.; Jiao, G.; Nevo, E.; Chen, G. Genetic diversity of wild barley (Hordeum vulgare ssp. spontaneum) and its utilization for barley improvement. Sci. Cold Arid Reg. 2012, 4, 0453–0461. [Google Scholar]
- Nevo, E.; Chen, G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 2010, 33, 670–685. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, F.; Arzani, A.; Rahimmalek, M.; Sun, D.; Peng, J. Salinity tolerance of wild barley Hordeum vulgare ssp. spontaneum. Plant Breed. 2020, 139, 304–316. [Google Scholar] [CrossRef]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M. Some important physiological selection criteria for salt tolerance in plants. Flora Morphol. Distrib. Funct. Ecol. Plants 2004, 199, 361–376. [Google Scholar] [CrossRef]
- Ahmadi, J.; Pour-Aboughadareh, A.; Fabriki Ourang, S.; Poczai, K.P.P. Unraveling salinity stress responses in ancestral and neglected wheat species at early growth stage: A baseline for utilization in future wheat improvement programs. Physiol. Mol. Biol. Plants 2020, 26, 537–549. [Google Scholar] [CrossRef]
- Ahmadi, A.; Emam, Y.; Pessarakli, M. Response of various cultivars of wheat and maize to salinity stress. J. Food Agric. Environ. 2009, 7, 123–128. [Google Scholar]
- Fricke, W.; Peters, W.S. The biophysics of leaf growth in salt stressed barley: A study at the cell level. Plant Physiol. 2002, 129, 374–388. [Google Scholar] [CrossRef]
- Pirasteh-Anosheh, H.; Ranjbar, G.; Pakniyat, H.; Emam, Y. Physiological mechanisms of salt stress tolerance in plants: An overview. In Plant Environment Interaction: Responses and Approaches to Mitigate Stress; Mahgoub-Azooz, M., Ahmad, P., Eds.; John Wiley & Sons, Ltd.: Jammu and Kashmir, India, 2016; Chapter 8; pp. 141–160. [Google Scholar]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Zeeshan, M.; Lu, M.; Sehar, S.; Holford, P.; Wu, F. Comparison of Biochemical, Anatomical, Morphological, and Physiological Responses to Salinity Stress in Wheat and Barley Genotypes Deferring in Salinity Tolerance. Agronomy 2020, 10, 127. [Google Scholar] [CrossRef]
- Thabet, S.G.; Moursi, Y.S.; Sallam, A.; Karam, M.A.; Alqudah, A.M. Genetic associations uncover candidate SNP markers and genes associated with salt tolerance during seedling developmental phase in barley. Environ. Exp. Bot. 2021, 188, 104499. [Google Scholar] [CrossRef]
- Mwando, E.; Angessa, T.T.; Han, Y.; Zhou, G.; Li, C. Quantitative trait loci mapping for vigour and survival traits of barley seedlings after germinating under salinity stress. Agronomy 2021, 11, 103. [Google Scholar] [CrossRef]
- Mwando, E.; Han, Y.; Angessa, T.T.; Zhou, G.; Hill, C.B.; Zhang, X.Q.; Li, C. Genome-Wide Association Study of Salinity Tolerance During Germination in Barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Maddur, A.M. The inheritance of salt tolerance in barley (Hordeum vulgare L.). Dissert. Abstr. Int. B 1977, 37, 5911 B. [Google Scholar]
- Mano, Y.; Nakazumi, H.; Takeda, K. Varietal variation in and effects of some major genes on salt tolerance at the germination stage in barley. Jpn. J. Breed. 1996, 46, 227–233. [Google Scholar] [CrossRef]
- Mano, Y.; Takeda, K. Genetical studies on salt tolerance at germination in recombinant inbred, isogenic, and doubled haploid lines of barley (Hordeum vulgare L.). Bull Res. Inst. Bioresour. Okayama Univ. 1996, 4, 79–88. [Google Scholar]
- Ullrich, S.E.; Hayes, P.M.; Dyer, W.E.; Blake, T.K.; Clancy, J.A. Quantitative trait locus analysis of seed dormancy in Steptoe barley. In Pre-Harvest in Cereals Walker-Simmons; Ried, J.L., Ed.; USDA-ARS Washington State University Pullman: Washington, DC, USA, 1992; pp. 136–145. [Google Scholar]
- Karahara, I.; Horie, T. Functions and structure of roots and their contributions to salinity tolerance in plants. Breed. Sci. 2021, 71, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Barati, A.; Moghaddam, M.; Mohammadi, S.A.; Ghazvini, H.A.; Sadeghzadeh, B. Identification of QTLs associated with agronomic and physiological traits under salinity stress in barley. J. Agric. Sci. Technol. 2017, 19, 185–200. [Google Scholar]
- Fan, Y.; Zhou, G.; Shabala, S.; Chen, Z.; Cai, S.; Li, C.; Zhou, M. Genome-wide association study reveals a new QTL for salinity tolerance in barley (Hordeum vulgare L.). Front. Plant Sci. 2016, 7, 946. [Google Scholar] [CrossRef]
- Angessa, T.T.; Zhang, X.Q.; Zhou, G.; Broughton, S.; Zhang, W.; Li, C. Early growth stages salinity stress tolerance in CM72 x Gairdner doubled haploid barley population. PLoS ONE 2017, 12, e0179715. [Google Scholar] [CrossRef]
