Comparative Analysis of Cell-Free miR-205-5p, let-7f-5p, and miR-483-5p Expression in Ovarian Cell Cultures and Plasma Samples of Patients with Ovarian Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
2.2. Plasma Collection and miRNA Isolation
2.3. Cell Culturing Conditions
2.4. MiRNA Detection
2.5. Bioinformatic Analysis
3. Results
3.1. The Basal Expression of miR-205-5p and let-7f-5p Is Higher in the Estrogen Sensitive PEO1 Cell Line Than in the Estrogen Non-Sensitive A2780
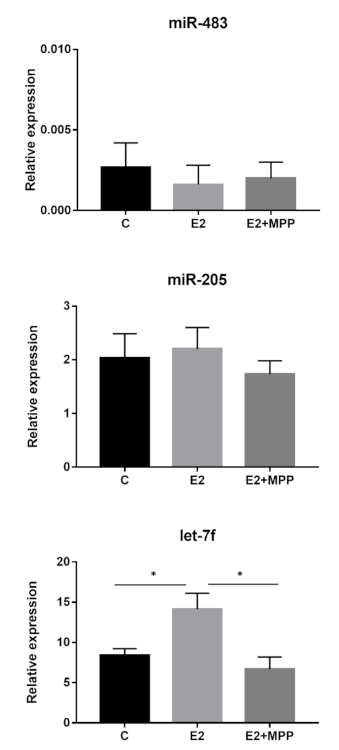
3.2. Let-7f-5p Is Upregulated in Response to Estradiol Exposure in PEO1
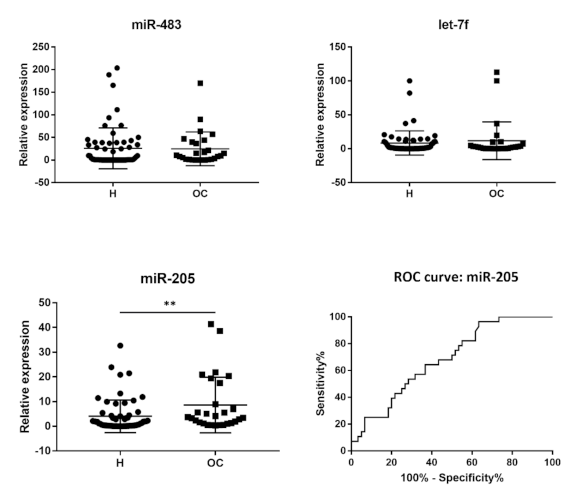
3.3. MiR-205-5p Is Overrepresented in the Plasma Samples of Patients with Malignant Ovarian Tumors
3.4. Bioinformatic Analysis of miR-483-5p, miR-205-5p, and let-7f-5p Target Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Szilágyi, M.; Pös, O.; Márton, É.; Buglyó, G.; Soltész, B.; Keserű, J.; Penyige, A.; Szemes, T.; Nagy, B. Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application. Int. J. Mol. Sci. 2020, 21, 6827. [Google Scholar] [CrossRef] [PubMed]
- Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.M.; Jemal, A. Cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Montagnana, M.; Benati, M.; Danese, E. Circulating biomarkers in epithelial ovarian cancer diagnosis: From present to future perspective. Ann. Transl. Med. 2017, 5, 276. [Google Scholar] [CrossRef]
- Sölétormos, G.; Duffy, M.J.; Hassan, S.O.A.; Verheijen, R.H.; Tholander, B.; Bast, R.C.; Gaarenstroom, K.N.; Sturgeon, C.M.; Bonfrer, J.M.; Petersen, P.H.; et al. Clinical use of cancer biomarkers in epithelial ovarian cancer: Updated guidelines from the European Group on Tumor Markers. Int. J. Gyn. Cancer 2016, 26, 43–51. [Google Scholar] [CrossRef]
- Márton, É.; Lukács, J.; Penyige, A.; Janka, E.; Hegedüs, L.; Soltész, B.; Méhes, G.; Póka, R.; Nagy, B.; Szilágyi, M. Circulating epithelial-mesenchymal transition-associated miRNAs are promising biomarkers in ovarian cancer. J. Biotechnol. 2019, 297, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Penyige, A.; Márton, É.; Soltész, B.; Szilágyi-Bónizs, M.; Póka, R.; Lukács, J.; Széles, L.; Nagy, B. Circulating miRNA profiling in plasma samples of ovarian cancer patients. Int. J. Mol. Sci. 2019, 20, 4533. [Google Scholar] [CrossRef]
- Márton, É.; Varga, A.; Széles, L.; Göczi, L.; Penyige, A.; Nagy, B.; Szilágyi, M. The Cell-Free Expression of MiR200 Family members correlates with estrogen sensitivity in human epithelial ovarian cells. Int. J. Mol. Sci. 2020, 21, 9725. [Google Scholar] [CrossRef]
- miRTargetLink. Available online: https://ccb-web.cs.uni-saarland.de/mirtargetlink/multinet.php (accessed on 20 January 2021).
