Abstract
The use of standard laboratory methods for trait evaluation is expensive and challenging, especially for low-resource breeding programs. For carotenoid assessment, rather than the standard HPLC method, these programs mostly rely on proxy approaches for quantitative total carotenoid content (TCC) assessment. To ensure data transferability and consistency, calibration models were developed using TCC iCheck and Chroma Meter proxy methods for the adoption of the alternative near-infrared phenotyping method in cassava. Calibration was developed for dry matter content (DMC) using a simple and inexpensive sampling procedure associated with the proxy TCC protocols. The partial least square (PLS) and random forest (RF) models were compared for the two traits, and the correlation (r) between the actual and predicted values in the training and validation (in bracket) sets of r = 0.85 (0.76) and r = 0.98 (0.82) with PLS and RF, respectively, for iCheck, and r = 0.99 (0.96) and r = 0.99 (0.96) with PLS and RF, respectively, for Chroma Meter, was obtained. The calibration result of r = 0.93 (0.83) and r = 0.99 (0.81) using PLS and RF, respectively, was obtained for DMC. This effort is valuable in carotenoids improvement and supports the ongoing effort in adopting portable spectrometers for rapid and cheap phenotyping in cassava.
1. Introduction
Cassava is a major food staple and a source of income for millions of people worldwide [1,2]. Inherently, cassava roots are a major starch sink and are thus used as a source of carbohydrate in human and animal diets. The crop has energy production of up to 1045 kJ/hectare, which places it higher than other starchy crops such as maize, sweet potato, sorghum, rice, and wheat [3]. The crop is rich in most important micronutrients, although in so many cases, the roots contain less zinc (14 ppm), iron (0.3 mg/100 g), and vitamin A (5–30 µg/100 g) than the leaves (vitamin A (8300 µg/100 g) and zinc (71 ppm)) [4]. Consequently, communities and countries that heavily depend on cassava roots as their staple food are highly vulnerable to micronutrient malnutrition, particularly vitamin A deficiency [5]. The routine intake of poor-quality diets that are characterized by low micronutrients results in a hidden hunger. Hidden hunger mainly affects populations that are economically challenged, and such communities can only access their staple foods and often do not have the means to grow or purchase more expensive micronutrient-rich foods [6]. Hidden hunger makes a significant contribution to the disease burden of children by limiting proper cognitive development, impairing physical development, and increasing susceptibility to infectious diseases [7]. Indeed, epidemiological investigations have found that low vitamin A status is frequently associated with increased disease incidence and is often associated with high mortality rates [8].
Since 2003, breeders across the Consultative Group on International Agricultural Research (CGIAR) and National Agricultural Research Systems (NARS) have devoted attention to developing food crop varieties with significant levels of bioavailable, critical micronutrients [6]. The micronutrients of focus are mainly vitamin A, iron, and zinc, which are recognized by the International Nutrition Community as most limiting in diets [7]. Biofortification provides a comparatively cost-effective, sustainable, and long-term means of delivering more of these critical micronutrients. It can be considered a home-grown therapy for addressing the key nutritional challenges that are commonplace in rural communities. It provides a feasible means of reaching malnourished rural populations who may have limited access to diverse diets, supplements, and commercially fortified foods [9].
Cassava is a major target crop for biofortification due to its significance in the diets and income of millions of people around the world. The current effort in increasing genetic gain from a selection in a large segregating cassava population using the standard high-performance liquid chromatography (HPLC) is not only challenging for the analyses of thousands of samples but very expensive, highly technical, and requires standard laboratory conditions and amenities which are often lacking in low-resource institutions and remote trial sites [10,11,12]. As such, investments in proxy options have been made to improve the evaluation of total carotenoid content (TCC) in cassava, including the use of iCheck [13], Chroma Meter [13], and spectrophotometer [14]. Alternatively, the use of near infra-red spectroscopy (NIRS), now available in a portable format, provides a rapid, accurate, and inexpensive alternative for both total and individual carotenoid evaluation in cassava [12,15,16,17].