- Sayed, M.A.; Nassar, S.M.; Moustafa, E.S.; Said, M.T.; Börner, A.; Hamada, A. Genetic Mapping Reveals Novel Exotic and Elite QTL Alleles for Salinity Tolerance in Barley. Agronomy 2021, 11, 1774. [Google Scholar] [CrossRef]
- Zhou, G.; Zhang, Q.; Tan, C.; Zhang, X.Q.; Li, C. Development of genome-wide InDel markers and their integration with SSR, DArT and SNP markers in single barley map. BMC Genom. 2015, 16, 804. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.T.; Yan, J.; Jiang, Y.; Zhan, Z.J.; Zhao, G.; Tondelli, A.; Cattivelli, L.; Cheng, J.P. Genetic dissection of winter barley seedling response to salt and osmotic stress. Mol. Breed. 2019, 39, 139. [Google Scholar] [CrossRef]
- Gill, M.B.; Zeng, F.; Shabala, L.; Zhang, G.P.; Yu, M.; Demidchik, V.; Shabala, S.; Zhou, M.X. Identification of QTL related to ROS formation under hypoxia and their association with waterlogging and salt tolerance in barley. Int. J. Mol. Sci. 2019, 20, 699. [Google Scholar] [CrossRef] [PubMed]
- Capo-chichi, L.J.A.; Eldridge, S.; Elakhdar, A.; Kubo, T.; Brueggeman, R.; Anyia, A.O. QTL Mapping and Phenotypic Variation for Seedling Vigour Traits in Barley (Hordeum vulgare L.). Plants 2021, 10, 1149. [Google Scholar] [CrossRef]
- Sayed, M.A.; Tarawneh, R.; Youssef, H.M.; Pillen, K.; Börner, A. Detection and Verification of QTL for Salinity Tolerance at Germination and Seedling Stages Using Wild Barley Introgression Lines. Plants 2021, 10, 2246. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fan, Y.; Mak, M.; Babla, M.; Holford, P.; Wang, F.; Chen, G.; Scott, G.; Wang, G.; Shabala, S.; et al. QTLs for stomatal and photosynthetic traits related to salinity tolerance in barley. BMC Genom. 2017, 18, 9. [Google Scholar] [CrossRef]
- Saade, S.; Brien, C.; Pailles, Y.; Berger, B.; Shahid, M.; Russell, J.; Waugh, R.; Negrão, S.; Tester, M. Dissecting new genetic components of salinity tolerance in two-row spring barley at the vegetative and reproductive stages. PLoS ONE 2020, 15, e0236037. [Google Scholar] [CrossRef]
- Wang, J.B.; Sun, G.L.; Ren, X.F.; Li, C.D.; Liu, L.P.; Wang, Q.F.; Du, B.B.; Sun, D.F. QTL underlying some agronomic traits in barley detected by SNP markers. BMC Genet. 2016, 17, 103. [Google Scholar] [CrossRef]
- Xue, D.; Huang, Y.; Zhang, X.; Wei, K.; Westcott, S.; Li, C.; Chen, M.; Zhang, G.; Lance, R. Identification of QTLs associated with salinity tolerance at late growth stage in barley. Euphytica 2009, 169, 187–196. [Google Scholar] [CrossRef]
- Sbei, H.; Sato, K.; Shehzad, T.; Harrabi, M.; Okuno, K. Detection of QTLs for salt tolerance in Asian barley (Hordeum vulgare L.) by association analysis with SNP markers. Breed Sci. 2014, 64, 378–388. [Google Scholar] [CrossRef]
- Xue, R.; Wang, J.; Li, C.; Johnson, P.; Lu, C.; Zhou, M. A single locus is responsible for salinity tolerance in a Chinese landrace barley (Hordeum vulgare L.). PLoS ONE 2012, 7, e43079. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shabala, S.; Li, C.; Liu, C.; Zhang, W.; Zhou, M. Quantitative Trait Loci for Salinity Tolerance Identified under Drained and Waterlogged Conditions and Their Association with Flowering Time in Barley (Hordeum vulgare. L). PLoS ONE 2015, 10, e0134822. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, J.A. A influência de características fenológicas na avaliação da tolerância à seca em sorgo. In Circular Técnica 165; Embrapa Milho e Sorgo: Sete Lagoas, Brazil, 2011. [Google Scholar]
- Khalili, M.; Naghavi, M.R.; Pour-Aboughadareh, A.R.; Talebzadeh, S.J. Evaluating of drought stress tolerance based on selection indices in spring canola cultivars (Brassica napus L.). J. Agric. Sci. 2012, 4, 78–85. [Google Scholar] [CrossRef]
- Cambraia, J. Aspectos bioquímicos, celulares e fisiológicos dos estresses nutricionais em plantas. In Estresses Ambientais: Danos e Benefícios em Plantas; Nogueira, U.M.T., Araújo, R.J.M.C., Willadino, E.L., Cavalcante, L.G., Eds.; Imprensa Universitária UFRPE: Recife, Brazil, 2005; pp. 95–105. [Google Scholar]
- Troyjack, C.; Carvalho, I.R.; Pimentel, J.R.; Junior, G.T.; Szareski, V.J.; Dubal, Í.T.P.; Conte, G.G. Envronmental stresses and its implications on breeding of brewing barley. Agron. Sci. Biotechnol. 2021, 7, 1–18. [Google Scholar] [CrossRef]
- Lopes, N.F.; Lima, M.G.S. Fisiologia da Produção; Viçosa, M.G., Ed.; Editora UFV: Vicosa, Brazil, 2015. [Google Scholar]