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Sun, J.; Huang, Y.R.; Harrington, W.R.; Sheng, S.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S. Antagonists selective for estrogen receptor α. Endocrinology 2002, 143, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.-Y.; Zhang, X.-W.; Liu, L.; Yu, J.-P.; Li, H.; Wang, S.-Z.E.; Ren, X.-B.; Cao, S. MiR-205 in cancer: An angel or a devil? Eur. J. Cell Biol. 2013, 92, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, N.; Dhasmana, A.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. miR-205: A Potential biomedicine for cancer therapy. Cells 2020, 9, 1957. [Google Scholar] [CrossRef]
- Dong, Y.; Si, J.-W.; Li, W.-T.; Liang, L.; Zhao, J.; Zhou, M.; Li, N.; Li, T. miR-200a/miR-141 and miR-205 upregulation might be associated with hormone receptor status and prognosis in endometrial carcinomas. Int. J. Clin. Exp. Pathol. 2015, 8, 2864–2875. [Google Scholar]
- Wu, H.; Zhu, S.; Mo, Y.-Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009, 19, 439–448. [Google Scholar] [CrossRef]
- Boll, K.; Reiche, K.; Kasack, K.; Mörbt, N.; Kretzschmar, A.K.; Tomm, J.M.; Verhaegh, J.M.; Schalken, J.; von Bergen, M.; Horn, F.; et al. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 2013, 32, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Lebanony, D.; Benjamin, H.; Gilad, S.; Ezagouri, M.; Dov, A.; Ashkenazi, K.; Gefen, N.; Izraeli, S.; Rechavi, G.; Pass, H.; et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J. Clin. Oncol. 2009, 27, 2030–2037. [Google Scholar] [CrossRef]
- Gottardo, F.; Liu, C.G.; Ferracin, M.; Calin, G.A.; Fassan, M.; Bassi, P.; Sevignani, C.; Byrne, D.; Negrini, M.; Pagano, F.; et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. Semin. Orig. Investig. 2007, 25, 387–392. [Google Scholar] [CrossRef]
- Eyking, A.; Reis, H.; Frank, M.; Gerken, G.; Schmid, K.W.; Cario, E. MiR-205 and MiR-373 are associated with aggressive human mucinous colorectal cancer. PLoS ONE 2016, 11, e0156871. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Jiao, K.; Zhao, C.; Liu, H.; Meng, Q.; Wang, Z.; Feng, C.; Li, Y. Effect of miR‑205 on proliferation and migration of thyroid cancer cells by targeting CCNB2 and the mechanism. Oncol. Lett. 2020, 19, 2568–2574. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.-G.; Alder, H.; et al. MicroRNA Signatures in Human Ovarian Cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, L.; Li, J.; Zhu, S.; Tai, M.; Mason, C.W.; Chapman, J.A.; Reynolds, E.A.; Weiner, C.P.; Zhou, H.H. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J. Ovarian Res. 2017, 10, 33. [Google Scholar] [CrossRef]
- Li, J.; Hu, K.; Gong, G.; Zhu, D.; Wang, Y.; Liu, H.; Wu, X. Upregulation of MiR-205 transcriptionally suppresses SMAD4 and PTEN and contributes to human ovarian cancer progression. Sci. Rep. 2017, 7, srep41330. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.; Liang, A.; Jiang, A.; Zong, L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol. Lett. 2018, 15, 7571–7578. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhu, W.; Chen, Q.; Yuan, Y.; Wang, Y.; Wang, J.; Wu, X. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics 2019, 9, 8206–8220. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, L.; Zhao, Y.; Yang, D.; Song, F.; Wen, Y.; Hao, Q.; Hu, Z.; Zhang, W.; Chen, K. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS ONE 2013, 8, e77853. [Google Scholar] [CrossRef]
