Transcriptional Profiling of Hippocampus Identifies Network Alterations in Alzheimer’s Disease
Abstract
:1. Introduction
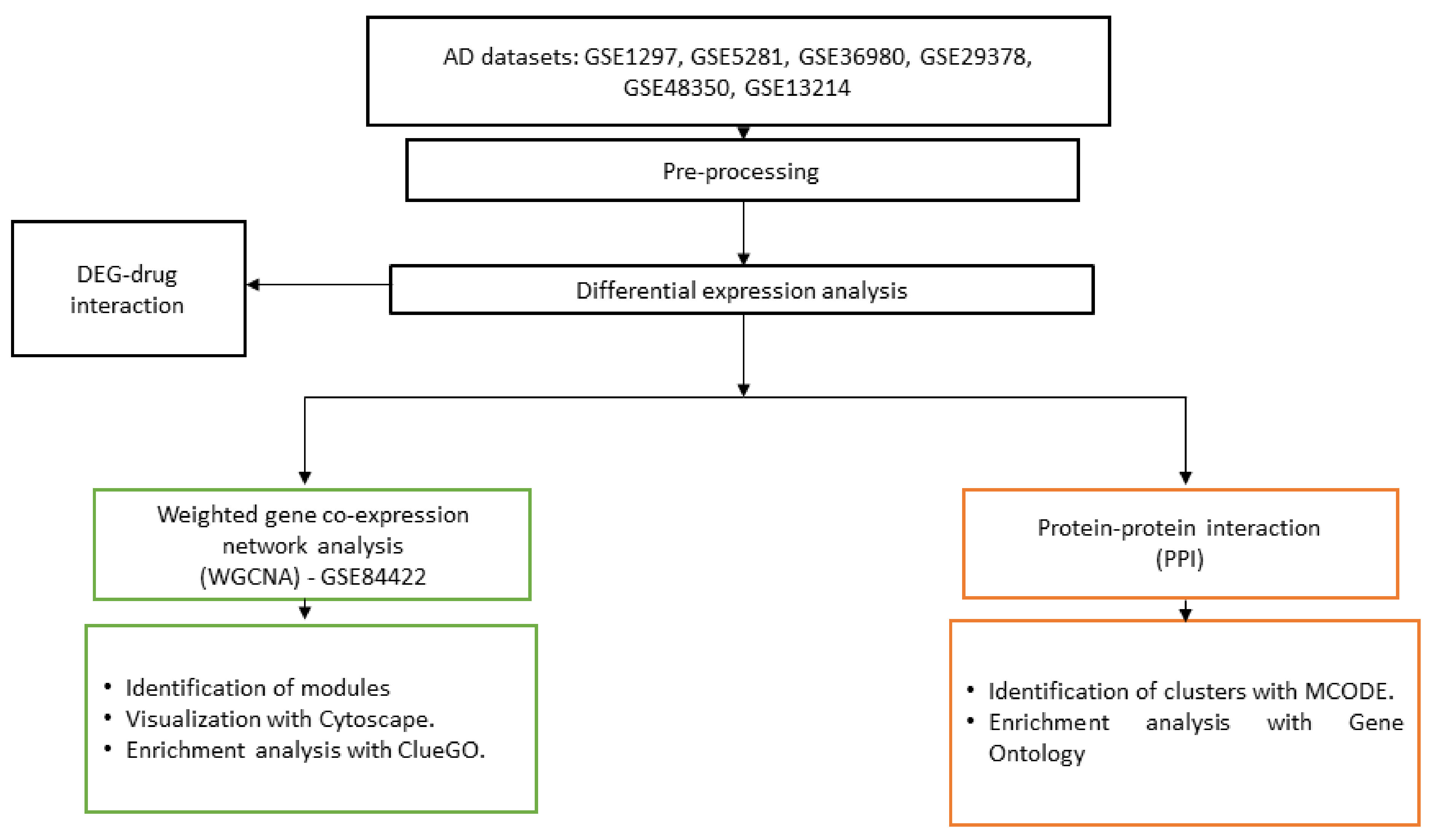
2. Materials and Methods
2.1. Datasets
2.2. Differential Expression Analysis
2.3. Differentially Expressed Gene–Drug Interactions
2.4. Co-Expression Network
2.5. Protein–Protein Interaction
2.6. Pathway and Gene Ontology Analysis
3. Results
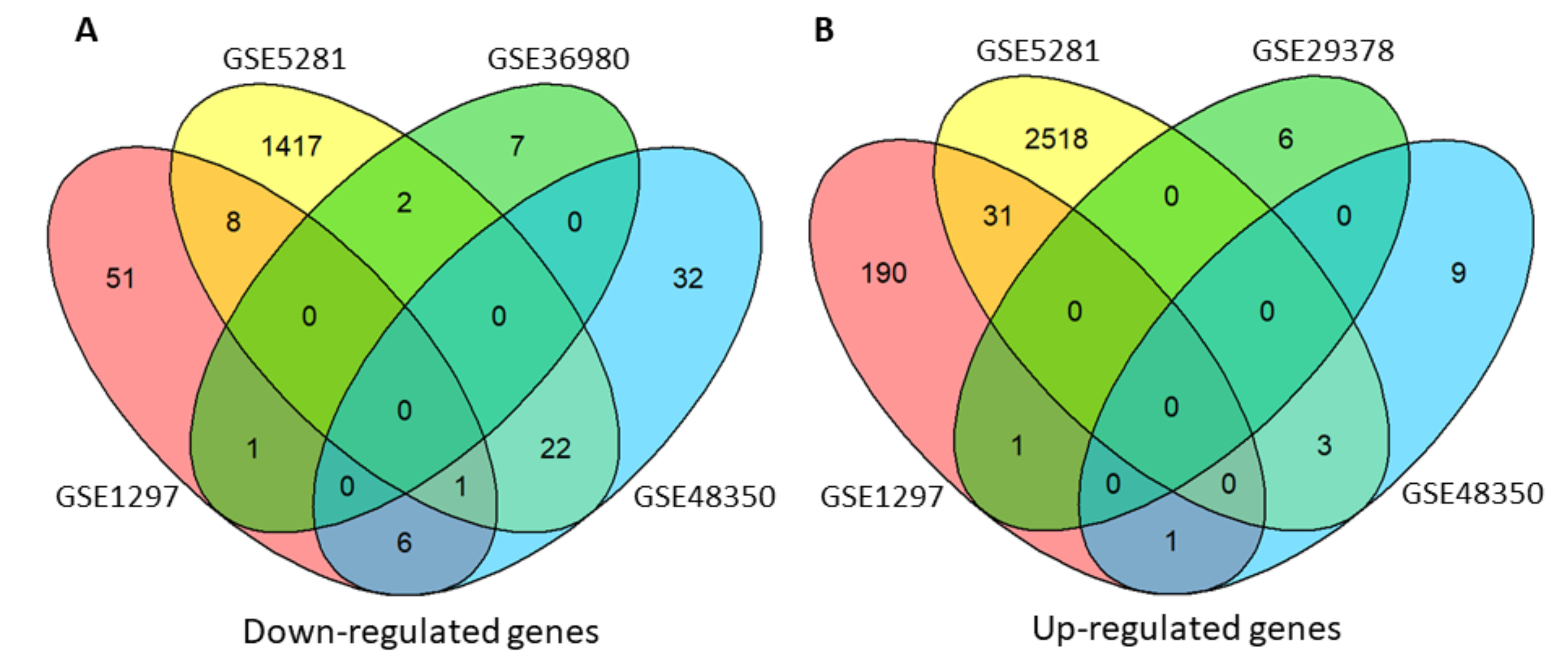
3.1. Differentially Expressed Genes
3.2. Drugs Interact with Differentially Expressed Genes in AD
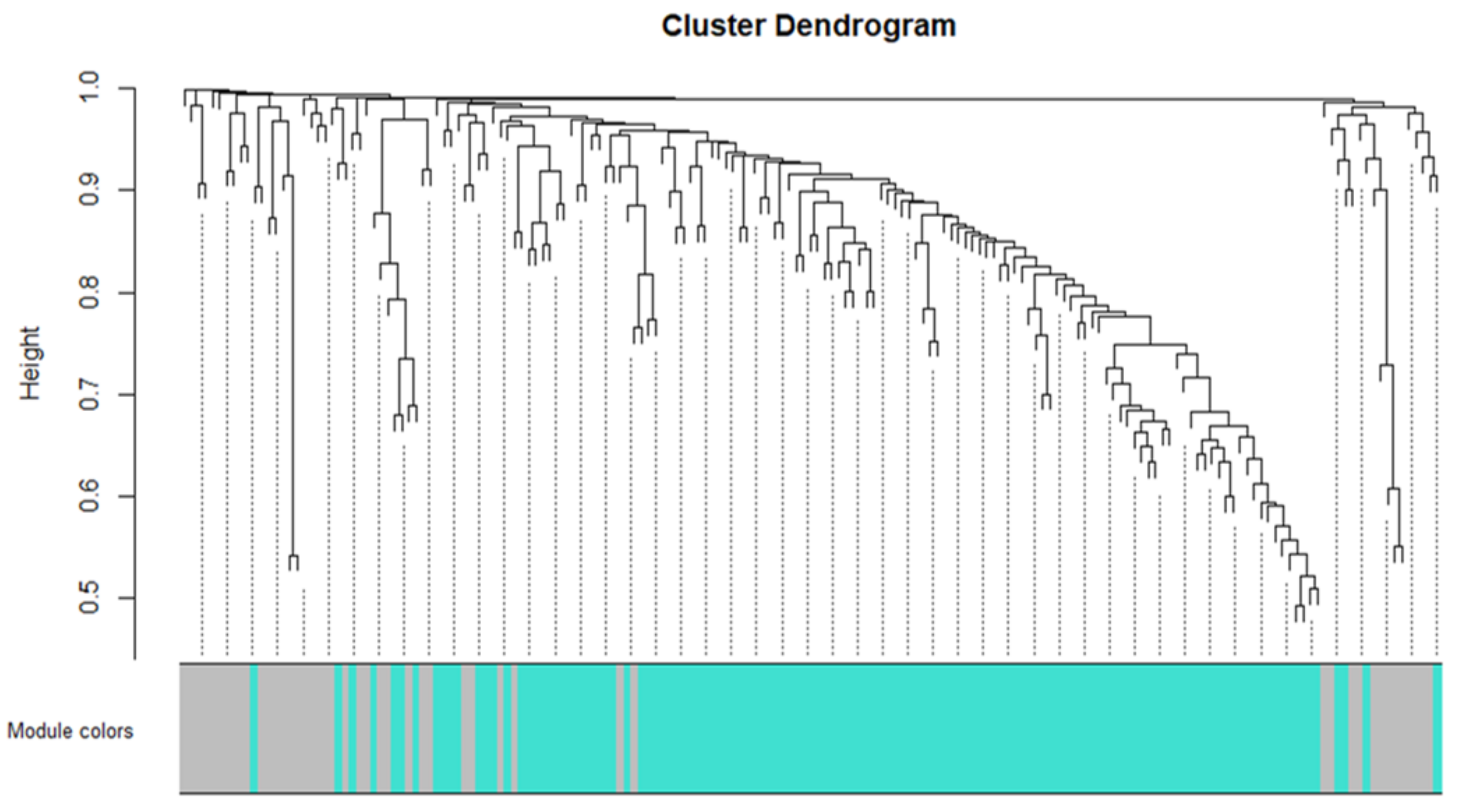
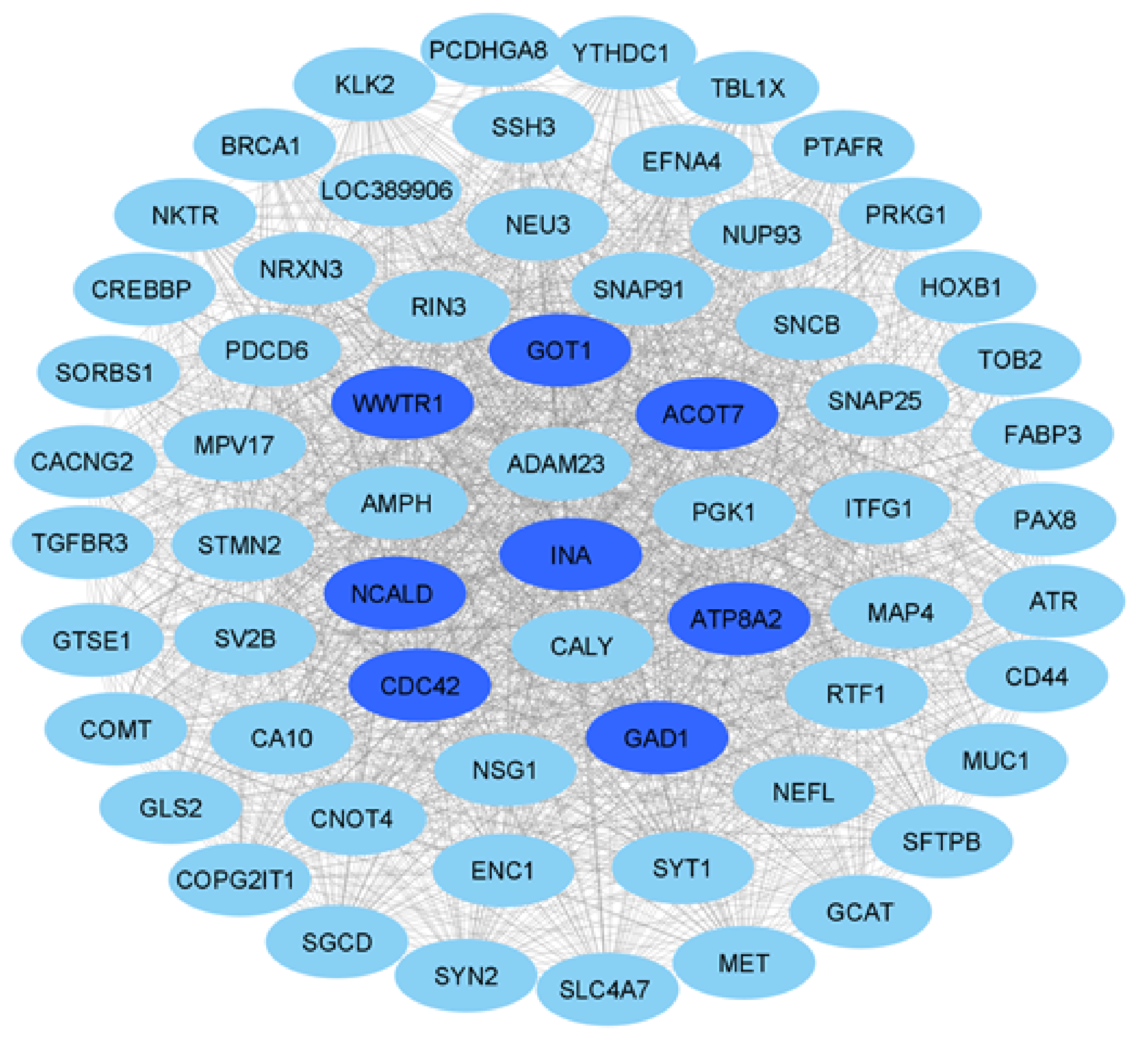
3.3. Network Construction and Module Detection
3.4. Enrichment Analysis
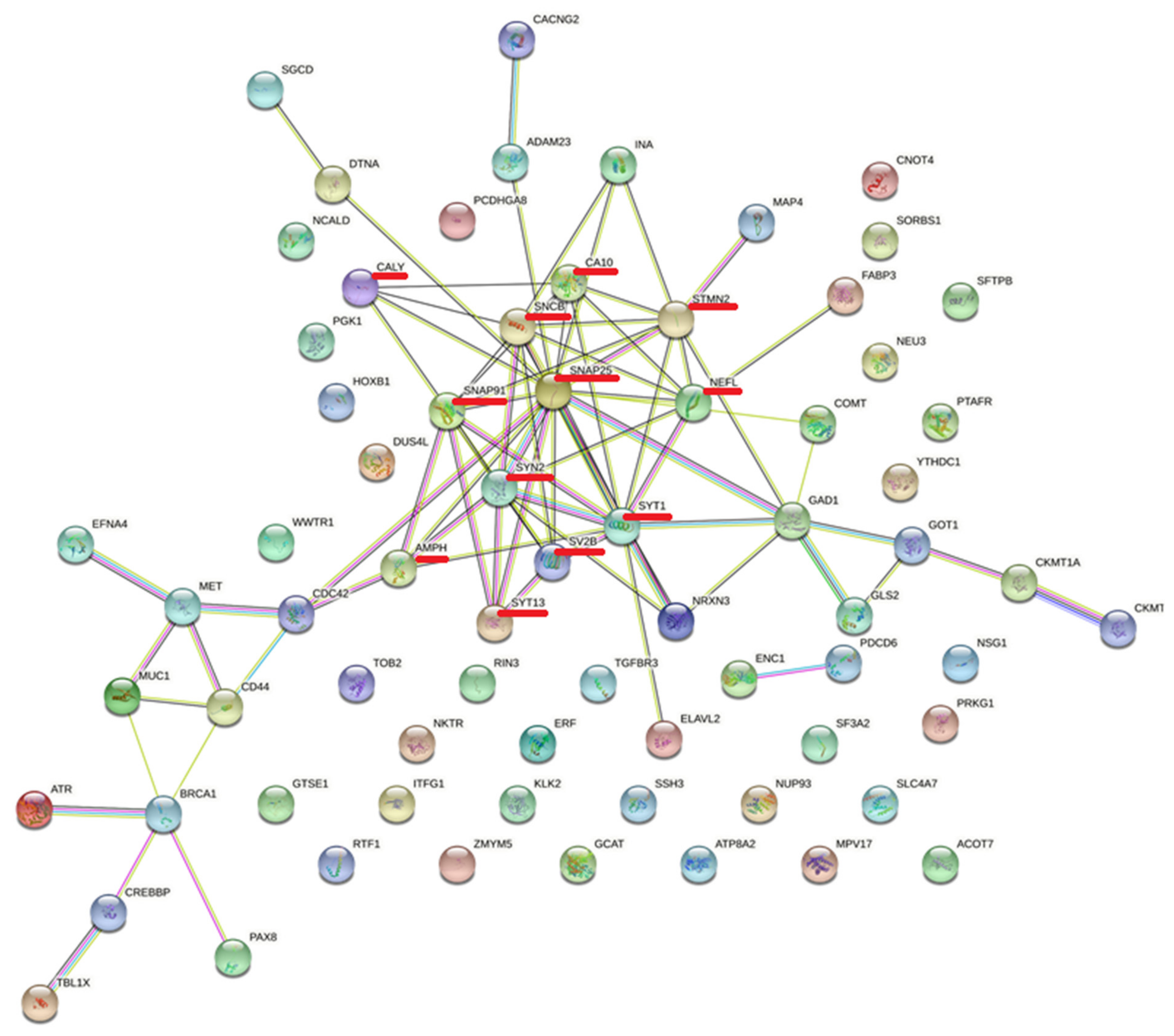
3.5. Protein–Protein Interactions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, G.; Liu, M.; Du, K.; Zhong, X.; Gong, S.; Jiao, L.; Wei, M. Differential Expression of mRNAs in the Brain Tissues of Patients with Alzheimer’s Disease Based on GEO Expression Profile and Its Clinical Significance. BioMed Res. Int. 2019, 2019, 8179145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Singh, A.; Ekavali. A review on Alzheimer’s disease pathophysiology and its management: An update. Pharmacol. Rep. 2015, 67, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Huang, W.J.; Feng, F.; Zhou, B.; Yao, H.X.; Guo, Y.E.; Wang, P.; Wang, L.; Shu, N.; Zhang, X. Abnormal characterization of dynamic functional con-nectivity in Alzheimer’s disease. Neural. Regen. Res. 2022, 17, 2014–2021. [Google Scholar] [PubMed]
- Boeve, B.F.; Boxer, A.L.; Kumfor, F.; Pijnenburg, Y.; Rohrer, J.D. Advances and controversies in frontotemporal dementia: Diagnosis, biomarkers, and therapeutic considerations. Lancet Neurol. 2022, 21, 258–272. [Google Scholar] [CrossRef]
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef]