- Oukarroum, A.; Madidi, S.; El Schansker, G.; Strasser, R.J. Probing the responses of barley cultivars (Hordeum vulgare L.) by chlorophyll a fluorescence OLKJIP under drought stress and re-watering. Environ. Exp. Bot. 2007, 60, 438–446. [Google Scholar] [CrossRef]
- Cantero, C.; Villar, J.M.; Fereres, E. Relaciones hídricas de genótipos de cereal de invierno en condiciones de secano semiárido em la Segarra. Riegos Dren. 1989, 29, 32–39. [Google Scholar]
- Mahmood, Y.; Hassan, H.N.; Mohammed, M.S. Yield Performance of Barley Hybrids (Hordeum vulgare L.) under Drought stress and non-stressed Conditions. Passer J. 2021, 3, 107–113. [Google Scholar] [CrossRef]
- Cai, K.; Chen, X.; Han, Z.; Wu, X.; Zhang, S.; Li, Q.; Nazir, M.M.; Zhang, G.; Zeng, F. Screening of worldwide barley collection for drought tolerance: The assessment of various physiological measures as the selection criteria. Front. Plant Sci. 2020, 11, 1159. [Google Scholar] [CrossRef]
- Naz, A.A.; Ehl, A.; Pillen, K.; Leon, J. Validation for root-related quantitative trait locus effects of wild origin in the cultivated background of barley (Hordeum vulgare L.). Plant Breed. 2012, 131, 392–398. [Google Scholar]
- Gahoonia, T.S.; Nielsen, N.E. Barley genotypes with long root hairs sustain high grain yields in low-P field. Plant Soil. 2004, 262, 55–62. [Google Scholar] [CrossRef]
- Mehrabi, A.A.; Pour-Aboughadareh, A.; Mansouri, S.; Hosseini, A. Genome-wide association analysis of root system architecture features and agronomic traits in durum wheat. Mol. Breed. 2020, 40, 55. [Google Scholar] [CrossRef]
- Siddiqui, N.; Léon, J.; Naz, A.A.; Ballvora, A. Genetics and genomics of root system variation in adaptation to drought stress in cereal crops. J. Exp. Bot. 2021, 72, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Marok, M.A.; Marok-Alim, D.; Rey, P. Contribution of functional genomics to identify the genetic basis of water-deficit tolerance in barley and the related molecular mechanisms. J. Agron. Crop Sci. 2021, 207, 913–935. [Google Scholar] [CrossRef]
- Fan, Y.; Shabala, S.; Ma, Y.; Xu, R.; Zhou, M. Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genom. 2015, 16, 43. [Google Scholar] [CrossRef]
- Mora, F.; Quitral, Y.A.; Matus, I.; Russell, J.; Waugh, R.; Del Pozo, A. SNP-based QTL mapping of 15 complex traits in barley under rain-fed and well-watered conditions by a mixed modeling approach. Front. Plant Sci. 2016, 7, 909. [Google Scholar] [CrossRef]
- Wójcik-Jagła, M.; Fiust, A.; Ko’scielniak, J.; Rapacz, M. Association mapping of drought tolerance-related traits in barley to complement a traditional biparental QTL mapping study. Theor. Appl. Genet. 2018, 131, 167–181. [Google Scholar] [CrossRef]
- Abed, A.; Belzile, F. Comparing single-SNP, multi-SNP, and haplotype-based approaches in association studies for major traits in barley. Plant Genome 2019, 12, 190036. [Google Scholar] [CrossRef]
- Tarawneh, R.A.; Alqudah, A.M.; Nagel, M.; Börner, A. Genome-wide association mapping reveals putative candidate genes for drought tolerance in barley. Environ. Exp. Bot. 2020, 180, 104237. [Google Scholar] [CrossRef]
- Dhanagond, S.; Liu, G.; Zhao, Y.; Chen, D.; Grieco, M.; Reif, J.; Kilian, B.; Graner, A.; Neumann, K. Non-invasive phenotyping reveals genomic regions involved in pre-anthesis drought tolerance and recovery in spring barley. Front. Plant Sci. 2019, 10, 1307. [Google Scholar] [CrossRef]
- Abdel-Ghani, A.H.; Sharma, R.; Wabila, C.; Dhanagond, S.; Owais, S.J.; Duwayri, M.A.; Al-Dalain, S.A.; Klukas, C.; Chen, D.; Lübberstedt, T. Genome-wide association mapping in a diverse spring barley collection reveals the presence of QTL hotspots and candidate genes for root and shoot architecture traits at seedling stage. BMC Plant Biol. 2019, 19, 216. [Google Scholar] [CrossRef]
- Pham, A.; Maurer, A.; Pillen, K.; Brien, C.; Dowling, K.; Berger, B.; Eglinton, J.K.; March, T.J. Genome-wide association of barley plant growth under drought stress using a nested association mapping population. BMC Plant Biol. 2019, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- Moualeu-Ngangue, D.; Dolch, C.; Schneider, M.; Leon, J.; Uptmoor, R.; Stutzel, H. Physiological and morphological responses of different spring barley genotypes to water deficit and associated QTLs. PLoS ONE 2020, 15, e0237834. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Malik, R.; Tikle, A.N.; Neeraj Budhlakoti, R.P.S. Microsatellite markers based association mapping for yield contributing traits in exotic barley (Hordeum vulgare L.) accessions from ICARDA. Indian J. Genet. Plant Breed. 2021, 81, 1–5. [Google Scholar]