Generally, the deployment of NIRS requires the development of reliable calibration equations from a standard reference analytical method. So far, the current effort in developing calibration for carotenoids using portable NIRS has relied on the collaborative effort between low-resource and more advanced resource institutions [16,18]. However, as most low-resource breeding programs continue to embrace the use of NIRS for carotenoid evaluations, most of the available reference values and genetic improvement studies were developed by the proxy iCheck and Chroma Meter protocols for TCC rather than HPLC values due to the lack of infrastructures and human capacity to run standard laboratories [19]. Therefore, for consistency and transferability of data, the need for alternative calibration using non-HPLC reference values becomes inevitable for the large-scale screening and breeding for TCC in cassava.
Consequently, this study was in response to the need to develop a direct NIRS calibration equation for TCC on reference values derived from low-cost and simple sampling proxy methods for TCC on wider African cassava breeding populations. We compared calibration performances from two evaluation protocols: iCheck and Chroma Meter, which have been previously deployed for TCC quantification in cassava roots [13,18]. The benefit of using a non-linear machine learning algorithm over the traditional partial least square (PLS) for carotenoids has been previously reported [16,20,21]; therefore, we compared calibrations from the random forest (RF) and PLS models. The effect of sample preparation on calibration performance has been reported where root mashing to obtain homogeneous samples was beneficial [16]. However, root mashing requires special devices and infrastructures which might not be available for off-site trials. Sample preparations for iCheck and Chroma Meter require relatively simple and low-cost chopping of roots with knives. Therefore, we equally developed calibration for root dry matter content (DMC) using the simple and cheap sampling method.
2. Materials and Methods
2.1. Study Population
A set of 190 clones with varying levels of visual root color was selected using a simple random sampling technique from the genetic gain population of the International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria. The genetic gain represents a diverse collection of clones developed at IITA over the years and has been fully described in many other studies [22,23]. This population was planted in the 2015 cropping season at Ubiaja, which is the IITA field station in Nigeria. Ubiaja has an altitude of 221 m and is located at a latitude of 6.6493° N and a longitude of 6.3918° E. This location has annual rainfall means ranging from 1800 mm to 2000 mm.
2.2. Sample Preparation
From a plot of ten plants, three were harvested, from which three sizable roots were selected for laboratory analyses. Within six hours, the selected sample roots were transported to the laboratory in IITA, Ibadan. Because of the distance between Ubiaja and Ibadan, the sampled roots were stored in a freezer at 0 °C for processing the next day. Before analyses, the roots were peeled, washed with tap water, and dried with a paper towel. During sample preparation, care was taken to prevent samples from being exposed directly to sunlight by harvesting early in the day and covering all laboratory openings with filters. Using a stainless kitchen knife, each root was cut longitudinally into two halves. The halves were then cut longitudinally into four quarters from each root, and the two opposite quarters of the two halves were selected and chopped into very tiny pieces (approximately <0.2 cm3) and later mixed uniformly. From this uniform sample, 150 g was packed into a transparent zipped polythene bag as a representative sample to be used for analyses.
2.3. iCheck TCC Assessment
A homogenized sample weighing 5 g was ground using a mortar and pestle. In the process of grinding, 10 mL of distilled water was added to make the grinding process easy. The sample was ground until it formed a fine paste. This paste was then transferred into a calibrated 50 mL falcon tube. Another 5 mL of distilled water was used to wash the mortar and then poured into the falcon tube. The volume of the falcon tube was adjusted to 25 mL. The contents of the falcon tube were then shaken thoroughly, and thereafter, 0.4 mL of the solution was injected into a disposable reagent vial using a needle and syringe. After shaking the reagent vial with its contents for 10 s and left to stand for 5 min, the sample TCC value was assessed with the iCheck device.
2.4. Chroma Meter TCC Measurement
A Chroma Meter (CR-400) was used to estimate TCC in cassava roots based on root color intensity (measured in b values). This device uses the LAB color space, which mathematically points out all perceivable colors in L*, a*, and b* dimensions (where the L* coordinate corresponds to a lightness coordinate, and the a* coordinate corresponds either to red (positive values) or to green (negative values)). TCC was measured in the b* dimension, where the negative values represent the blue color and the positive values represent the yellow color. Therefore, positive values in the b* dimension were used for the measurement of TCC [18,24].
