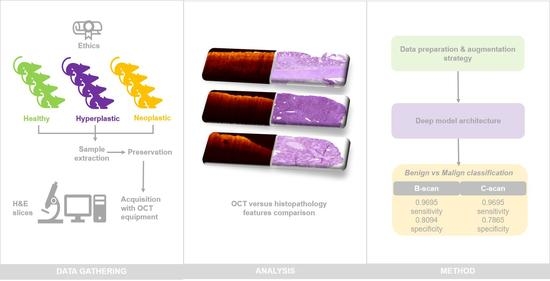
Characterization of Optical Coherence Tomography Images for Colon Lesion Differentiation under Deep Learning
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Models
2.2. Equipment
2.3. Acquisition Procedure
2.3.1. Sample Acquisition Procedure
2.3.2. Image Acquisition Protocol
2.3.3. Dataset Summary
2.4. Ethical Considerations
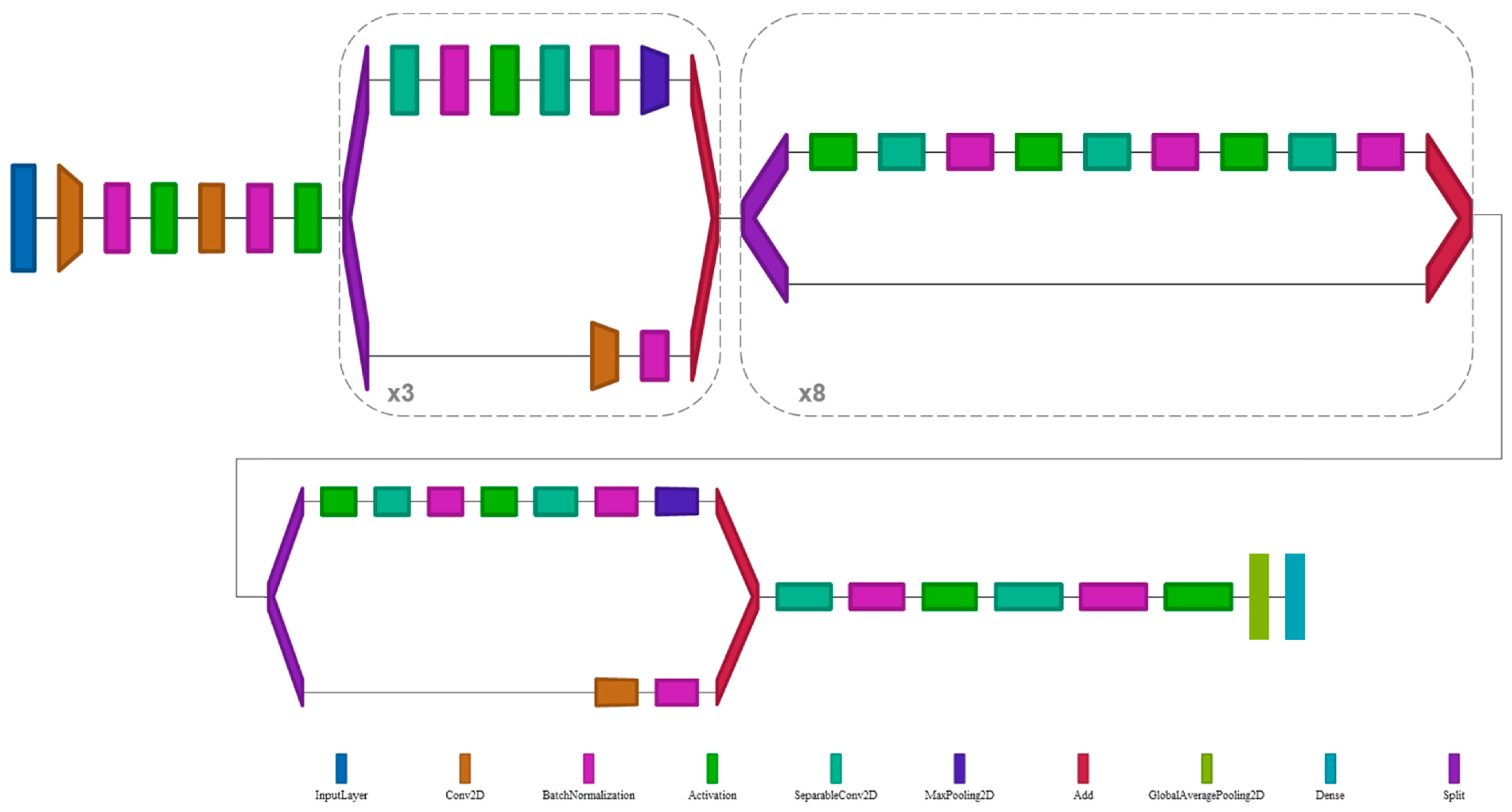
2.5. Deep Learning Architecture
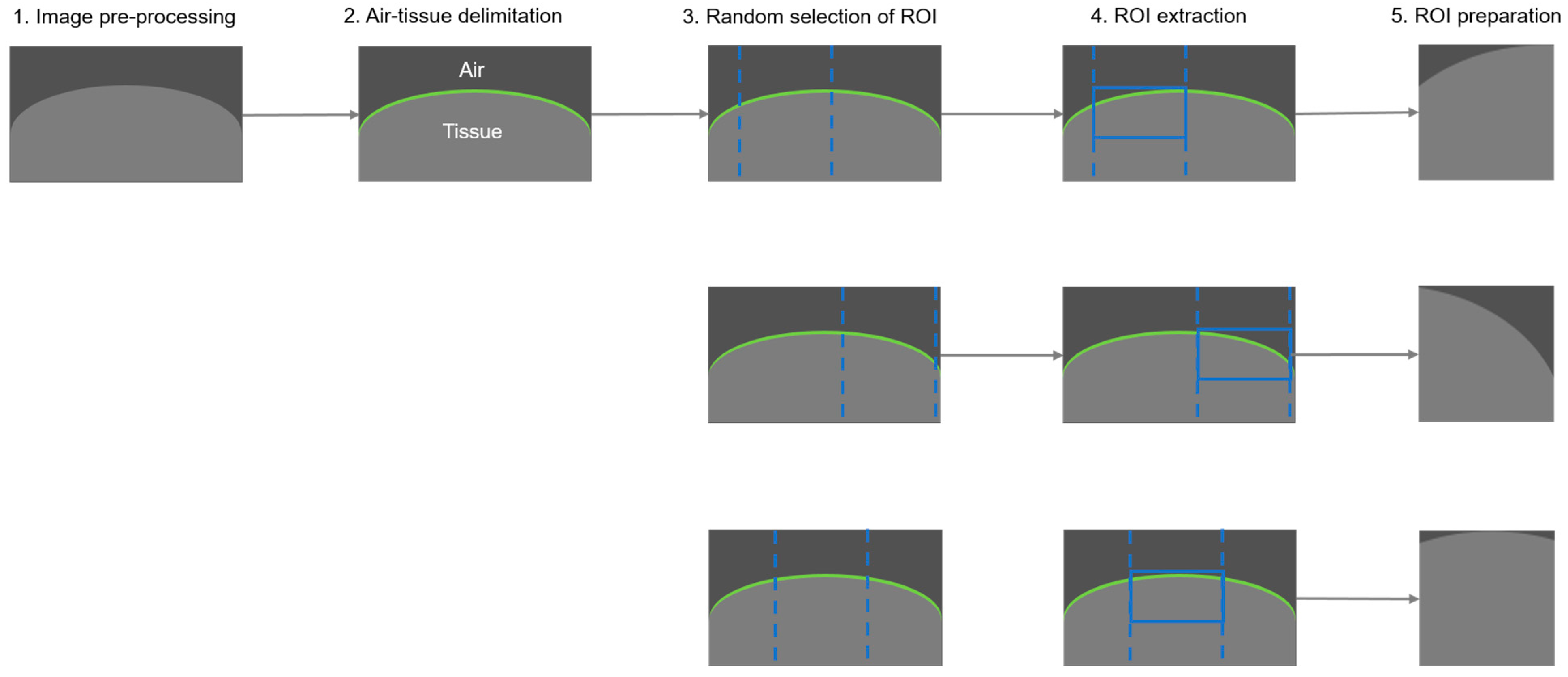
2.5.1. Data Preparation and Augmentation
- Image pre-processing
- 2.
- Air-tissue delimitation
- 3.
- Random selection of region of interest
- 4.
- ROI extraction
- 5.
- ROI preparation (post-processing)
2.5.2. Data Imbalance Management
2.5.3. Training Process
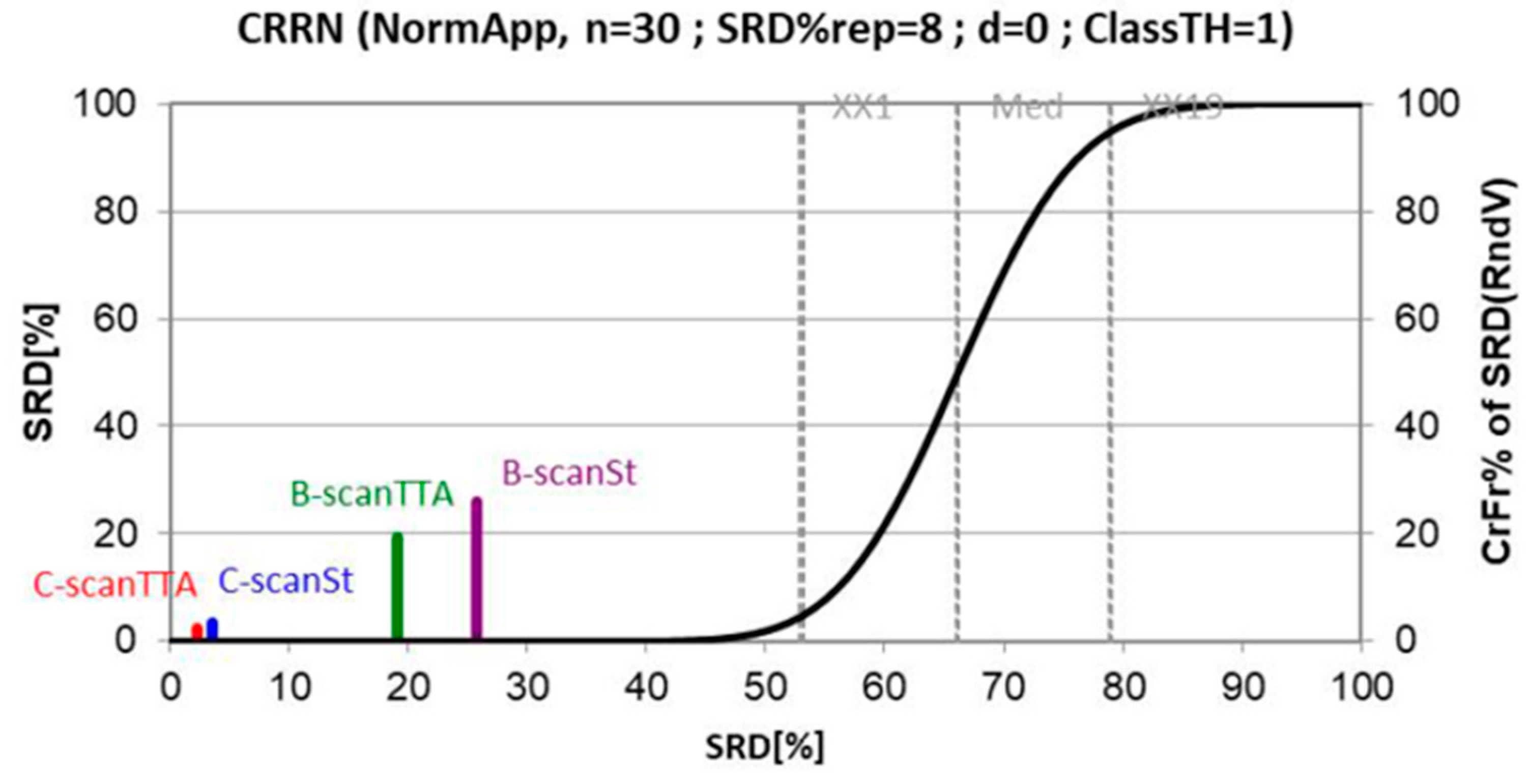
2.5.4. Data Evaluation and Test-Time Augmentation
3. Results
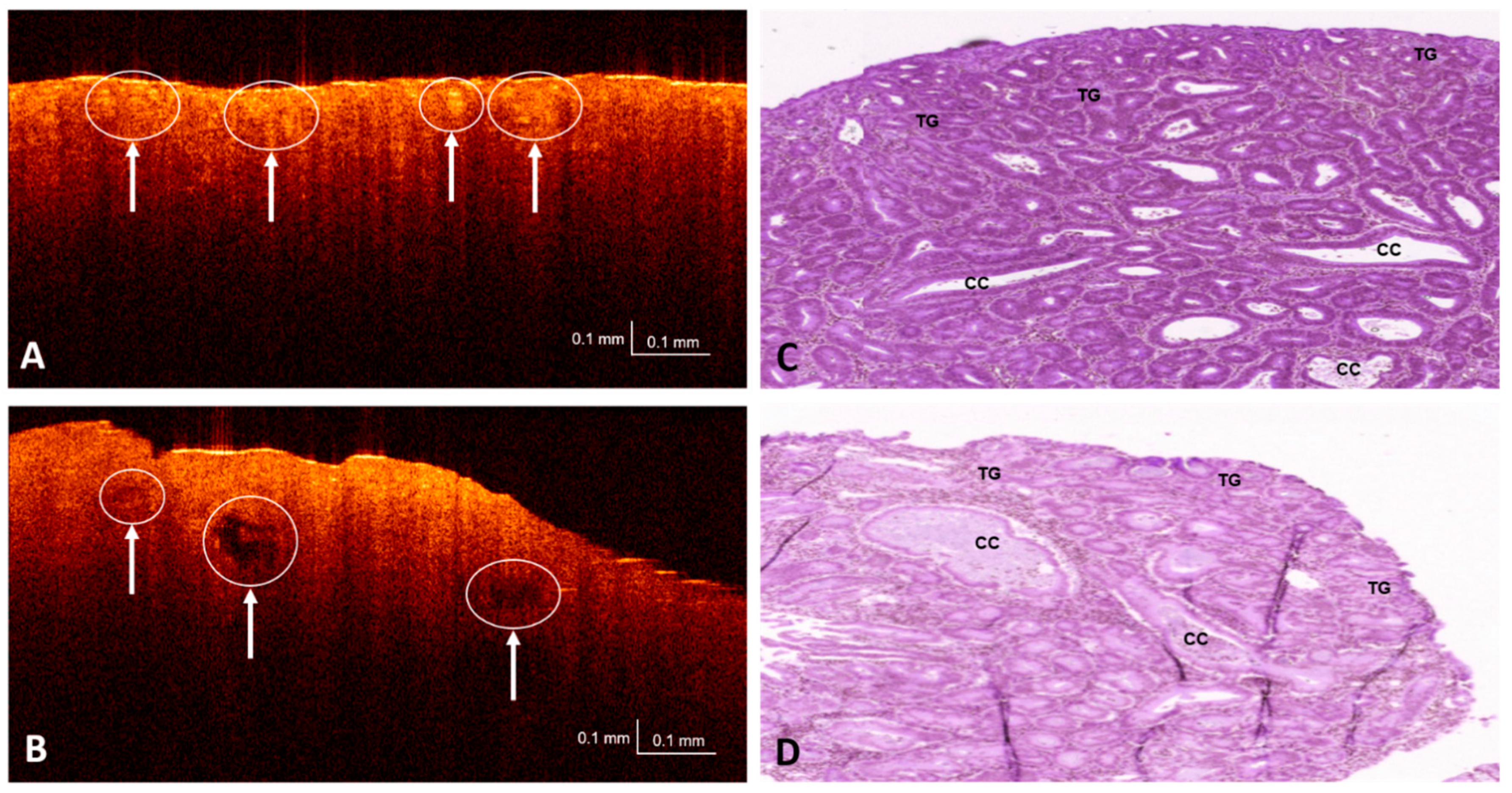
3.1. OCT and H&E Histology Comparative Analysis
3.2. Dataset Partitioning and Testing
3.3. Performance Metrics and Evaluation
- True positive (TP): Malignant lesion correctly identified as malignant.
- False positive (FP): Benign lesion incorrectly identified as malignant.
- True negative (TN): Benign lesion correctly identified as benign.
- False negative (FN): Malignant lesion incorrectly identified as benign.
- Sensitivity. Also known as the true positive rate (TPR). Number of true/all positive assessments. TPR = TP/(TP + FN) = number of malign lesions with positive test/total number of malign lesions.
- Specificity. Also known as the true negative rate (TNR). Number of true/all negative assessments. TNR = TN/(FP + TN) = number of benign lesions with negative test/total number of benign lesions.
- Positive predictive value (PPV). In case of a malignant prediction, probability that the lesion is actually malignant. PPV = TP/(TP + FP) = Number of true positives/number of positive calls.
- Negative predictive value (NPV). In case of a benign prediction, probability that the lesion is actually benign. NPV = TN/(TN + FN) = Number of true negatives/number of negative calls.
- Balanced accuracy (BAC). Measures the number of samples that were correctly classified in the expected class considering class frequencies. Number of correct/all assessments considering class frequencies. BAC = (TPR + TNR)/2 = (Sensitivity + Specificity)/2.
3.4. Thresholds