- Zhang, M.; Fu, M.M.; Qiu, C.W.; Cao, F.; Chen, Z.H.; Zhang, G.; Wu, F. Response of Tibetan wild barley genotypes to drought stress and identification of quantitative trait loci by genome-wide association analysis. Int. J. Mol. Sci. 2019, 20, 791. [Google Scholar] [CrossRef] [PubMed]
- Moursi, Y.S.; Thabet, S.G.; Amro, A.; Dawood, M.F.A.; Baenziger, P.S.; Sallam, A. Detailed genetic analysis for identifying QTLs associated with drought tolerance at seed germination and seedling stages in barley. Plants 2020, 9, 1425. [Google Scholar] [CrossRef]
- Gudys, K.; Guzy-Wrobelska, J.; Janiak, A.; Dziurka, M.A.; Ostrowska, A.; Hura, K.; Jurczyk, B.; Żmuda, K.; Grzybkowska, D.; Śróbka, J.; et al. Prioritization of candidate genes in QTL regions for physiological and biochemical traits underlying drought response in barley (Hordeum vulgare L.). Front. Plant Sci. 2018, 9, 769. [Google Scholar] [CrossRef]
- Arifuzzaman, M.; Sayed, M.A.; Muzammil, S.; Pillen, K.; Schumann, H.; Naz, A.A.; Leon, J. Detection and validation of novel QTL for shoot and root traits in barley (Hordeum vulgare L.). Mol. Breed. 2014, 34, 1373–1387. [Google Scholar] [CrossRef]
- Szücs, P.; Blake, V.C.; Bhat, P.R.; Chao, S.; Close, T.J.; Cuesta-Marcos, A.; Muehlbauer, G.J.; Ramsay, L.; Waugh, R.; Hayes, P.M. An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome 2009, 2, 134–140. [Google Scholar] [CrossRef]
- Berger, G.L.; Liu, S.; Hall, M.D.; Brooks, W.S.; Chao, S.; Muehlbauer, G.J.; Baik, B.K.; Steffenson, B.; Griffey, C.A. Marker-trait associations in Virginia Tech winter barley identified using genome-wide mapping. Theor. Appl. Genet. 2012, 126, 693–710. [Google Scholar] [CrossRef]
- Gawenda, I.; Thorwarth, P.; Günther, T.; Ordon, F.; Schmid, K.J. Genome-wide association studies in elite varieties of German winter barley using single-marker and haplotype-based methods. Plant Breed. 2015, 134, 28–39. [Google Scholar] [CrossRef]
- Honsdorf, N.; March, T.J.; Pillen, K. QTL controlling grain filling under terminal drought stress in a set of wild barley introgression lines. PLoS ONE 2017, 12, e0185983. [Google Scholar] [CrossRef] [PubMed]
- Inostroza, L.; Del Pozo, A.; Matus, I.; Castillo, D.; Hayes, P.; Machado, S.; Corey, A. Association mapping of plant height, yield, and yield stability in recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. Spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Mol. Breed. 2009, 23, 365–376. [Google Scholar]
- Elakhdar, A.; Kumamaru, T.; Qualset, C.O.; Brueggeman, R.S.; Amer, K.; Capo-chichi, L. Assessment of genetic diversity in Egyptian barley (Hordeum vulgare L.) genotypes using SSR and SNP markers. Genet. Resour. Crop. Evol. 2018, 65, 1937–1951. [Google Scholar] [CrossRef]
- Diab, A.A.; Teulat-Merah, B.; This, D.; Ozturk, N.Z.; Benscher, D.; Sorrells, M.E. Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor. Appl. Genet. 2004, 109, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Peighambari, S.A.; Samadi, B.Y.; Nabipour, A.W.C.I.T.; Charmet, G.; Sarrafi, A. QTL analysis for agronomic traits in a barley doubled haploids population grown in Iran. Plant Sci. 2005, 169, 1008–1013. [Google Scholar] [CrossRef]
- Cattivelli, L.; Ceccarelli, S.; Romagosa, I.; Stanca, M. Abiotic stresses in Barley: Problems and solutions. In Barley: Production, Improvement, and Uses; Steven, E., Ed.; Blackwell Publishing Ltd.: Oxford, UK, 2010; pp. 282–306. [Google Scholar]
- Andleeb, T.; Shah, T.; Nawaz, R.; Munir, I.; Munsif, F.; Jalal, A. QTL mapping for drought stress tolerance in plants. In Salt and Drought Stress Tolerance in Plants; Springer: Cham, Switzerland, 2020; pp. 383–403. [Google Scholar]
- Gous, P.W.; Hickey, L.; Christopher, J.T.; Franckowiak, J.; Fox, G.P. Discovery of QTL for stay-green and heat-stress in barley (Hordeum vulgare) grown under simulated abiotic stress conditions. Euphytica 2016, 207, 305–317. [Google Scholar] [CrossRef]
- Zhou, H.; Luo, W.; Gao, S.; Ma, J.; Zhou, M.; Wei, Y.; Zheng, Y.; Liu, Y.; Liu, C. Identification of loci and candidate genes controlling kernel weight in barley based on a population for which whole genome assemblies are available for both parents. Crop J. 2020, 9, 854–861. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Palczak, J.; Wojciechowski, T.; Lynch, J.P.; Naz, A.A.; Léon, J.; Ballvora, A. Genetic components of root architecture and anatomy adjustments to water-deficit stress in spring barley. Plant Cell Environ. 2020, 43, 692–711. [Google Scholar] [CrossRef]