2.5. Repeatability Check: iCheck and Chroma Meter
The repeatability precision (reproducibility) of the iCheck and Chroma Meter results were evaluated accordingly using the Horwitz equation [25], where the predicted relative standard deviation (PRSD) was compared with the actual relative standard deviation (RSD). On each genotype of 10 randomly selected sub-samples, TCC was measured five times on the same sample using both iCheck and Chroma Meter. PRSD was calculated by Horwitz’s equation: PRSD = 2C^ (−0.15), where C is the concentration of the analyte (mean) expressed as a decimal fraction. Herein, iCheck values were measured in µg/g (parts per million), and therefore to acquire C, these values were converted to µg/106 µg. On the other hand, Chroma Meter values were measured out of 100, and thus, to acquire C, these values were divided by 100. RSD was computed using the following formula:
RSD = (Standard deviation/Mean) × 100
Repeatability precision of a method is accepted when PRSD > RSD [26]. The repeatability (reproducibility) of these methods was further evaluated using the HorRat equation. This is one of the chemical methods that has been adopted by the Association of Analytical Chemists (AOAC) to test the repeatability of methods employed in food analyses [25]. Accordingly, a method is fully acceptable with high precision or reproducibility when 0.3 ≤ HorRat ≤ 1 [25,27]. The HorRat equation was calculated using the following formula:
HorRat = RSD/PRSD
2.6. Dry Matter Content Oven-Drying Reference
An electric-based large oven was used to determine root DMC, the percentage of dry weight relative to a given fresh weight of the root samples. Two replicates of about 100 g of chopped and homogenized roots were oven-dried at a constant temperature of 105 °C for 24 h at IITA, Ibadan, Nigeria. Samples were weighed before and after drying, and the average DMC of the two replications was used for analyses.
2.7. Spectra Collection
A portable NIRS device (QualitySpec Trek: S-10016) was used to collect spectrum data. The chopped and uniformly mixed cassava root samples were fed into NIRS sampling cups. The sampling cups with the chopped and homogenized cassava root sample was placed against the portable NIRS device window for spectra data collection. With 50 count average (how many times the entire wavelength range is scanned and then averaged together to produce a spectrum), the spectral acquisition time was 5 s.
2.8. NIRS Calibration
The spectrum data were first pre-treated using standard normal variate and detrending (SNVD) (D = 2, G = 5, S1 = 2, S2 = 1) to correct for external interferences on the spectrum data. D indicates the derivative order number (0 indicates no derivation, 1 means the first derivative, and so on), G indicates the gap (the number of data points over which derivation is computed), S1 indicates the number of data points in the first smoothing (1 means no smoothing), and S2 indicates the number of data points in the second smoothing (1 means no smoothing) [15,16].
Calibration equations were developed in the R statistical platform using the PLS and RF models from the caret package. The datasets were first screened for outliers using the Mahalanobis distance method [28,29], and the retained set (n = 177) was divided into calibration and validation sets in a ratio of 3:1, respectively. The repeated cross-validation method was used in developing equations for TCC using iCheck and Chroma Meter as well as for DMC. The performance of the cross-validated (10-fold cross-validation repeated 25×) equations was used in predicting the values of the validation set. The entire process from the data division into calibration and validation sets to the prediction of validation set values was repeated five times, and the reported statistics (the average of five by twenty-five cross-validation replications) include the root mean square error of cross-validation (RMSECV), the root mean square error of prediction (RMSEP), and the coefficient of correlation (r) in both the calibration and validations sets.
The root mean square error of cross-validation/prediction (RMSECV/RMSEP) was calculated as follows:
where is the predicted value of each sample in the calibration or validation sets, yi is the measured/actual value of each sample, and N is the sample number in the calibration and prediction sets.
3. Results
3.1. Summary Statistics and Correlation between TCC (iCheck and Chroma Meter Measures) and DMC
The summary statistics (Table 1) of the parameters showed that the value of TCC measured with iCheck ranged from 0.12 µg/g to 10.18 µg/g, and Chroma Meter ranged between 11.70 and 39.62. Similarly, the minimum and maximum values of DM were 7.56 and 42.70, respectively. The standard deviation values of TCC measured by iCheck and Chroma Meter were 2.47 and 6.78, respectively, whereas that of DMC was 7.08.

Table 1.
Summary statistics of traits.
There was a negative correlation between DMC and TCC measured by both iCheck and Chroma Meter. The correlation between DMC and iCheck TCC values was r = −0.37, and r = −0.33 between DMC and Chroma Meter TCC values. There was a high correlation (r = 0.72) between iCheck TCC values with Chroma Meter TCC values (Figure 1).