- Golde, T.E.; Eckman, C.B.; Younkin, S.G. Biochemical detection of Abeta isoforms: Implications for pathogenesis, diagnosis, and treatment of Alzheimer’s disease. Biochim. Biophys. Acta 2000, 1502, 172–187. [Google Scholar] [CrossRef] [Green Version]
- Lacour, A.; Espinosa, A.; Louwersheimer, E.; Heilmann-Heimbach, S.; Hernández, I.; Wolfsgruber, S.; Fernández, V.; Wagner, H.; Rosende-Roca, M.; Mauleón, A.; et al. Genome-wide significant risk factors for Alzheimer’s disease: Role in progression to dementia due to Alzheimer’s disease among subjects with mild cognitive impairment. Mol. Psychiatry 2016, 22, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Lanoiselée, H.M.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.-C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 mu-tations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef] [Green Version]
- Sukjamnong, S.; Thongkorn, S.; Kanlayaprasit, S.; Saeliw, T.; Hussem, K.; Warayanon, W.; Hu, V.W.; Tencomnao, T.; Sarachana, T. Prenatal exposure to bisphenol A alters the transcriptome-interactome profiles of genes associated with Alzheimer’s disease in the offspring hippocampus. Sci. Rep. 2020, 10, 9487. [Google Scholar] [CrossRef]
- Podtelezhnikov, A.A.; Tanis, K.Q.; Nebozhyn, M.; Ray, W.J.; Stone, D.J.; Loboda, A.P. Molecular Insights into the Pathogenesis of Alzheimer’s Disease and Its Relationship to Normal Aging. PLoS ONE 2011, 6, e29610. [Google Scholar] [CrossRef] [Green Version]
- Kwok, M.K.; Lin, S.L.; Schooling, C.M. Re-thinking Alzheimer’s disease therapeutic targets using gene-based tests. eBioMedicine 2018, 37, 461–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.A.; Woltjer, R.L.; Goodenbour, J.M.; Horvath, S.; Geschwind, D.H. Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med. 2013, 5, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.R.T.; Grinberg, L.T.; Farfel, J.M.; Diniz, B.S.; Lima, L.A.; Silva, P.J.S.; Ferretti, R.E.L.; Rocha, R.M.; Filho, W.J.; Carraro, D.M.; et al. Transcriptional Alterations Related to Neuropathology and Clinical Manifestation of Alzheimer’s Disease. PLoS ONE 2012, 7, e48751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blalock, E.M.; Geddes, J.W.; Chen, K.C.; Porter, N.M.; Markesbery, W.R.; Landfield, P.W. Incipient Alzheimer’s disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. USA 2004, 101, 2173–2178. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.S.; Dunckley, T.; Beach, T.G.; Grover, A.; Mastroeni, D.; Ramsey, K.; Caselli, R.J.; Kukull, W.A.; McKeel, D.; Morris, J.C.; et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: A reference data set. Physiol. Genom. 2008, 33, 240–256. [Google Scholar] [CrossRef] [Green Version]
- Berchtold, N.C.; Coleman, P.D.; Cribbs, D.H.; Rogers, J.; Gillen, D.L.; Cotman, C.W. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol. Aging 2012, 34, 1653–1661. [Google Scholar] [CrossRef] [Green Version]
- Hokama, M.; Oka, S.; Leon, J.; Ninomiya, T.; Honda, H.; Sasaki, K.; Iwaki, T.; Ohara, T.; Sasaki, T.; LaFerla, F.M.; et al. Altered Expression of Diabetes-Related Genes in Alzheimer’s Disease Brains: The Hisayama Study. Cereb. Cortex 2013, 24, 2476–2488. [Google Scholar] [CrossRef]
- Lee, T.; Lee, H. Shared Blood Transcriptomic Signatures between Alzheimer’s Disease and Diabetes Mellitus. Biomedicines 2021, 9, 34. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [Green Version]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef]
- Avram, S.; Bologa, C.G.; Holmes, J.; Bocci, G.; Wilson, T.B.; Nguyen, D.T.; Curpan, R.; Halip, L.; Bora, A.; Yang, J.J.; et al. DrugCentral 2021 supports drug discovery and repo-sitioning. Nucleic Acids Res. 2021, 49, D1160–D1169. [Google Scholar] [CrossRef] [PubMed]
- Corsello, S.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-generation drug library and information resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Cava, C.; Bertoli, G.; Castiglioni, I. A protein interaction map identifies existing drugs targeting SARS-CoV-2. BMC Pharmacol Toxicol. 2020, 21, 65. [Google Scholar] [CrossRef]