3.5. Classification Results
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Feature | Human | Rats |
|---|---|---|
| Anatomy of the large intestine compared macroscopically | ||
| Cecum to rectum | ~100–150 cm | ~25 cm |
| Taenia coli and haustra | Present | None; smooth serosa; may have fecal pellets |
| Appendix | Present, vermiform, ~9 cm | Absent |
| Functional cecum | Absent | Present; fermentation, vitamins K and B |
| Proximal/ascending/right | Colon from ileocecal valve to the hepatic flexure | Transverse folds in the mucosa, from cecum to mid colon; Rat: folds are visible through serosa |
| Mid/transverse | Connects the hepatic to the splenic flexure | Very short; lumen narrows; no mucosal folds |
| Distal/descending/left | Splenic flexure to left lower quadrant; S-shaped sigmoid colon extends from descending colon to rectosigmoid junction; sigmoid colon may be redundant | Fecal pellets may be seen |
| Rectum | 12–15 cm curved; proximal two-thirds of rectum has a mesothelial covering within the peritoneal cavity, whereas the distal third of rectum is extraperitoneal, lying within the deep pelvis, surrounded by adventitia, fascia, and fat | Indistinct from distal colon; Rat: ~50–80 mm, prolapse is rare |
| Large intestine anatomy compared at histological level | ||
| Mucosa | Transverse folds at all regions | Mucosal folds vary by region. Cecum and proximal colon: transverse; mid colon: flat with no folds; distal colon and rectum: longitudinal |
| Absorptive colonocytes | Similar to rodent | Present |
| Mucous/goblet cells | Similar to rodent | Present |
| Enteroendocrine cells | Similar to rodent | Present |
| Paneth cells | Cecum and appendix | Absent |
| Microfold (M) cells | Similar to rodent | Present |
| Lamina propria | Similar to rodent | Lymphocytes, plasma cells, macrophages, eosinophils, mast cells |
| Muscularis mucosae | Variable thickness; traversed by lymphoid follicles; poorly developed in appendix | Thin |
| Submucosa | Contains adipose tissue, arterioles, venules, lymphatics, and Meissner’s plexus | Rodents thinner than humans |
| Muscular tunics | Auerbach’s plexus between the two muscle bands | Muscular tunics thicken distally |
| Proximal colon | Transverse folds | Transverse mucosal folds |
| Transverse colon | Transverse folds | Flat mucosa |
| Distal colon | Transverse folds | Longitudinal mucosal folds |