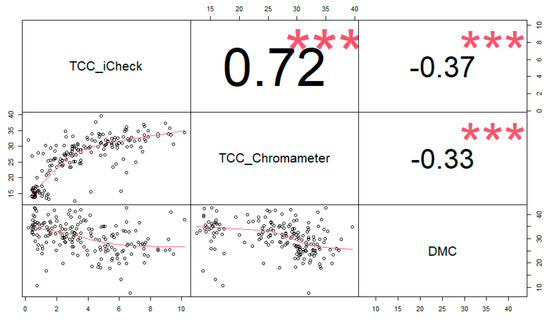
Figure 1.
Trait association between total carotenoid content (TCC) (measured with iCheck (TCC_iCheck) and Chroma Meter (TCC_Chroma Meter)) and root dry matter content (DMC).
3.2. iCheck and Chroma Meter Repeatability Assessment
The two proxy iCheck and Chroma Meter TCC values showed high repeatability precision, where both methods had high values of predicted relative standard deviation (PRSD) compared to the actual relative standard deviation (RSD) (Figure 2).
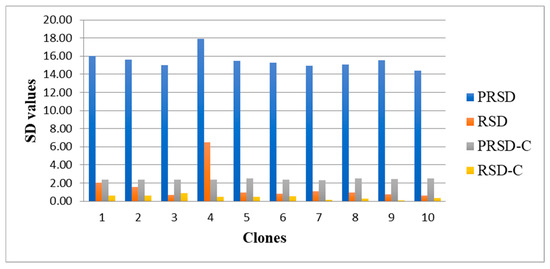
Figure 2.
Predicted relative standard deviation (PRSD) and relative standard deviation (RSD) for iCheck values; PRSD-C = predicted relative standard deviation; RSD-C = relative standard deviation for Chroma Meter values; and SD = standard deviation.
From the ten samples selected for repeatability precision analysis, the result showed that iCheck and Chroma Meter had values that fall within the acceptable AOAC requirement (Table 2).

Table 2.
HorRat values for iCheck and Chroma Meter.
3.3. Calibration Performance of TCC and DMC Using PLS and RF Models
The RMSECV and RMSEP values were generally higher with PLS than the RF models for all the traits (Table 3). The correlation between the actual and the predicted values ranged from 0.85 to 0.99 in training and 0.76 to 0.96 across the traits. In the training set, the mean correlation between the actual and predicted iCheck TCC values using RF and PLS models was r = 0.98 and r = 0.85, respectively (Figure 3a). In the validation set, the correlation between actual and predicted iCheck TCC values using the RF model and PLS model was r = 0.82 and r = 0.76, respectively (Figure 3b). In general, for TCC calibration from iCheck, the mean correlation between actual TCC (iCheck) and predicted values was higher using RF than PLS models.

Table 3.
The root mean square error of cross-validation (RMSECV), and the root mean square error of prediction (RMSEP) of TCC using iCheck and Chroma Meter evaluation methods and DMC under different calibration methods: partial least square (PLS) and random forest (RF).
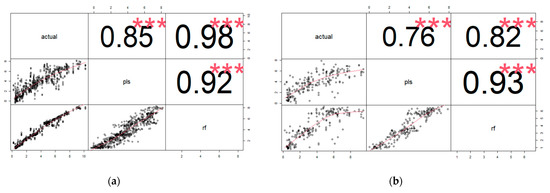
Figure 3.
Correlation between actual and predicted iCheck TCC values using PLS and RF calibration models in the training (a) and validation sets (b).
For TCC Chroma Meter, the correlation between the actual and predicted Chroma Meter values in the training set using the PLS model was equal to that of the RF model, r = 0.99 (Figure 4a). In the validation set, the correlation between actual and predicted Chroma Meter TCC values using the PLS model was r = 0.96, while that of the RF model was also r = 0.96 (Figure 4b).
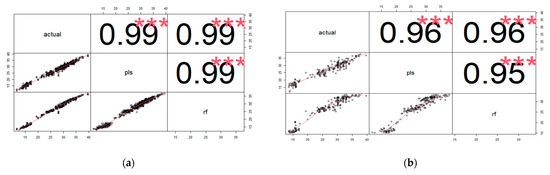
Figure 4.