- Wehner, G.; Balko, C.; Humbeck, K.; Zyprian, E.; Ordon, F. Expression profiling of genes involved in drought stress and leaf senescence in juvenile barley. BMC Plant Biol. 2016, 16, 3. [Google Scholar] [CrossRef]
- Craufurd, P.Q.; Flower, D.J.; Peacock, J.M. Effect of heat and drought stress on sorghum (Sorghum bicolor). I. Panicle development and leaf appearance. Exp. Agric. 1993, 29, 61–76. [Google Scholar] [CrossRef]
- Prasad, P.V.V.; Pisipati, S.R.; Momčilović, I.; Ristic, Z. Independent and Combined Effects of High Temperature and Drought Stress During Grain Filling on Plant Yield and Chloroplast EF-Tu Expression in Spring Wheat. J. Agron. Crop Sci. 2011, 197, 430–441. [Google Scholar] [CrossRef]
- Savin, R.; Nicolas, M.E. Effects of short periods of drought and high temperature on grain growth and starch accumulation of two malting barley cultivars. Funct. Plant Biol. 1996, 23, 201. [Google Scholar] [CrossRef]
- Dellagostin, S.M.; Martinazzo, E.G.; Pimentel-Junior, R.; Troyjack, C.; Pedó, T. Temperaturas extremas e qualidade fisiológica de sementes de plantas de lavoura. In Estresses Ambientais e a Produção de Sementes: Ciência e Aplicação; Aumonde, F.A., Pedó, T.Z., Martinazzo, T., Villela, E.G., Pelotas, R.S., Eds.; Cópias Santa Cruz: Pelotas, Brazil, 2017; pp. 171–198. [Google Scholar]
- Essemine, J.; Ammar, S.; Bouzid, S. Impact of heat stress on germination and growth in higher plants: Physiological, biochemical and molecular repercussions and mechanisms of defence. J. Biol. Sci. 2010, 10, 565–572. [Google Scholar] [CrossRef]
- Brestic, M.; Zivcak, M. PSII fluorescence techniques for measurement of drought and high temperature stress signal in crop plants: Protocols and applications. In Molecular Stress Physiology of Plants; Springer: Berlin, Germany, 2013; pp. 87–131. [Google Scholar]
- Wang, X.; Dinler, B.S.; Vignjevic, M.; Jacobsen, S.; Wollenweber, B. Physiological and proteome studies of responses to heat stress during grain filling in contrasting wheat cultivars. Plant Sci. 2015, 230, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Harris, P.J. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Oukarroum, A.; El Madidi, S.; Strasser, R.J. Differential heat sensitivity index in barley cultivars (Hordeum vulgare L.) monitored by chlorophyll a fluorescence OKJIP. Plant Physiol. Biochem. 2016, 105, 102–108. [Google Scholar] [CrossRef]
- Gupta, N.K.; Khan, A.; Maheshwari, A.; Narayan, S.; Chhapola, O.P.; Arora, A.; Singh, G. Effect of post anthesis high temperature stress on growth, physiology and antioxidative defense mechanisms in contrasting wheat genotypes. Indian J. Plant Physiol. 2015, 20, 103–110. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Bahrami, F.; Arzani, A.; Rahimmalek, M. Photosynthetic and yield performance of wild barley (Hordeum vulgare ssp. spontaneum) under terminal heat stress. Photosynthetica 2019, 57, 9–17. [Google Scholar] [CrossRef]
- Guerra, D.; Morcia, C.; Badeck, F.; Rizza, F.; Delbono, S.; Francia, E.; Milc, J.A.; Monostori, I.; Galiba, G.; Cattivelli, L.; et al. Extensive allele mining discovers novel genetic diversity in the loci controlling frost tolerance in barley. Theor. Appl. Genet. 2022, 135, 553–569. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Hernandez, J.; Herb, D.; Baenziger, P.S.; Bochard, A.M.; Capettini, F.; Casas, A.; Cuesta-Marcos, A.; Einfeldt, C.; Fisk, S.; et al. Perspectives on low temperature tolerance and vernalization sensitivity in barley: Prospects for facultative growth habit. Front. Plant Sci. 2020, 11, 585927. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.F.A.; Moursi, Y.S.; Amro, A.; Baenziger, S.; Sallam, A. Investigation of Heat-Induced Changes in the Grain Yield and Grains Metabolites, with Molecular Insights on the Candidate Genes in Barley. Agronomy 2020, 10, 1730. [Google Scholar] [CrossRef]
- Tondelli, A.; Pagani, D.; Ghafoori, I.N.; Rahimi, M.; Ataei, R.; Rizza, F.; Flavell, A.J.; Cattivelli, L. Allelic variation at Fr-H1/Vrn-H1 and Fr-H2 loci is the main determinant of frost tolerance in spring barley. Environ. Exp. Bot. 2014, 106, 148–155. [Google Scholar] [CrossRef]