| Rectum | Transverse folds | Indistinguishable from distal colon |
Appendix B
| Fold | Data Type | Evaluation | BAC | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|
| 1 | B-scan | Standard | 0.8967 | 0.9472 | 0.8462 | 0.9207 | 0.8948 |
| TTA | 0.9024 | 0.9553 | 0.8494 | 0.9229 | 0.9098 | ||
| C-scan | Standard | 0.8736 | 0.9615 | 0.7857 | 0.8929 | 0.9167 | |
| TTA | 0.8736 | 0.9615 | 0.7857 | 0.8929 | 0.9167 | ||
| 2 | B-scan | Standard | 0.9000 | 0.9583 | 0.8417 | 0.9362 | 0.8928 |
| TTA | 0.9060 | 0.9616 | 0.8505 | 0.9398 | 0.9012 | ||
| C-scan | Standard | 0.8988 | 0.9643 | 0.8333 | 0.9310 | 0.9091 | |
| TTA | 0.8988 | 0.9643 | 0.8333 | 0.9310 | 0.9091 | ||
| 3 | B-scan | Standard | 0.7568 | 0.9549 | 0.5588 | 0.8455 | 0.8305 |
| TTA | 0.7590 | 0.9585 | 0.5594 | 0.8461 | 0.8421 | ||
| C-scan | Standard | 0.6917 | 0.9667 | 0.4167 | 0.8056 | 0.8333 | |
| TTA | 0.7333 | 0.9667 | 0.5000 | 0.8286 | 0.8571 | ||
| 4 | B-scan | Standard | 0.8311 | 0.9657 | 0.6966 | 0.9037 | 0.8732 |
| TTA | 0.8398 | 0.9790 | 0.7007 | 0.9048 | 0.9189 | ||
| C-scan | Standard | 0.8500 | 1.0000 | 0.7000 | 0.9032 | 1.0000 | |
| TTA | 0.8000 | 1.0000 | 0.6000 | 0.8750 | 1.0000 | ||
| 5 | B-scan | Standard | 0.9437 | 0.9642 | 0.9233 | 0.9733 | 0.8988 |
| TTA | 0.9587 | 0.9705 | 0.9468 | 0.9815 | 0.9172 | ||
| C-scan | Standard | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| TTA | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | ||
| 6 | B-scan | Standard | 0.9553 | 0.9905 | 0.9201 | 0.9812 | 0.9584 |
| TTA | 0.9709 | 0.9922 | 0.9495 | 0.9881 | 0.9666 | ||
| C-scan | Standard | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| TTA | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
References
- Office World Health Organization Europe. Colorectal Cancer. Available online: https://www.euro.who.int/en/health-topics/noncommunicable-diseases/cancer/news/news/2012/2/early-detection-of-common-cancers/colorectal-cancer (accessed on 15 December 2020).
- World Cancer Research Fund International. Colorectal Cancer Statistics. Available online: https://www.wcrf.org/dietandcancer/cancer-trends/colorectal-cancer-statistics, (accessed on 15 December 2020).
- Society, A.C. Can Colorectal Polyps and Cancer Be Found Early? Available online: https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/detection.html (accessed on 15 December 2020).