Correlation between actual and predicted Chroma Meter TCC values using PLS and RF calibration models in the training (a) and validation sets (b).
In the training set, the correlation between actual DMC and predicted DMC values using the PLS model was r = 0.93 and r = 0.99 with RF (Figure 5a). In the validation set, the correlation was r = 0.83 for PLS and r = 0.81 for RF (Figure 5b). The correlation between PLS and RF models was 0.97.

Figure 5.
Correlation between actual and predicted DMC values using PLS and RF calibration models in the training (a) and validation sets (b).
4. Discussion
This study is important in sustaining the ongoing effort in TCC improvement, especially in low-resource breeding programs and off-site breeding stations. While the generation of the adequate calibration set from African germplasm that has trait values derived from a standard reference method such as HPLC might not be visible in the near future, the use of long-existing references from proxy alternatives for the calibration of the newly adopted near-infrared spectrometers will benefit the ongoing effort in TCC evaluation and genetic improvement. This study helps in developing strategies for the efficient phenotyping transition to NIRS technology, particularly the adoption of the portable NIRS. The early sampling approach needed for adequate sample homogeneity relies on the use of an electric-based blender [14,15] or a special construction for field-based mashing of roots [16].
The calibration results, especially the correlation between the predicted and actual values using a validation set from both iCheck and Chroma Meter for TCC evaluation as well as DMC, with r values ranging from 0.76 in iCheck to 0.97 in Chroma Meter using PLS in both cases, are robust enough for the assessment of these traits based on the portable NIRS [12,30,31]. The results are comparable to earlier calibration efforts in cassava roots for these traits, although calibrations from fine-ground and homogenized samples provided higher calibration performance [15,16,24]. The use of random forest, a non-linear model for calibration, has been of benefit in some cases [20,21], but the benefit over the conventional PLS model was not obvious in this study. The correlation between predicted values from the two models was highly correlated (r > 0.92), especially the correlation between the predicted DMC values in the validation set (r = 0.97).
The range of values for both TCC and DMC was similar to those of previous studies and represents the range of values in most of the base germplasm collection of the African Consultative Group on International Agricultural Research (CGIAR) and National Agricultural Research Systems (NARS) centers [19,23,32]. For advanced populations with higher trait values, updating the models is always recommendable. There was an adequate mix of white- and yellow-fleshed roots in the calibration dataset, although iCheck provides more quantitative values. Quantitative evaluation techniques for both total and individual carotenoids are very important in the biofortification and genetic advancement efforts in cassava [17,33,34].
The negative correlation between TCC from the two evaluation methods and DMC has been reported several times in the African cassava germplasm compared to what has been reported in the South American germplasm [19,34,35]. There are ongoing efforts to understand and design an appropriate breeding approach in reversing this trend, as this has implications on cassava variety adoption [35,36]. An ideal variety needs to meet both the target high carotenoids levels as well as the desired quality attributes sought by cassava consumers. Meeting this goal will require extensive and well-designed crosses, evaluation, and selection schemes which further highlight the importance of cheap but rapid and standardized evaluation protocols.
The two proxy methods for TCC had high repeatability precision, with PRSD values higher than the corresponding RSD (Figure 2) and HorRat values falling within the fully acceptable 0.3 ≤ HorRat ≤ 1 region [25,27]. Therefore, these proxy methods, according to AOAC standards, are acceptable and repeatable since repeatability precision is inversely proportional to random errors [25]. Moreover, these methods have been shown to have a high correlation with the standard HPLC method [13,18,36]. The use of the full range portable spectroscopy (350–2500 nm) provides a rapid, flexible, field-based, and simultaneous evaluation of many traits. The sampling process used in the proxy TCC evaluation methods depends on low-cost and available procedures; therefore, the calibration of the portable NIRS from these proxy methods could be sustainable in generating adequate calibration sets with wide application to support previous and future research efforts on these traits.
5. Conclusions
Using a cheap and simple sampling approach, robust results were obtained from the calibration of a portable NIRS device for TCC from iCheck and Chroma Meter, and DMC from the oven-drying method. On average, higher calibration performance was obtained using Chroma Meter than iCheck for TCC evaluation. By leveraging on the high repeatability and the already established correlation with the standard HPLC method, the development of calibration from the two TCC assessment methods as well as DMC from low-cost sampling methods holds many benefits for the resource-constrained research institutes that cannot afford HPLC and other expensive reference methods for NIRS calibration. This work supports the existing effort in the deployment of the portable NIRS device for rapid, accurate, and flexible phenotyping alternatives in cassava.