- Lee, T.; Lee, H.; Initiative, T.A.D.N. Identification of Disease-Related Genes That Are Common between Alzheimer’s and Cardiovascular Disease Using Blood Genome-Wide Transcriptome Analysis. Biomedicines 2021, 9, 1525. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable pro-tein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2–27. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Yu, W.; Yang, Y.; Lü, Y. Exploring the Key Genes and Identification of Potential Diagnosis Biomarkers in Alzheimer’s Disease Using Bioinformatics Analysis. Front. Aging Neurosci. 2021, 13, 602781. [Google Scholar] [CrossRef]
- Cui, J.J.; Tran-Dubé, M.; Shen, H.; Nambu, M.; Kung, P.P.; Pairish, M.; Jia, L.; Meng, J.; Funk, L.; Botrous, I.; et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J. Med. Chem. 2011, 54, 6342–6363. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kim, S.K.; Choi, S.Y.; Kim, D.H.; Gadhe, C.G.; Lee, H.N.; Kim, H.-J.; Kim, J.; Cho, S.J.; Hwang, H.; et al. Identification of crizotinib derivatives as potent SHIP2 inhibitors for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2018, 157, 405–422. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Hu, J.; Wu, S.; Xiaoe, Z.; Xu, H.; Zhang, Y.; Zhang, J.; Yang, Y. Structural bioinformatics-based identification of EGFR inhibitor gefitinib as a putative lead compound for BACE. Chem. Biol. Drug Des. 2014, 83, 81–88. [Google Scholar] [CrossRef]
- Javidnia, M.; Hebron, M.L.; Xin, Y.; Kinney, N.G.; Moussa, C.E.-H. Pazopanib Reduces Phosphorylated Tau Levels and Alters Astrocytes in a Mouse Model of Tauopathy. J. Alzheimer’s Dis. 2017, 60, 461–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, T.; Hoang, T.; Jelokhani-Niaraki, M.; Rao, P.P. Tau-derived-hexapeptide 306VQIVYK311 aggregation inhibitors: Ni-trocatechol moiety as a pharmacophore in drug design. ACS Chem. Neurosci. 