- Axon, A.; Diebold, M.D.; Fujino, M.; Fujita, R.; Genta, R.M.; Gonvers, J.J.; Guelrud, M.; Inoue, H.; Jung, M.; Kashida, H.; et al. Update on the Paris classification of superficial neoplastic lesions in the digestive tract. Endoscopy 2005, 37, 570–578. [Google Scholar]
- Hewett, D.G.; Kaltenbach, T.; Sano, Y.; Tanaka, S.; Saunders, B.P.; Ponchon, T.; Soetikno, R.; Rex, D.K. Validation of a simple classification system for endoscopic diagnosis of small colorectal polyps using narrow-band imaging. Gastroenterology 2012, 143, 599–607. [Google Scholar] [CrossRef]
- Kavic, S.M.; Basson, M.D. Complications of endoscopy. Am. J. Surg. 2001, 181, 319–332. [Google Scholar] [CrossRef]
- Reumkens, A.; Rondagh, E.J.A.; Bakker, C.M.; Winkens, B.; Masclee, A.A.M.; Sanduleanu, S. Post-colonoscopy complications: A systematic review, time trends, and meta-analysis of population-based studies. Am. J. Gastroenterol. 2016, 111, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Kandel, P.; Wallace, M.B. Should we resect and discard low risk diminutive colon polyps. Clin. Endosc. 2019, 52, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Glover, B.; Teare, J.; Patel, N. The Status of Advanced Imaging Techniques for Optical Biopsy of Colonic Polyps. Clin. Transl. Gastroenterol. 2020, 11, e00130. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Markowitz, O. Introduction to reflectance confocal microscopy and its use in clinical practice. JAAD Case Rep. 2018, 4, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Iftimia, N.V. Overview of supercontinuum sources for multiphoton microscopy and optical biopsy. In Neurophotonics and Biomedical Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2018; pp. 329–351. [Google Scholar]
- Drexler, W.; Fujimoto, J.G. Optical Coherence Tomography-Technology and Applications; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Mason, S.E.; Poynter, L.; Takats, Z.; Darzi, A.; Kinross, J.M. Optical Technologies for Endoscopic Real-Time Histologic Assessment of Colorectal Polyps: A Meta-Analysis. Am. J. Gastroenterol. 2019, 114, 1219–1230. [Google Scholar] [CrossRef]
- Taunk, P.; Atkinson, C.D.; Lichtenstein, D.; Rodriguez-Diaz, E.; Singh, S.K. Computer-assisted assessment of colonic polyp histopathology using probe-based confocal laser endomicroscopy. Int. J. Colorectal Dis. 2019, 34, 2043–2051. [Google Scholar] [CrossRef]