Author Contributions
Conceptualization, W.A. and U.N.I.; methodology, W.A., U.N.I., R.K., C.E., I.R., E.P., P.K., R.E., P.G., and B.-E.O.; data collection and analyses, W.A. and U.N.I.; writing—original draft preparation, W.A. and U.N.I.; writing—review and editing, W.A., U.N.I., I.R., and P.K.; supervision, R.K., C.E., I.R., E.P., P.K., R.E., P.G., and B.-E.O.; funding acquisition, R.K., C.E., I.R., and P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Bill and Melinda Gates Foundation (BMGF) and UKAID (Grant 1048542, http://www.gatesfoundation.org) as part of the Next Generation Cassava Breeding project.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset for this study can be found using the link: ftp://ftp.cassavabase.org/manuscripts/Abincha_et_al_2021_NIRS_Calibration_Set.csv.
Acknowledgments
We acknowledge the contributions and support of the staff of the International Institute of Tropical Agriculture, IITA, Ibadan, for their efforts in data collection and processing of samples.
Conflicts of Interest
The authors declare no competing interests.
References
- Burns, A.; Gleadow, R.; Cliff, J.; Zacarias, A.; Cavagnaro, T. Cassava: The drought, war and famine crop in a changing world. Sustainability 2010, 2, 3572–3607. [Google Scholar] [CrossRef]
- Nweke, F. New Challenges in the Cassava Transformation in Nigeria and Ghana; International Food Policy Research Institute EPTD: Washington, DC, USA, 2004. [Google Scholar]
- Montagnac, J.A.; Davis, C.R.; Tanumihardjo, S.A. Nutritional value of cassava for use as a staple food and recent advances for improvement. Compr. Rev. Food Sci. Food Saf. 2009, 8, 181–194. [Google Scholar] [CrossRef]
- Salvador, E.M.; Steenkamp, V.; McCrindle, C.M.E. Production, consumption and nutritional value of cassava (Manihot esculenta, Crantz) in Mozambique: An overview. J. Agric. Biotechnol. Sustain. Dev. 2014, 6, 29–38. [Google Scholar] [CrossRef]
- Gegios, A.; Amthor, R.; Maziya-Dixon, B.; Egesi, C.; Mallowa, S.; Nungo, R.; Gichuki, S.; Mbanaso, A.; Manary, M.J. Children consuming cassava as a staple food are at risk for inadequate zinc, iron, and vitamin A intake. Plant Foods Hum. Nutr. 2010, 65, 64–70. [Google Scholar] [CrossRef]
- Asare-marfo, D.; Gonzalez, C.; Perez, S.; Schwarz, J.; Zeller, M. Prioritizing Countries for Biofortification Interventions Using Country-Level Data Dorene. In Harvestplus Working Paper; Harvest Plus: Washington, DC, USA, 2013. [Google Scholar]
- Black, R.E.; Victora, C.G.; Walker, S.P.; Bhutta, Z.A.; Christian, P.; de Onis, M.; Ezzati, M.; Grantham-McGregor, S.; Katz, J.; Martorell, R.; et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet 2013, 382, 427–451. [Google Scholar] [CrossRef]
- McCormick, D.B. The Vitamins: Fundamental Aspects in Nutrition and Health. Am. J. Clin. Nutr. 1999, 70, 426. [Google Scholar] [CrossRef]
- Saltzman, A.; Birol, E.; Bouis, H.E.; Boy, E.; de Moura, F.F.; Islam, Y.; Pfeiffer, W.H. Biofortification: Progress toward a more nourishing future. Glob. Food Secur. 2013, 2, 9–17. [Google Scholar] [CrossRef]
- Boligon, A.A.; Athayde, L.; Athayde, M.L. Importance of HPLC in Analysis of Plants Extracts. Austin Chromatogr. 2014, 1, 2–33. [Google Scholar]