- Tóth, B.; Francia, E.; Rizza, F.; Stanca, A.M.; Galiba, G.; Pecchioni, N. Development of PCR-based markers on chromosome 5H for assisted selection of frost-tolerant genotypes in barley. Mol. Breed. 2004, 14, 265–273. [Google Scholar] [CrossRef]
- Akar, T.; Francia, E.; Tondelli, A.; Rizza, F.; Stanca, A.M.; Pecchioni, N. Marker-assisted characterization of frost tolerance in barley (Hordeum vulgare L.). Plant Breed. 2009, 128, 381–386. [Google Scholar] [CrossRef]
- Ahres, M.; Gierczik, K.; Boldizsár, Á.; Vítámvás, P.; Galiba, G. Temperature and light-quality-dependent regulation of freezing tolerance in barley. Plants 2020, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Templer, S.E.; Ammon, A.; Pscheidt, D.; Ciobotea, O.; Schuy, C.; McCollum, C.; Sonnewald, U.; Hanemann, A.; Förster, J.; Ordon, F.; et al. Metabolite profiling of barley flag leaves under drought and combined heat and drought stress reveals metabolic QTLs for metabolites associated with antioxidant defense. J. Exp. Bot. 2017, 68, 1697–1713. [Google Scholar] [CrossRef]
- Fisk, S.P.; Cuesta-Marcos, A.; Cistue, L.; Russell, J.; Smith, K.P.; Baenziger, S.; Bedo, Z.; Corey, A.; Filichkin, T.; Karsai, I.; et al. FR-H3: A new QTL to assist in the development of fall-sown barley with superior low temperature tolerance. Theor. Appl. Genet. 2013, 126, 335–347. [Google Scholar] [CrossRef]
- Hayes, P.M.; Chen, F.Q.; Corey, A.; Pan, A.; Chen, T.H.H.; Baird, E.; Powell, W.; Thomas, W.; Waugh, R.; Bedo, Z.; et al. The Dicktoo × Morex population: A model for dissecting components of winter hardiness in barley. In Plant Cold Hardiness; Li, P.H., Chen, T.H.H., Eds.; Springer: Boston, MA, USA, 1997; pp. 77–87. [Google Scholar]
- Francia, E.; Rizza, F.; Cattivelli, L.; Stanca, A.M.; Galiba, G.; Toth, B.; Pecchioni, N. Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ‘Tremois’ (spring) barley map. Theor. Appl. Genet. 2004, 108, 670–680. [Google Scholar] [CrossRef]
- Saade, S.; Negrão, S.; Plett, D.; Garnett, T.; Tester, M. Genomic and Genetic Studies of Abiotic Stress Tolerance in Barley. In The Barley Genome, Compendium of Plant Genomes; Springer International Publishing, Part of Springer Nature; Stein, A.G., Muehlbauer, G.J., Eds.; Springer: Cham, Switzerland, 2018; p. 259. [Google Scholar]
- Troyjack, C.; Szarescki, V.J.; Martinazzo, E.G.; Aumonde, T.Z.; Pedó, T. Ecofisiologia, produção e qualidade de sementes de plantas de lavoura em resposta ao alagamento do solo e a restrição hídrica. In Estresses Ambientais e a Produção de Sementes: Cência e Aplicação; Aumonde, F.A., Pedó, T.Z., Martinazzo., T., Villela, E.G., Pelotas, R.S., Eds.; Cópias Santa Cruz: Pelotas, Brazil, 2017; pp. 139–169. [Google Scholar]
- Wang, M.; Jiang, N.; Jia, T.; Leach, L.; Cockram, J.; Waugh, R.; Ramsay, L.; Thomas, B.; Luo, Z. Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor. Appl. Genet. 2012, 124, 233–246. [Google Scholar] [CrossRef]
- Sachs, M.; Vartapetian, B. Plant anaerobic stress I. Metabolic adaptation to oxygen deficiency. Plant Stress 2007, 1, 123–135. [Google Scholar]
- Amarante, L.; do Badinelli Colares, D.; Oliveira, M.L.A.; Zenzen, I.; Badinelli, P.G.; Bernardi, E. Teores de Clorofilas em Soja Associada Simbioticamente com Diferentes Estirpes de. Rev. Bras. Biociênc. 2007, 5 (Suppl. 2), 906–908. [Google Scholar]
- Yordanova, R.Y.; Popova, L.P. Photosynthetic response of barley plants to soil flooding. Photosynthetica 2001, 39, 515–520. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Chang, R. Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann. Bot. 2005, 96, 507–518. [Google Scholar] [CrossRef]
- Ahmadi, J.; Pour-Aboughadareh, A.; Fabriki Ourang, S.; Mehrabi, A.A.; Siddique, K.H.M. Wild relatives of wheat: Aegilops-Triticum accessions disclose different antioxidative and physiological response to water stress. Acta Physiol Plant. 2018, 40, 90. [Google Scholar] [CrossRef]
- Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 2014, 5, 685. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 5th ed.; Oxford University Press: New York, NY, USA, 2015; pp. 1–29. [Google Scholar]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid Redox Sign. 2009, 11, 861–905. [Google Scholar] [CrossRef]
- Hossain, M.A.; da Silva, J.A.T.; Fujita, M. Glyoxalase system and reactive oxygen species detoxification system in plant abiotic stress response and tolerance. In Abiotic Stress in Plants-Mechanisms and Adaptations; Shankar, A.K., Venkateswarlu, B., Eds.; INTECH-Open Access Publisher: Rijeka, Croatia, 2011; pp. 235–266. [Google Scholar]