- Ussui, V.M.; Wallace, M.B. Confocal endomicroscopy of colorectal polyps. Gastroenterol. Res. Pract. 2012, 2012, 545679. [Google Scholar] [CrossRef]
- Cicchi, R.; Sturiale, A.; Nesi, G.; Kapsokalyvas, D.; Alemanno, G.; Tonelli, F.; Pavone, F.S. Multiphoton morpho-functional imaging of healthy colon mucosa, adenomatous polyp and adenocarcinoma. Biomed. Opt. Express 2013, 4, 1204–1213. [Google Scholar] [CrossRef]
- He, K.; Zhao, L.; Chen, Y.; Huang, X.; Ding, Y.; Hua, H.; Liu, L.; Wang, X.; Wang, M.; Zhang, Y.; et al. Label-free multiphoton microscopic imaging as a novel real-time approach for discriminating colorectal lesions: A preliminary study. J. Gastroenterol. Hepatol. 2019, 34, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Pfau, P.R.; Sivak, M.V.; Chak, A.; Kinnard, M.; Wong, R.C.K.; Isenberg, G.A.; Izatt, J.A.; Rollins, A.; Westphal, V. Criteria for the diagnosis of dysplasia by endoscopic optical coherence tomography. Gastrointest. Endosc. 2003, 58, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Zagaynova, E.; Gladkova, N.; Shakhova, N.; Gelikonov, G.; Gelikonov, V. Endoscopic OCT with forward-looking probe: Clinical studies in urology and gastroenterology | Natalia Shakhova-Academia.edu. J. Biophotonics 2008, 1, 114–128. [Google Scholar] [CrossRef]
- Iftimia, N.; Iyer, A.K.; Hammer, D.X.; Lue, N.; Mujat, M.; Pitman, M.; Ferguson, R.D.; Amiji, M. Fluorescence-guided optical coherence tomography imaging for colon cancer screening: A preliminary mouse study. Biomed. Opt. Express 2012, 3, 178–191. [Google Scholar] [CrossRef]
- Ding, Q.; Deng, Y.; Yu, X.; Yuan, J.; Zeng, Z.; Mu, G.; Wan, X.; Zhang, J.; Zhou, W.; Huang, L.; et al. Rapid, high-resolution, label-free, and 3-dimensional imaging to differentiate colorectal adenomas and non-neoplastic polyps with micro-optical coherence tomography. Clin. Transl. Gastroenterol. 2019, 10, e00049. [Google Scholar] [CrossRef]
- Kudo, S.E.; Tamura, S.; Nakajima, T.; Yamano, H.O.; Kusaka, H.; Watanabe, H. Diagnosis of colorectal tumorous lesions by magnifying endoscopy. Gastrointest. Endosc. 1996, 44, 8–14. [Google Scholar] [CrossRef]