- Su, N.; Qiu, H.; Chen, Y.; Yang, T.; Yan, Q.; Wan, X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol. Rep. 2013, 29, 2297–2302. [Google Scholar] [CrossRef]
- Gulei, D.; Magdo, L.; Jurj, A.; Raduly, L.; Cojocneanu-Petric, R.; Moldovan, A.; Moldovan, C.; Florea, A.; Pasca, S.; Pop, L.-A.; et al. The silent healer: miR-205-5p up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Boyerinas, B.; Park, S.-M.; Hau, A.; E Murmann, A.; E Peter, M. The role of let-7 in cell differentiation and cancer. Endocrine-Related Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; García, V.; Zaballos, A.; Provencio, M.; Lombardía, L.; Almonacid, L.; Garcia, J.M.; Domínguez, G.; Pena, C.; Diaz, R.; et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 2011, 37, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Yu, D.-C.; Li, Q.-G.; Chen, X.; Zhang, C.-Y.; Ding, Y.-T. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin. Lab. 2014, 60, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; He, L.; Zhao, X.; Miao, Y.; Gu, Y.; Guo, C.; Xue, Z.; Dou, W.; Hu, F.; Wu, K.; et al. MicroRNA Let-7f Inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS ONE 2011, 6, e18409. [Google Scholar] [CrossRef]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef]
- Bhat-Nakshatri, P.; Wang, G.; Collins, N.R.; Thomson, M.J.; Geistlinger, T.R.; Carroll, J.S.; Brown, M.; Hammond, S.; Srour, E.F.; Liu, Y.; et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009, 37, 4850–4861. [Google Scholar] [CrossRef]
- Fiorillo, A.A.; Heier, C.R.; Huang, Y.-F.; Tully, C.B.; Punga, T.; Punga, A.R. Estrogen Receptor, Inflammatory, and FOXO Transcription Factors Regulate Expression of Myasthenia Gravis-Associated Circulating microRNAs. Front. Immunol. 2020, 11, 151. [Google Scholar] [CrossRef]
- Yang, Z.-G.; Ma, X.-D.; He, Z.-H.; Guo, Y.-X. miR–483-5p promotes prostate cancer cell proliferation and invasion by targeting RBM5. Int. Braz. J. Urol. 2017, 43, 1060–1067. [Google Scholar] [CrossRef]
- Korzeniewski, N.; Tosev, G.; Pahernik, S.; Hadaschik, B.; Hohenfellner, M.; Duensing, S. Identification of cell-free microRNAs in the urine of patients with prostate cancer. Urol. Oncol. 2015, 33, 16.e17–16.e22. [Google Scholar] [CrossRef]
- Xue, L.; Nan, J.; Dong, L.; Zhang, C.; Li, H.; Na, R.; He, H.; Wang, Y. Upregulated miR-483-5p expression as a prognostic biomarker for esophageal squamous cell carcinoma. Cancer Biomarkers 2017, 19, 193–197. [Google Scholar] [CrossRef]
- Chabre, O.; Libé, R.; Assie, G.; Barreau, O.; Bertherat, J.; Bertagna, X.; Feige, J.-J.; Cherradi, N. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocrine-Related Cancer 2013, 20, 579–594. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, Y.; Zhao, H.; Yang, X.; Luo, Y.; Ren, Y.; Liu, W.; Li, N. Serum miR-483-5p: A novel diagnostic and prognostic biomarker for patients with oral squamous cell carcinoma. Tumor Biol. 2016, 37, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, A.; Wang, Q.; Gurvich, I.; Siegel, A.B.; Remotti, H.; Santella, R.M. Exploration of Genome-Wide Circulating MicroRNA in Hepatocellular Carcinoma: MiR-483-5p as a Potential Biomarker. Cancer Epidemiol. Biomarkers Prev. 2013, 22, 2364–2373. [Google Scholar] [CrossRef]
- Patel, D.; Boufraqech, M.; Jain, M.; Zhang, L.; He, M.; Gesuwan, K.; Gulati, N.; Nilubol, N.; Fojo, T.; Kebebew, E. MiR-34a and miR-483-5p are candidate serum biomarkers for adrenocortical tumors. Surgery 2013, 154, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, X.; Bi, T.; Ding, Y.; Zhao, J.; Wang, C.; Jia, T.; Han, D.; Guo, G.; Wang, B.; et al. MiRNA expression signature for potentially predicting the prognosis of ovarian serous carcinoma. Tumor Biol. 2013, 34, 3501–3508. [Google Scholar] [CrossRef]