- Tong, C.; Hill, C.B.; Zhou, G.; Zhang, X.-Q.; Jia, Y.; Li, C. Opportunities for Improving Waterlogging Tolerance in Cereal Crops-Physiological Traits and Genetic Mechanisms. Plants 2021, 10, 1560. [Google Scholar] [CrossRef]
- Jia, W.; Ma, M.; Chen, J.; Wu, S. Plant morphological, physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int. J. Mol. Sci. 2021, 22, 1088. [Google Scholar] [CrossRef]
- Romina, P.; Abeledo, L.G.; Miralles, D.J. Identifying the critical period for waterlogging on yield and its components in wheat and barley. Plant Soil. 2014, 378, 265–277. [Google Scholar]
- Wei, W.L.; Li, D.H.; Wang, L.H.; Wang, L.H.; Ding, X.; Zhang, Y.X.; Gao, Y.; Zhang, X.R. Morpho-anatomical and physiological responses to waterlogging of sesame (Sesamum indicum L.). Plant Sci. 2013, 208, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Shabala, S.; Koutoulis, A.; Shabala, L.; Johnson, P.; Hayes, D.; Nichols, D.S.; Zhou, M.X. Waterlogging tolerance in barley is associated with faster aerenchyma formation in adventitious roots. Plant Soil. 2015, 394, 355–372. [Google Scholar] [CrossRef]
- Luan, H.; Guo, B.; Pan, Y.; Lv, C.; Shen, H.; Xu, R. Morpho-anatomical and physiological responses to waterlogging stress in different barley (Hordeum vulgare L.) genotypes. Plant Growth Regul. 2018, 85, 399–409. [Google Scholar] [CrossRef]
- Liu, M.; Li, Y.; Ma, Y.; Zhao, Q.; Stiller, J.; Feng, Q.; Tian, Q.; Liu, D.; Han, B.; Liu, C. The draft genome of a wild barley genotype reveals its enrichment in genes related to biotic and abiotic stresses compared to cultivated barley. Plant Biotechnol. J. 2020, 18, 443–456. [Google Scholar] [CrossRef]
- Li, H.; Vaillancourt, R.; Mendham, N.; Zhou, M. Comparative mapping of quantitative trait loci associated with waterlogging tolerance in barley (Hordeum vulgare L.). BMC Genom. 2008, 9, 401. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, G.; Shabala, S.; Koutoulis, A.; Shabala, L.; Johnson, P.; Li, C.H.; Zhou, M. Identification of aerenchyma formation related QTL in barley that can be effective in breeding for waterlogging tolerance. Theor. Appl. Genet. 2016, 129, 1167–1177. [Google Scholar] [CrossRef]
- Broughton, S.; Zhou, G.; Teakle, N.; Matsuda, R.; Zhou, M.; O’Leary, R.; Colmer, T.; Li, C. Waterlogging tolerance is associated with root porosity in barley (Hordeum vulgare L.). Mol. Breed. 2015, 35, 27. [Google Scholar] [CrossRef]
- Huang, X.; Shabala, S.; Shabala, L.; Rengel, Z.; Wu, X.; Zhang, G.; Zhou, M. Linking waterlogging tolerance with Mn2+ toxicity: A case study for barley. Plant Biol. 2015, 17, 26–33. [Google Scholar] [CrossRef]
- Borrego-Benjumea, A.; Carter, A.; Zhu, M.; Tucker, J.R.; Zhou, M.; Badea, A. Genome-Wide Association Study of Waterlogging Tolerance in Barley (Hordeum vulgare L.) Under Controlled Field Conditions. Front. Plant Sci. 2021, 12, 711654. [Google Scholar] [CrossRef]
- Zhang, X.; Fan, Y.; Shabala, S.; Koutoulis, A.; Shabala, L.; Johnson, P.; Hu, H.; Zhou, M. A new major-effect QTL for waterlogging tolerance in wild barley (H. spontaneum). Theor. Appl. Genet. 2017, 130, 1559–1568. [Google Scholar] [CrossRef]
- Zhou, M. Accurate phenotyping reveals better QTL for waterlogging tolerance in barley. Plant Breed. 2011, 130, 203–208. [Google Scholar] [CrossRef]
- Xue, D.W.; Zhou, M.X.; Zhang, X.Q.; Chen, S.; Wei, K.; Zeng, F.R.; Mao, Y.; Wu, F.B.; Zhang, G.P. Identification of QTLs for yield and yield components of barley under different growth conditions. J. Zhejiang Univ. Sci. B. 2010, 11, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, P.H.; Rashid, Z.; Vinayan, M.T.; Almeida, G.D.; Phagna, R.K.; Babu, R. QTL mapping of agronomic waterlogging tolerance using recombinant inbred lines derived from tropical maize (Zea mays L) germplasm. PLoS ONE 2015, 10, e0124350. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Johnson, P.; Zhou, G.; Li, C.; Lance, R. Quantitative trait loci for waterlogging tolerance in a barley cross of Franklin × YuYaoXiangTian Erleng and the relationship between waterlogging and salinity tolerance. Crop Sci. 2012, 52, 2082–2088. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, B.; Yu, F.; Li, L.; Wang, M.; Xue, Y.; Zhang, Z.; Yan, J.; Yue, B.; Zheng, Y. Identification of major QTL for waterlogging tolerance using genome-wide association and linkage mapping of maize seedlings. Plant Mol. Biol. Rep. 2013, 31, 594. [Google Scholar] [CrossRef]