- Adler, D.C.; Zhou, C.; Tsai, T.-H.; Schmitt, J.; Huang, Q.; Mashimo, H.; Fujimoto, J.G. Three-dimensional endomicroscopy of the human colon using optical coherence tomography. Opt. Express 2009, 17, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Ahsen, O.O.; Lee, H.C.; Liang, K.; Wang, Z.; Figueiredo, M.; Huang, Q.; Potsaid, B.; Jayaraman, V.; Fujimoto, J.G.; Mashimo, H. Ultrahigh-speed endoscopic optical coherence tomography and angiography enables delineation of lateral margins of endoscopic mucosal resection: A case report. Therap. Adv. Gastroenterol. 2017, 10, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Ahsen, O.O.; Wang, Z.; Lee, H.-C.; Liang, W.; Potsaid, B.M.; Tsai, T.-H.; Giacomelli, M.G.; Jayaraman, V.; Mashimo, H.; et al. Endoscopic forward-viewing optical coherence tomography and angiography with MHz swept source. Opt. Lett. 2017, 42, 3193–3196. [Google Scholar] [CrossRef]
- Zeng, Y.; Rao, B.; Chapman, W.C.; Nandy, S.; Rais, R.; González, I.; Chatterjee, D.; Mutch, M.; Zhu, Q. The Angular Spectrum of the Scattering Coefficient Map Reveals Subsurface Colorectal Cancer. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Picón Ruiz, A.; Alvarez Gila, A.; Irusta, U.; Echazarra Huguet, J. Why deep learning performs better than classical machine learning engenering. Dyn. Ing. Ind. 2020, 95, 119–122. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object recognition with gradient-based learning. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 1999; Volume 1681, pp. 319–345. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; The MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- LeCun, Y.A.; Bengio, Y.; Hinton, G.E. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, 271–297. [Google Scholar] [CrossRef]
- Wei, J.W.; Suriawinata, A.A.; Vaickus, L.J.; Ren, B.; Liu, X.; Lisovsky, M.; Tomita, N.; Abdollahi, B.; Kim, A.S.; Snover, D.C.; et al. Evaluation of a Deep Neural Network for Automated Classification of Colorectal Polyps on Histopathologic Slides. JAMA Netw. Open 2020, 3, e203398. [Google Scholar] [CrossRef]
- Medela, A.; Picon, A. Constellation loss: Improving the efficiency of deep metric learning loss functions for the optimal embedding of histopathological images. J. Pathol. Inform. 2020, 11, 38. [Google Scholar]
- Terradillos, E.; Saratxaga, C.L.; Mattana, S.; Cicchi, R.; Pavone, F.S.; Andraka, N.; Glover, B.J.; Arbide, N.; Velasco, J.; Echezarraga, M.C.; et al. Analysis on the characterization of multiphoton microscopy images for malignant neoplastic colon lesion detection under deep learning methods. in press.