- Shi, L.; Liu, S.; Zhao, W.; Shi, J. miR-483–5p and miR-486–5p are down-regulated in cumulus cells of metaphase II oocytes from women with polycystic ovary syndrome. Reprod. Biomed. Online 2015, 31, 565–572. [Google Scholar] [CrossRef] [PubMed]

| let7f | miR205 | ||||
|---|---|---|---|---|---|
| GAD DISEASE | |||||
| Category | Count | p-value | Category | Count | p-value |
| prostate cancer ES: 2.59 | 5 | 6.25 × 10−4 | prostate cancer ES: 2.94 | 6 | 0.011 |
| ovarian cancer ES: 2.59 | 4 | 0.003 | breast cancer ES: 2.37 | 10 | 6.72 × 10−6 |
| epithelial ovarian cancer ES: 2.59 | 3 | 0.008 | endometrial cancer ES: 1.91 | 4 | 2.36 × 10−4 |
| esophageal adenocarcinoma ES: 1.41 | 3 | 0.034 | esophageal adenocarcinoma ES: 1.91 | 7 | 3.14 × 10−4 |
| epithelial ovarian cancer ES: 1.91 | 5 | 0.001 | |||
| lung cancer ES: 1.91 | 7 | 0.002 | |||
| leukemia ES: 1.91 | 3 | 0.005 | |||
| colorectal CancerES: 1.91 | 5 | 0.026 | |||
| GOTERM Biological process | |||||
| Category | Count | p-value | Category | Count | p-value |
| GO:0000715 nucleotide-excision repair, DNA damage recognitionES: 3.98 | 3 | 1.62 × 10−4 | GO:0008285 negative regulation of cell proliferation ES: 3.73 | 9 | 1.51 × 10−6 |
| GO:0043066 negative regulation of apoptotic processES: 2.93 | 6 | 0.002 | |||
| GO:0042127 regulation of cell proliferationES: 1.92 | 4 | 0.007 | |||
| GO:0008284 positive regulation of cell proliferationES: 1.91 | 6 | 0.003 | |||
| GO:0030336 negative regulation of cell migrationES: 1.76 | 3 | 0.002 | |||
| KEGG Pathway | |||||
| Category | Count | p-value | Category | Count | p-value |
| hsa05215:Prostate cancer ES: 2.83 | 5 | 5.32 × 10−4 | |||
| hsa05222:Small cell lung cancer ES: 2.83 | 4 | 0.006 | |||
| hsa05200:Pathways in cancer ES: 2.37 | 9 | 2.03 × 10−4 | |||
| hsa05215:Prostate cancer ES: 1.91 | 5 | 5.32 × 10−4 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Márton, É.; Varga, A.; Soltész, B.; Penyige, A.; Lukács, J.; Póka, R.; Nagy, B.; Szilágyi, M. Comparative Analysis of Cell-Free miR-205-5p, let-7f-5p, and miR-483-5p Expression in Ovarian Cell Cultures and Plasma Samples of Patients with Ovarian Cancer. Appl. Sci. 2021, 11, 1735. https://doi.org/10.3390/app11041735
Márton É, Varga A, Soltész B, Penyige A, Lukács J, Póka R, Nagy B, Szilágyi M. Comparative Analysis of Cell-Free miR-205-5p, let-7f-5p, and miR-483-5p Expression in Ovarian Cell Cultures and Plasma Samples of Patients with Ovarian Cancer. Applied Sciences. 2021; 11(4):1735. https://doi.org/10.3390/app11041735
Chicago/Turabian StyleMárton, Éva, Alexandra Varga, Beáta Soltész, András Penyige, János Lukács, Róbert Póka, Bálint Nagy, and Melinda Szilágyi. 2021. "Comparative Analysis of Cell-Free miR-205-5p, let-7f-5p, and miR-483-5p Expression in Ovarian Cell Cultures and Plasma Samples of Patients with Ovarian Cancer" Applied Sciences 11, no. 4: 1735. https://doi.org/10.3390/app11041735
APA StyleMárton, É., Varga, A., Soltész, B., Penyige, A., Lukács, J., Póka, R., Nagy, B., & Szilágyi, M. (2021). Comparative Analysis of Cell-Free miR-205-5p, let-7f-5p, and miR-483-5p Expression in Ovarian Cell Cultures and Plasma Samples of Patients with Ovarian Cancer. Applied Sciences, 11(4), 1735. https://doi.org/10.3390/app11041735