- Bhatnagar-Panwar, M.; Bhatnagar-Mathur, P.; Bhaaskarla, V.V.; Dumbala, S.R.; Sharma, K.K. Rapid, accurate and routine HPLC method for large-scale screening of pro-vitamin A carotenoids in oilseeds. J. Plant Biochem. Biotechnol. 2013, 24, 84–92. [Google Scholar] [CrossRef]
- Lebot, V. Near-Infrared Spectroscopy for Quality Evaluation of Root Crops: Practical Constraints, Preliminary Studies, and Future Prospects. J. Root Crop. 2012, 38, 3–14. [Google Scholar]
- Kulakow, P.; Parkes, E.Y.; Friedrich, S.; Schweigert, F.J.; Alamu, E.O.; Maroya, N.G.; Maziya-Dixon, B. Linearity, Reproducibility, and Comparison of iCheckTM CAROTENE with Spectrophotometer and HPLC for Evaluation of Total Carotenoids in Cassava Roots. Eur. J. Nutr. Food Saf. 2015, 5, 1040–1041. [Google Scholar] [CrossRef]
- Belalcazar, J.; Dufour, D.; Andersson, M.S.; Pizarro, M.; Luna, J.; Londoño, L.; Morante, N.; Jaramillo, A.M.; Pino, L.; López-Lavalle, L.A.B. High-throughput phenotyping and improvements in breeding cassava for increased carotenoids in the roots. Crop Sci. 2016, 56, 2916–2925. [Google Scholar] [CrossRef]
- Davrieux, F.; Dufour, D.; Dardenne, P.; Belalcazar, J.; Pizarro, M.; Luna, J.; Londoño, L.; Jaramillo, A.; Sanchez, T.; Morante, N.; et al. LOCAL regression algorithm improves near infrared spectroscopy predictions when the target constituent evolves in breeding populations. J. Near Infrared Spectrosc. 2016, 24, 109–117. [Google Scholar] [CrossRef]
- Ikeogu, U.N.; Davrieux, F.; Dufour, D.; Ceballos, H.; Egesi, C.N.; Jannink, J.L. Rapid analyses of dry matter content and carotenoids in fresh cassava roots using a portable visible and near infrared spectrometer (Vis/NIRS). PLoS ONE 2017, 12, e0188918. [Google Scholar] [CrossRef] [PubMed]
- Ikeogu, U.N.; Akdemir, D.; Wolfe, M.D.; Okeke, U.G.; Chinedozi, A.; Jannink, J.; Egesi, C.N. Genetic Correlation, Genome-Wide Association and Genomic Prediction of Portable NIRS Predicted Carotenoids in Cassava Roots. Front. Plant Sci. 2019, 10, 1570. [Google Scholar] [CrossRef]
- Abincha, W.; Kawuki, R.; Ikeogu, U.; Obara, J.; Hussin, Y.; Egesi, C.; Kulakow, P.; Rabbi, I.; Owor, B.E.; Parkes, E. Comparison of Near-infrared Spectroscopy with other options for total carotenoids content phenotyping in fresh cassava roots. J. Sci. Agric. 2020, 4, 6196. [Google Scholar] [CrossRef]
- Rabbi, I.Y.; Udoh, L.I.; Wolfe, M.; Parkes, E.Y.; Gedil, M.A.; Dixon, A.; Ramu, P.; Jannink, J.L.; Kulakow, P. Genome-wide association mapping of correlated traits in Cassava: Dry matter and total carotenoid content. Plant Genome 2017, 10, 94. [Google Scholar] [CrossRef]
- Ghasemi, J.B.; Tavakoli, H. Application of random forest regression to spectral multivariate calibration. Anal. Methods 2013, 5, 1863. [Google Scholar] [CrossRef]
- Sanguk, L.; Hangseok, C.; Kyungjoon, C.; Kyeong, K.M.; Soo, K.J.; Hee, Y.C.; Lee, S.H.; Hoeil, C. Random forest as a non-parametric algorithm for near-infrared (NIR) spectroscopic discrimination for geographical origin of agricultural samples. Bull. Korean Chem. Soc. 2012, 33, 4267–4270. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Rabbi, I.Y.; Egesi, C.; Hamblin, M.; Kawuki, R.; Kulakow, P.; Lozano, R.; del Carpio, D.P.; Ramu, P.; Jannink, J.L. Genome-Wide Association and Prediction Reveals Genetic Architecture of Cassava Mosaic Disease Resistance and Prospects for Rapid Genetic Improvement. Plant Genome 2016, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.D.; del Carpio, D.P.; Alabi, O.; Ezenwaka, L.C.; Ikeogu, U.N.; Kayondo, I.S.; Lozano, R.; Okeke, U.G.; Ozimati, A.A.; Williams, E.; et al. Prospects for genomic selection in cassava breeding. Plant Genome 2017, 10, 15. [Google Scholar] [CrossRef]