- Sánchez-Peralta, L.F.; Picón, A.; Sánchez-Margallo, F.M.; Pagador, J.B. Unravelling the effect of data augmentation transformations in polyp segmentation. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1975–1988. [Google Scholar] [CrossRef]
- Sánchez-Peralta, L.F.; Pagador, J.B.; Picón, A.; Calderón, Á.J.; Polo, F.; Andraka, N.; Bilbao, R.; Glover, B.; Saratxaga, C.L.; Sánchez-Margallo, F.M. PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets. Appl. Sci. 2020, 10, 8501. [Google Scholar] [CrossRef]
- Sánchez-Peralta, L.F.; Bote-Curiel, L.; Picón, A.; Sánchez-Margallo, F.M.; Pagador, J.B. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif. Intell. Med. 2020, 108, 101923. [Google Scholar] [CrossRef] [PubMed]
- Picon, A.; Medela, A.; Sanchez-Peralta, L.F.; Cicchi, R.; Bilbao, R.; Alfieri, D.; Elola, A.; Glover, B.; Saratxaga, C.L. Autofluorescence image reconstruction and virtual staining for in-vivo optical biopsying. IEEE Access 2021, 9, 32081–32093. [Google Scholar] [CrossRef]
- Yanagihara, R.T.; Lee, C.S.; Ting, D.S.W.; Lee, A.Y. Methodological challenges of deep learning in optical coherence tomography for retinal diseases: A review. Transl. Vis. Sci. Technol. 2020, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Tong, Y.; Yu, Y.; Xing, Y.; Chen, C.; Shen, Y. Deep learning-based automated classification of multi-categorical abnormalities from optical coherence tomography images. Transl. Vis. Sci. Technol. 2018, 7, 41. [Google Scholar] [CrossRef]
- Jiang, Z.; Huang, Z.; Qiu, B.; Meng, X.; You, Y.; Liu, X.; Liu, G.; Zhou, C.; Yang, K.; Maier, A.; et al. Comparative study of deep learning models for optical coherence tomography angiography. Biomed. Opt. Express 2020, 11, 1580–1597. [Google Scholar] [CrossRef]
- Singla, N.; Dubey, K.; Srivastava, V. Automated assessment of breast cancer margin in optical coherence tomography images via pretrained convolutional neural network. J. Biophotonics 2019, 12, e201800255. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Xu, S.; Chapman, W.C.; Li, S.; Alipour, Z.; Abdelal, H.; Chatterjee, D.; Mutch, M.; Zhu, Q. Real-time colorectal cancer diagnosis using PR-OCT with deep learning. Theranostics 2020, 10, 2587–2596. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef]
- Amos-Landgraf, J.M.; Kwong, L.N.; Kendziorski, C.M.; Reichelderfer, M.; Torrealba, J.; Weichert, J.; Haag, J.D.; Chen, K.S.; Waller, J.L.; Gould, M.N.; et al. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 4036–4041. [Google Scholar] [CrossRef]
- Irving, A.A.; Yoshimi, K.; Hart, M.L.; Parker, T.; Clipson, L.; Ford, M.R.; Kuramoto, T.; Dove, W.F.; Amos-Landgraf, J.M. The utility of Apc-mutant rats in modeling human colon cancer. DMM Dis. Model. Mech. 2014, 7, 1215–1225. [Google Scholar] [CrossRef]
- Bote-Chacón, J.; Moreno-Lobato, B.; Sanchez-Margallo, F.M. Pilot study for the characterization of a murine model of hyperplastic growth in colon. In Proceedings of the 27th International Congress European Association of Endoscopic Surgery, Seville, Spain, 12–15 June 2019. [Google Scholar]
- Bote-Chacón, J.; Ortega-Morán, J.F.; Pagador, B.; Moreno-Lobato, B.L.; Saratxaga, C.; Sánchez-Margallo, F.M. Validation of murine hyperplastic model of the colon. In Proceedings of the Abstracts of the first virtual Congres of the Spanish Society of Surgical Research. Br. J. Surg. 2022. to be published. [Google Scholar]
- Thorlabs CAL110C1-Spectral Domain OCT System. Available online: https://www.thorlabs.com/thorproduct.cfm?partnumber=CAL110C1 (accessed on 15 September 2020).