2013, 4, 1559–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsaqati, M.; Thomas, R.S.; Kidd, E.J. Proteins Involved in Endocytosis Are Upregulated by Ageing in the Normal Human Brain: Implications for the Development of Alzheimer’s Disease. J. Gerontol. Ser. A 2017, 73, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tate, B.A.; Mathews, P.M. Targeting the Role of the Endosome in the Pathophysiology of Alzheimer’s Disease: A Strategy for Treatment. Sci. Aging Knowl. Environ. 2006, 2006, re2. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Matsui, H.; Tomizawa, K. Amphiphysin I and regulation of synaptic vesicle endocytosis. Acta Med. Okayama 2009, 63, 305–323. [Google Scholar]
- Di Paolo, G.; Sankaranarayanan, S.; Wenk, M.R.; Daniell, L.; Perucco, E.; Caldarone, B.J.; Flavell, R.; Picciotto, M.R.; Ryan, T.A.; Cremona, O.; et al. Decreased synaptic vesicle recycling efficiency and cognitive deficits in amphiphysin 1 knockout mice. Neuron 2022, 33, 789–804. [Google Scholar] [CrossRef] [Green Version]
- De Jesús-Cortés, H.J.; Nogueras-Ortiz, C.J.; Gearing, M.; Arnold, S.E.; Vega, I.E. Amphiphysin-1 protein level changes associated with tau-mediated neurodegeneration. Neuroreport 2012, 23, 942–946. [Google Scholar] [CrossRef] [Green Version]
- Tao, B.; Ling, Y.; Zhang, Y.; Li, S.; Zhou, P.; Wang, X.; Li, B.; Jun, Z.; Zhang, W.; Xu, C.; et al. CA 10 and CA 11 negatively regulate neuronal activity-dependent growth of gliomas. Mol. Oncol. 2019, 13, 1018–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, D.; Li, R.; Huang, X.; Chen, G.; Liu, Y.; Meng, Y.; Wang, Y.; Wu, Y.; Mao, Y. Identification of molecular correlations of RBM8A with autophagy in Alzheimer’s disease. Aging 2019, 11, 11673–11685. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, N.; Chen, Y.-J.; Yin, D.