- Van Kleunen, M.; Fischer, M. Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytol. 2005, 166, 49–60. [Google Scholar] [CrossRef]
- Mano, Y.; Omori, F.; Muraki, M.; Takamizo, T. QTL mapping of adventitious root formation under flooding conditions in tropical maize (Zea mays L.) seedlings. Breed. Sci. 2005, 55, 343–347. [Google Scholar] [CrossRef]
- Gill, M.B.; Zeng, F.; Shabala, L.; Zhang, G.; Fan, Y.; Shabala, S.; Zhou, M. Cell-Based Phenotyping Reveals QTL for Membrane Potential Maintenance Associated with Hypoxia and Salinity Stress Tolerance in Barley. Front. Plant Sci. 2017, 8, 1941. [Google Scholar] [CrossRef]
- Smith, S.R. A critical review of the bioavailability and impacts of heavy metals in municipal solid waste composts compared to sewage sludge. Environ Int. 2009, 35, 142–156. [Google Scholar] [CrossRef]
- Kabata-Pendias, A. Trace Elements in Soils and Plants, 4th ed.; Taylor & Francis Group: London, UK, 2010; p. 548. [Google Scholar]
- Rahmani, A.; Asghari, A.; Jafari, H.; Sofalian, O. QTL mapping for physiological traits affecting lead tolerance in the Hordeum vulgare L. Environ. Stresses Crop Sci. 2021, 14, 849–860. [Google Scholar]
- Lwalaba, J.L.W.; Louis, L.T.; Zvobgo, G.; Richmond, M.E.A.; Fu, L.; Naz, S.; Mwamba, M.; Mundende, R.P.M.; Zhang, G. Physiological and molecular mechanisms of cobalt and copper interaction in causing phyto-toxicity to two barley genotypes differing in Co tolerance. Ecotoxicol. Environ. Saf. 2020, 187, 109866. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.A. Genetic potential for solving problems of soil mineral stress: Aluminum and manganese toxicities in cereal grains. In Plant Adaptation to Mineral Stress in Problem Soils; Wright, M.J., Ed.; Cornell University Press: Ithaca, NY, USA, 1976; pp. 5–64. [Google Scholar]
- Siedlecka, A.; BaszyńAski, T. Inhibition of electron flow around photosystem I in chloroplasts of Cd treated maize plants is due to Cd-induced iron deficiency. Physiol. Plant. 1993, 87, 199–202. [Google Scholar] [CrossRef]
- Vassilev, A.; Yordanov, I. Reductive analysis of factors limiting growth of cadmium-treated plants: A review. Bulg. J. Plant Physiol. 1997, 23, 114–133. [Google Scholar]
- Hall, J.L. Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 2002, 53, 1–11. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal tolerance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef]
- Ueno, D.; Koyama, E.; Yamaji, N.; Ma, J.F. Physiological, genetic, and molecular characterization of a high Cd-accumulating rice cultivar, Jarjan. J. Exp Bot. 2011, 62, 2265–2272. [Google Scholar] [CrossRef]
- Yuan, L.; Yang, S.; Liu, B.; Zhang, M.; Wu, K. Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 2012, 31, 67–79. [Google Scholar] [CrossRef]
- Derakhshani, B.; Jafary, H.; Zanjani, B.M.; Hasanpur, K.; Mishina, K.; Tanaka, T.; Kawahara, Y.; Oono, Y. Combined QTL mapping and RNA-Seq profiling reveals candidate genes associated with cadmium tolerance in barley. PLoS ONE 2020, 15, e0230820. [Google Scholar] [CrossRef]
- Wu, D.Z.; Sato, K.; Ma, J.F. Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol. 2015, 208, 817–829. [Google Scholar] [CrossRef]
- Huang, X.; Fan, Y.; Shabala, L.; Rengel, Z.; Shabala, S.; Zhou, M. A major QTL controlling the tolerance to manganese toxicity in barley (Hordeum vulgare L.). Mol. Breed. 2018, 38, 16. [Google Scholar] [CrossRef]
- Koochakpour, Z.; Solouki, M.; Fakheri, B.A.; Aghnoum, R.; Mahdi Nezhad, N. Genome wide association analysis of plant height, spike and awn length in Barley (Hordeum vulgare L.) Exposed to Mn Stress. J. Animal Plant Sci. 2020, 30, 384–390. [Google Scholar]
- Navakode, S.; Weidner, A.; Varshney, R.K.; Lohwasser, U.; Scholz, U.; Borner, A. A QTL analysis of aluminium tolerance in barley, using gene-based markers. Cereal Res. Commun. 2009, 37, 531–540. [Google Scholar] [CrossRef]
- Reuscher, S.; Kolter, A.; Hoffmann, A.; Pillen, K.; Krämer, U. Quantitative Trait Loci and Inter-Organ Partitioning for Essential Metal and Toxic Analogue Accumulation in Barley. PLoS ONE 2016, 11, e0153392. [Google Scholar] [CrossRef] [PubMed]
- Bian, M.; Jin, X.; Broughton, S.; Zhang, X.-Q.; Zhou, G.; Zhou, M.; Zhang, G.; Sun, D.; Li, C. A new allele of acid soil tolerance gene from a malting barley variety. BMC Genet. 2015, 16, 92. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).