- Gleed, R.D.; Ludders, J.W. Recent Advances in Veterinary Anesthesia and Analgesia: Companion Animals; International Veterinary Information Service: Ithaca, NY, USA, 2008. [Google Scholar]
- Abreu, M.; Aguado, D.; Benito, J.; Gómez de Segura, I.A. Reduction of the sevoflurane minimum alveolar concentration induced by methadone, tramadol, butorphanol and morphine in rats. Lab. Anim. 2012, 46, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Flecknell, P. Laboratory Animal Anaesthesia; Elsevier: Amsterdam, The Netherlands, 1996. [Google Scholar]
- Gabrecht, T.; Andrejevic-Blant, S.; Wagnières, G. Blue-Violet Excited Autofluorescence Spectroscopy and Imaging of Normal and Cancerous Human Bronchial Tissue after Formalin Fixation. Photochem. Photobiol. 2007, 83, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2016; Institute of Electrical and Electronics Engineers Inc.: New York, NY, USA, 2017; pp. 1800–1807. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Bäuerle, A.; van Onzenoodt, C.; Ropinski, T. Net2Vis-A Visual Grammar for Automatically Generating Publication-Tailored CNN Architecture Visualizations. IEEE Trans. Vis. Comput. Graph. 2019, 1. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man. Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Treuting, P.M.; Dintzis, S.M. Lower Gastrointestinal Tract. In Comparative Anatomy and Histology; Elsevier Inc.: Amsterdam, The Netherlands, 2012; pp. 177–192. [Google Scholar]
- Kollár-Hunek, K.; Héberger, K. Method and model comparison by sum of ranking differences in cases of repeated observations (ties). Chemom. Intell. Lab. Syst. 2013, 127, 139–146. [Google Scholar] [CrossRef]


| Actual Condition | |||
|---|---|---|---|
| Malignant | Benign | ||
| Predicted Condition | Malignant | TP | FP |
| Benign | FN | TN | |
| Data Type | Evaluation | BAC | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| B-scan | Standard | 0.8806 ± 0.0748 | 0.9635 ± 0.0148 | 0.7978 ± 0.1431 | 0.9268 ± 0.0498 | 0.8914 ± 0.0415 |
| TTA | 0.8895 ± 0.0792 | 0.8094 ± 0.1524 | 0.8094 ± 0.1524 | 0.9305 ± 0.0526 | 0.9093 ± 0.0400 | |
| C-scan | Standard | 0.8857 ± 0.1143 | 0.7893 ± 0.2180 | 0.7893 ± 0.2180 | 0.9221 ± 0.0735 | 0.9432 ± 0.0687 |
| TTA | 0.8843 ± 0.1068 | 0.7865 ± 0.2050 | 0.7865 ± 0.2050 | 0.9212 ± 0.0693 | 0.9472 ± 0.0614 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saratxaga, C.L.; Bote, J.; Ortega-Morán, J.F.; Picón, A.; Terradillos, E.; del Río, N.A.; Andraka, N.; Garrote, E.; Conde, O.M. Characterization of Optical Coherence Tomography Images for Colon Lesion Differentiation under Deep Learning. Appl. Sci. 2021, 11, 3119. https://doi.org/10.3390/app11073119
Saratxaga CL, Bote J, Ortega-Morán JF, Picón A, Terradillos E, del Río NA, Andraka N, Garrote E, Conde OM. Characterization of Optical Coherence Tomography Images for Colon Lesion Differentiation under Deep Learning. Applied Sciences. 2021; 11(7):3119. https://doi.org/10.3390/app11073119
Chicago/Turabian StyleSaratxaga, Cristina L., Jorge Bote, Juan F. Ortega-Morán, Artzai Picón, Elena Terradillos, Nagore Arbide del Río, Nagore Andraka, Estibaliz Garrote, and Olga M. Conde. 2021. "Characterization of Optical Coherence Tomography Images for Colon Lesion Differentiation under Deep Learning" Applied Sciences 11, no. 7: 3119. https://doi.org/10.3390/app11073119
APA StyleSaratxaga, C. L., Bote, J., Ortega-Morán, J. F., Picón, A., Terradillos, E., del Río, N. A., Andraka, N., Garrote, E., & Conde, O. M. (2021). Characterization of Optical Coherence Tomography Images for Colon Lesion Differentiation under Deep Learning. Applied Sciences, 11(7), 3119. https://doi.org/10.3390/app11073119