-M.; Mei, L.; Bergson, C. Complementary roles of the neuron-enriched endosomal proteins NEEP21 and calcyon in neuronal vesicle trafficking. J. Neurochem. 2014, 132, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.; Teunissen, C.E.; Otto, M.; Piehl, F.; Sormani, M.P.; Gattringer, T.; Barro, C.; Kappos, L.; Comabella, M.; Fazekas, F.; et al. Neurofilaments as biomarkers in neurological dis-orders. Nat. Rev. Neurol. 2018, 14, 577–589. [Google Scholar] [CrossRef]
- Jahn, R.; Südhof, T.C. Membrane Fusion and Exocytosis. Annu. Rev. Biochem. 1999, 68, 863–911. [Google Scholar] [CrossRef]
- Brinkmalm, A.; Brinkmalm, G.; Honer, W.G.; Frölich, L.; Hausner, L.; Minthon, L.; Hansson, O.; Wallin, A.; Zetterberg, H.; Blennow, K.; et al. SNAP-25 is a promising novel cerebrospinal fluid biomarker for synapse degeneration in Alzheimer’s disease. Mol. Neurodegener. 2014, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- Akila Parvathy Dharshini, S.; Taguchi, Y.H.; Michael Gromiha, M. Exploring the selective vulnerability in Alzheimer disease using tissue specific variant analysis. Genomics 2019, 111, 936–949. [Google Scholar] [CrossRef]
- Al Yemni, E.; Monies, D.; Alkhairallah, T.; Bohlega, S.; Abouelhoda, M.; Magrashi, A.; Mustafa, A.; AlAbdulaziz, B.; Alhamed, M.; Baz, B.; et al. Integrated Analysis of Whole Exome Sequencing and Copy Number Evaluation in Parkinson’s Disease. Sci. Rep. 2019, 9, 3344. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Xiao, Y.; Ravid, R.; Guan, Z.-Z. Changed Clathrin Regulatory Proteins in the Brains of Alzheimer’s Disease Patients and Animal Models. J. Alzheimer’s Dis. 2010, 22, 329–342. [Google Scholar] [CrossRef]
- Oeckl, P.; Halbgebauer, S.; Anderl-Straub, S.; Von Arnim, C.A.F.; Diehl-Schmid, J.; Froelich, L.; Grimmer, T.; Hausner, L.; Denk, J.; Jahn, H.; et al. Targeted Mass Spectrometry Suggests Beta-Synuclein as Synaptic Blood Marker in Alzheimer’s Disease. J. Proteome Res. 2020, 19, 1310–1318. [Google Scholar] [CrossRef]