- Sánchez, T.; Ceballos, H.; Dufour, D.; Ortiz, D.; Morante, N.; Calle, F.; Felde, T.Z.; Domínguez, M.; Davrieux, F. Prediction of carotenoids, cyanide and dry matter contents in fresh cassava root using NIRS and Hunter color techniques. Food Chem. 2014, 151, 444–451. [Google Scholar] [CrossRef]
- Horwitz, W.; Albert, R. The Horwitz Ratio (HorRat)A Useful Index of Method Performance with Respect to Precision.PDF. J. Assoc. Anal. Chem. 2006, 89, 1095–1109. [Google Scholar]
- Pojic, M.; Mastilovi, J.; Majcen, N. The Application of Near Infrared Spectroscopy in Wheat Quality Control. In Infrared Spectroscopy-Life and Biomedical Sciences, 1st ed.; Theophile, P.T., Ed.; In Tech: London, UK, 2012; pp. 167–185. [Google Scholar]
- Horwitz, W.; Kamps, L.R.; Boyer, K.W. Quality assurance in the analysis of foods and trace constituents. J. Assoc. Off. Anal. Chem. 1980, 63, 1344–1354. [Google Scholar] [CrossRef]
- Kim, M.G. Multivariate outliers and decompositions of mahalanobis distance. Commun. Stat. Theory Methods 2000, 29, 1511–1526. [Google Scholar] [CrossRef]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D.L. The Mahalanobis distance. Chemom. Intell. Lab. Syst. 2000, 50, 1–18. [Google Scholar] [CrossRef]
- Duan, J.; Huang, Y.; Zheng, Z.L.B.; Li, Q.; Xiong, Y.; Wu, L.; Min, S. Determination of 27 chemical constituents in Chinese southwest tobacco by FT-NIR spectroscopy. Ind. Crops Prod. 2012, 40, 21–26. [Google Scholar] [CrossRef]
- Fox, G.P.; O’Donnell, N.H.; Stewart, P.N.; Gleadow, R.M. Estimating hydrogen cyanide in forage sorghum (Sorghum bicolor) by near-infrared spectroscopy. J. Agric. Food Chem. 2012, 60, 6183–6187. [Google Scholar] [CrossRef] [PubMed]
- Esuma, W.; Rubaihayo, P.; Pariyo, A.; Kawuki, R.; Wanjala, B.; Nzuki, I.; Harvey, J.; Baguma, Y. Genetic Diversity of Provitamin A Cassava in Uganda. J. Plant Stud. 2012, 1, 60. [Google Scholar] [CrossRef]
- Ceballos, H.; Davrieux, F.; Talsma, E.F.; Belalcazar, J.; Chavarriaga, P.; Andersson, M.S. Carotenoids in Cassava Roots. In Carotenoids; IntechOpen: London, UK, 2017; Volume 3. [Google Scholar]
- Ceballos, H.; Morante, N.; Sánchez, T.; Ortiz, D.; Aragón, I.; Chávez, A.L.; Pizarro, M.; Calle, F.; Dufour, D. Rapid cycling recurrent selection for increased carotenoids content in cassava roots. Crop Sci. 2013, 53, 2342–2351. [Google Scholar] [CrossRef]
- Njoku, D.N.; Vernon, G.; Egesi, C.N.; Asante, I.; Offei, S.K.; Okogbenin, E.; Kulakow, P.; Eke-okoro, O.N.; Ceballos, H. Breeding for enhanced β-carotene content in cassava: Constraints and accomplishments. J. Crop Improv. 2011, 25, 560–571. [Google Scholar] [CrossRef]
- Jaramillo, A.M.; Londoño, L.F.; Orozco, J.C.; Patiño, G.; Belalcazar, J.; Davrieux, F.; Talsma, E.F. A comparison study of five different methods to measure carotenoids in biofortified yellow cassava (Manihot esculenta). PLoS ONE 2018, 13, e0209702. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).