- Prudencio, M.; Humphrey, J.; Pickles, S.; Brown, A.-L.; Hill, S.E.; Kachergus, J.M.; Shi, J.; Heckman, M.G.; Spiegel, M.R.; Cook, C.; et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J. Clin. Investig. 2020, 130, 6080–6092. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Xia, J.; Li, H.; Zhang, Z.; Yang, Y.; Huang, X.; He, Z.; Liu, J.; Yang, X. Ginsenoside Rg1 Ameliorates Behavioral Abnormalities and Modulates the Hippocampal Proteomic Change in Triple Transgenic Mice of Alzheimer’s Disease. Oxidative Med. Cell. Longev. 2017, 2017, 6473506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishioka, H.; Tooi, N.; Isobe, T.; Nakatsuji, N.; Aiba, K. BMS-708163 and Nilotinib restore synaptic dysfunction in human embryonic stem cell-derived Alzheimer’s disease models. Sci. Rep. 2016, 6, 33427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Ture, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila, J.; Perry, G. A Multilevel View of the Development of Alzheimer’s Disease. Neuroscience 2020, 457, 283–293. [Google Scholar] [CrossRef]

| Down-Regulated Genes | Drugs |
|---|---|
| CA10 | (1) ZONISAMIDE |
| CACNG2 | (1) GABAPENTIN |
| CALY | (1) APOMORPHINE (2) CLOZAPINE (3) TRIFLUOPERAZINE |
| CKMT1A | (1) CREATINE |
| GAD1 | (1) GLUTAMIC ACID |
| GLS2 | (1) GLUTAMIC ACID |
| GOT1 | (1) ASPARTIC ACID (2) CYSTEINE (3) GLUTAMIC ACID |
| MAP4 | (1) DOCETAXEL (2) PACLITAXEL |
| MET | (1) CRIZOTINIB (2) ERLOTINIB (3) CABOZANTINIB (4) GEFITINIB (5) PAZOPANIB (6) AFATINIB (7) ALECTINIB (8) ALTIRATINIB (9) AMG-208 (10) AMG-337 (11) AMUVATINIB (12) BMS-777607 (13) BMS-817378 (14) FORETINIB (15) GOLVATINIB (16) JNJ-38877605 (17) MGCD-265 (18) MK-2461 (19) MK-8033 (20) PF-04217903 (21) PHA-665752 (22) SAVOLITINIB (23) TIVANTINIB |
| Up-Regulated Genes | Drugs |
|---|---|
| COMT | (1) OPICAPONE (2) TOLCAPONE (3) ENTACAPONE (4) NIALAMIDE (5) 2-METHOXYESTRADIOL |
| PRKG1 | (1) GSK-690693 |
| PTAFR | (1) RUPATADINE (2) APAFANT (3) FOROPAFANT |
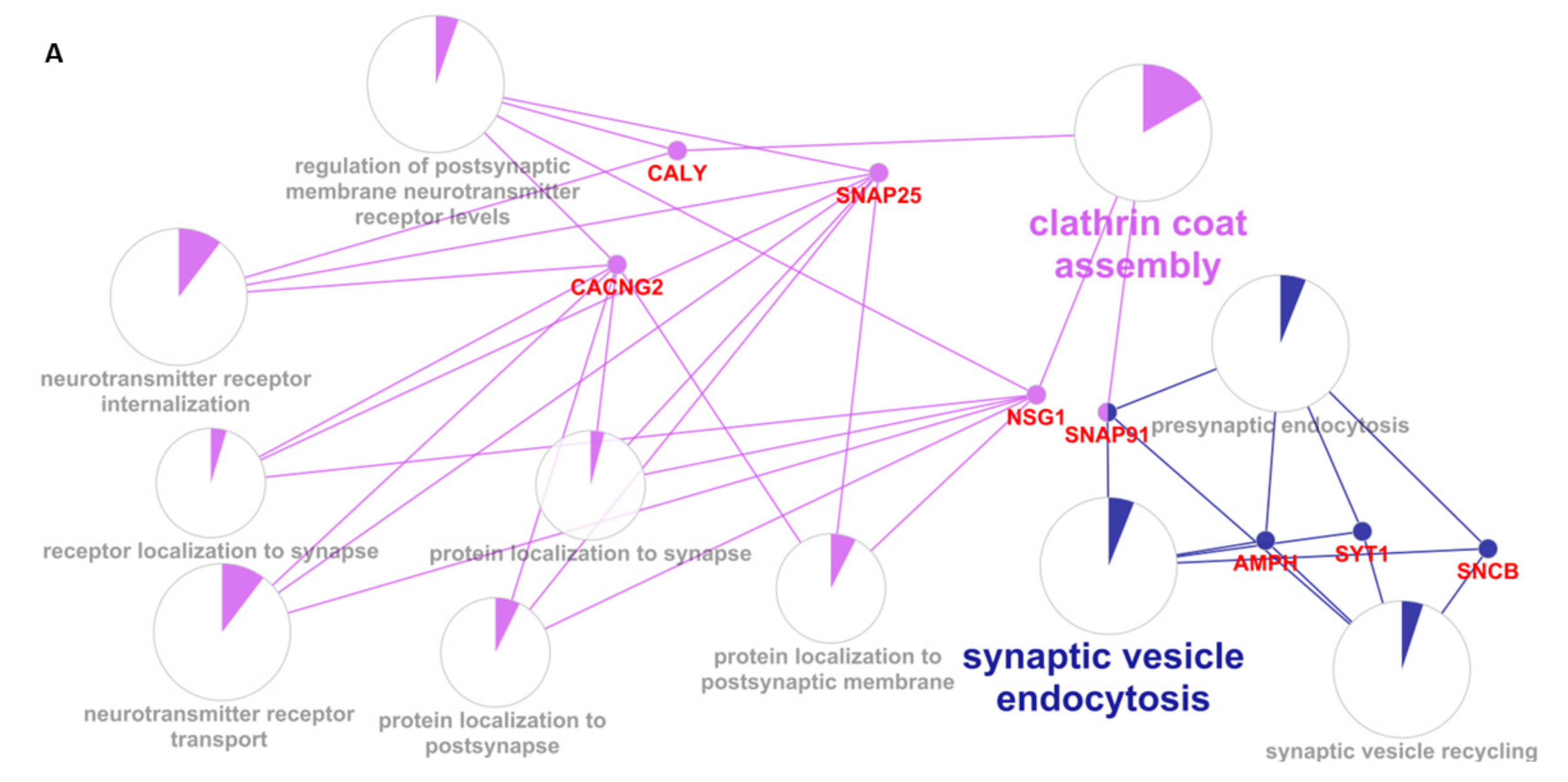
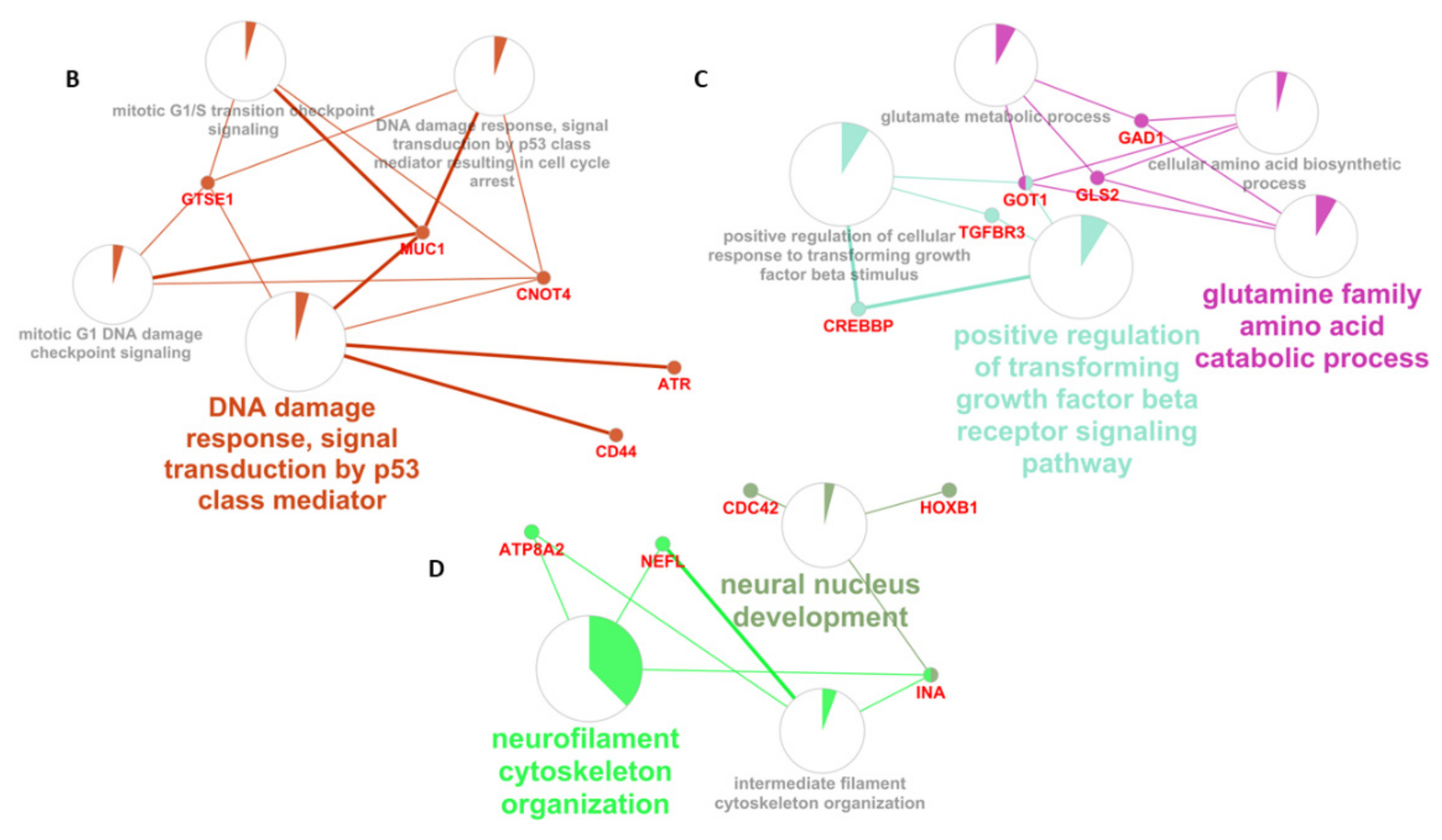
| GO: Biological Processes | # Tot Genes | Genes Obtained with MCODE | FDR |
|---|---|---|---|
| neurotransmitter receptor internalization | 9 | CALY SNAP25 | 0.0111 |
| clathrin coat assembly | 18 | CALY SNAP91 | 0.0329 |
| synaptic vesicle endocytosis | 52 | SNCB AMPH SYT1 SNAP91 | 0.0000933 |
| calcium-ion-regulated exocytosis | 43 | SYT1 SYT13 SYN2 | 0.00221 |
| neurotransmitter secretion | 81 | SYT1 SYN2 SNAP25 | 0.00997 |
| organelle localization | 475 | NEFL SYT1 SYN2 SNAP25 SNAP91 | 0.00405 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quarato, V.; D’Antona, S.; Battista, P.; Zupo, R.; Sardone, R.; Castiglioni, I.; Porro, D.; Frasca, M.; Cava, C. Transcriptional Profiling of Hippocampus Identifies Network Alterations in Alzheimer’s Disease. Appl. Sci. 2022, 12, 5035. https://doi.org/10.3390/app12105035
Quarato V, D’Antona S, Battista P, Zupo R, Sardone R, Castiglioni I, Porro D, Frasca M, Cava C. Transcriptional Profiling of Hippocampus Identifies Network Alterations in Alzheimer’s Disease. Applied Sciences. 2022; 12(10):5035. https://doi.org/10.3390/app12105035
Chicago/Turabian StyleQuarato, Veronica, Salvatore D’Antona, Petronilla Battista, Roberta Zupo, Rodolfo Sardone, Isabella Castiglioni, Danilo Porro, Marco Frasca, and Claudia Cava. 2022. "Transcriptional Profiling of Hippocampus Identifies Network Alterations in Alzheimer’s Disease" Applied Sciences 12, no. 10: 5035. https://doi.org/10.3390/app12105035
APA StyleQuarato, V., D’Antona, S., Battista, P., Zupo, R., Sardone, R., Castiglioni, I., Porro, D., Frasca, M., & Cava, C. (2022). Transcriptional Profiling of Hippocampus Identifies Network Alterations in Alzheimer’s Disease. Applied Sciences, 12(10), 5035. https://doi.org/10.3390/app12105035







