Synthesize and Segment: Towards Improved Catheter Segmentation via Adversarial Augmentation
Abstract
:1. Introduction
- −
- Synthetic X-ray from labels: we propose to generate synthetic X-ray from in-painted catheter masks via adversarial learning with CycleGAN as data augmentation for segmentation.
- −
- Improved generation with perceptual losses: to achieve more realistic generation from in-painted catheter masks, we incorporate a perceptual loss alongside the standard cycle loss.
- −
- Enforcing semantic similarity: we further propose a similarity loss to alleviate large deviations in the semantic quality of the generated images from the original.
- −
- Empirical results and several ablations show the effectiveness of the proposed training scheme with segmentation performance improving as synthetic augmentation is increased.
2. Related Work
2.1. Learning Based Methods for Segmentation and Detection
2.2. Image Translation in Medical Imaging
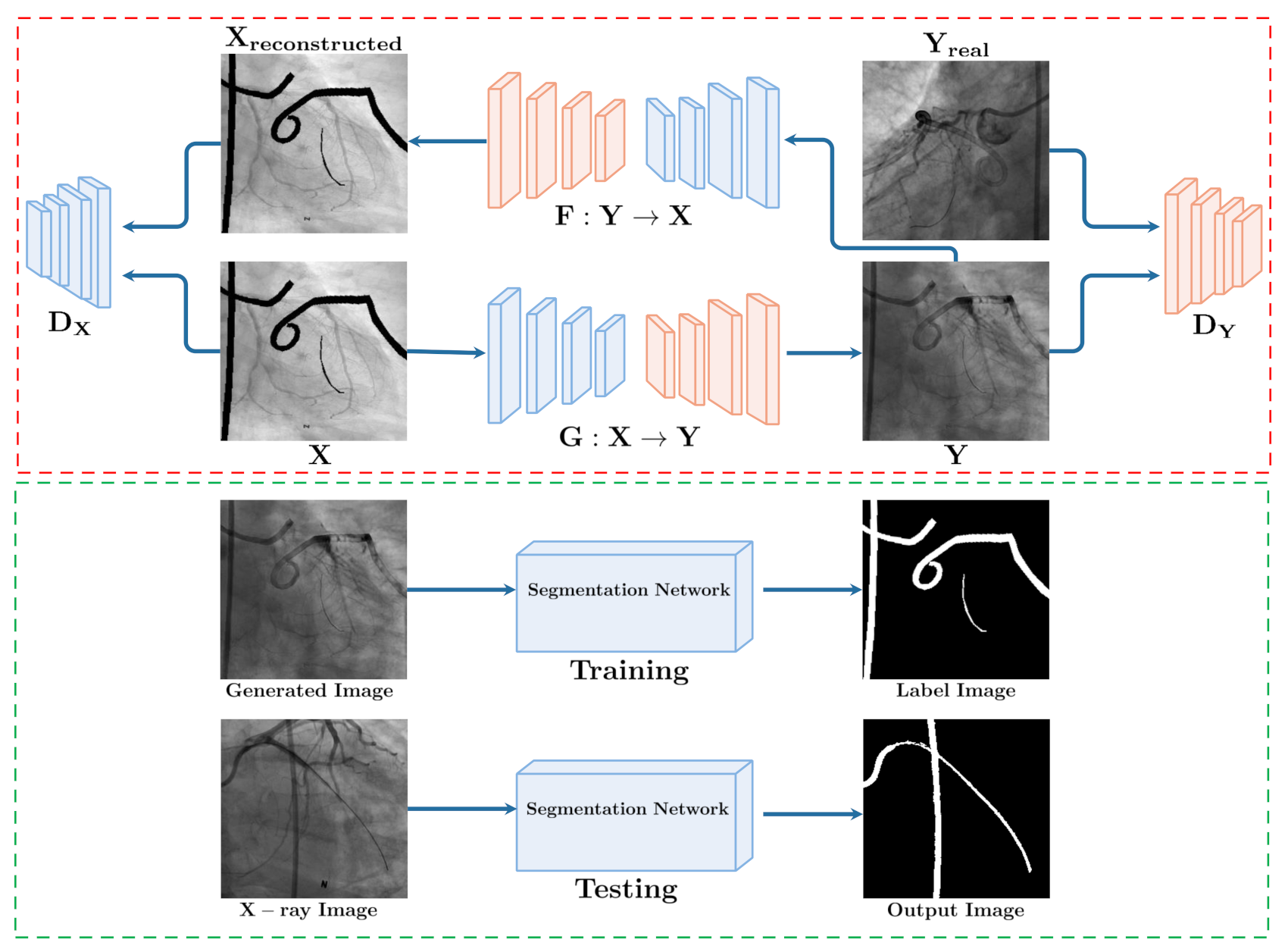
3. Methods
3.1. Synthesize: GAN Based X-ray Translation
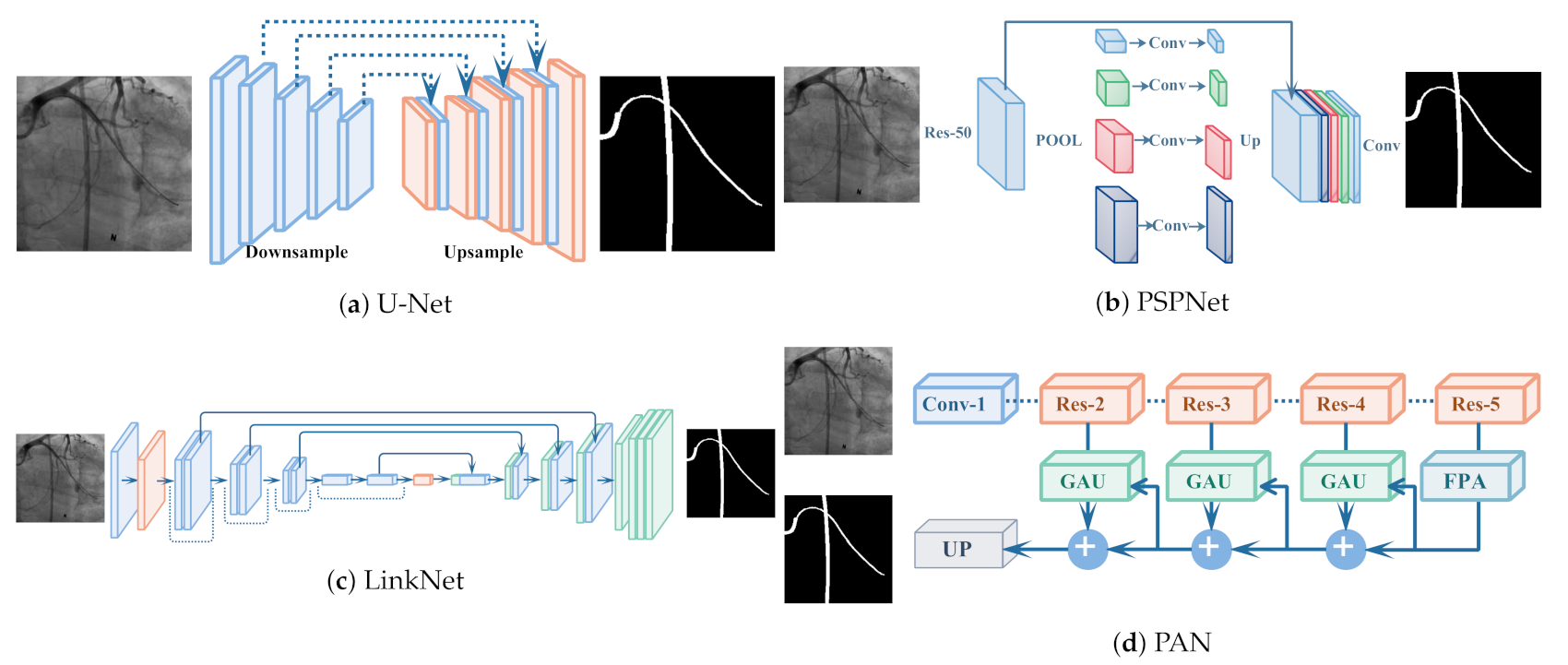
3.2. Segment: From Synthesis to Segmentation
4. Experiments
4.1. Datasets
4.2. Experimental Setup
4.3. Quantitative Results
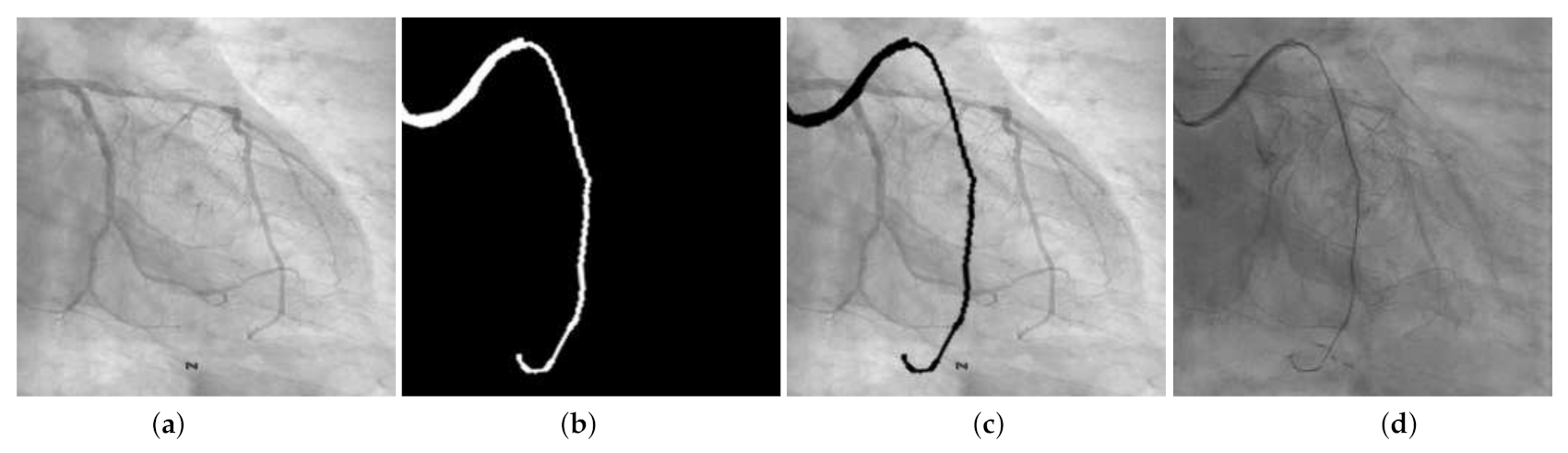
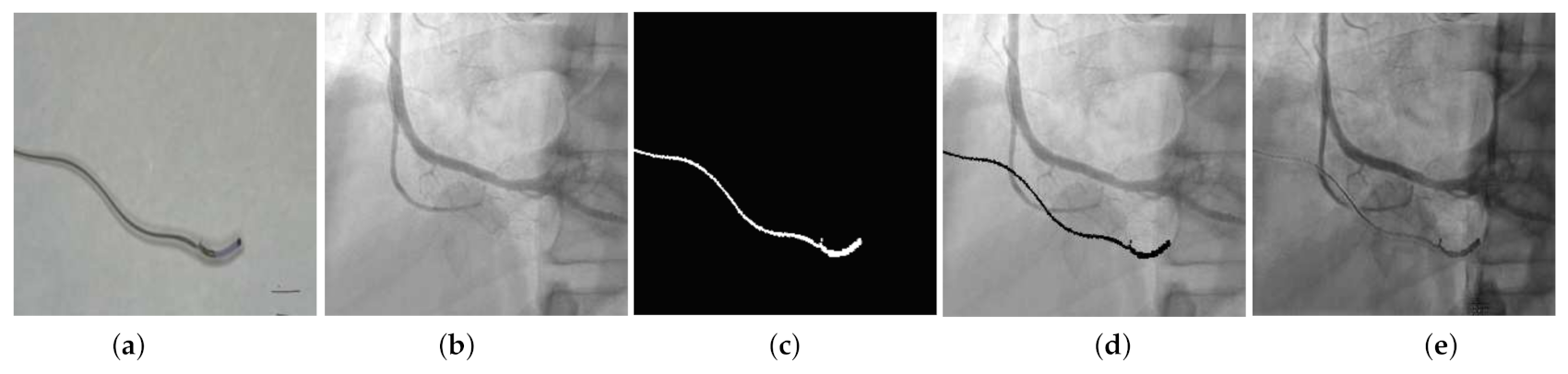
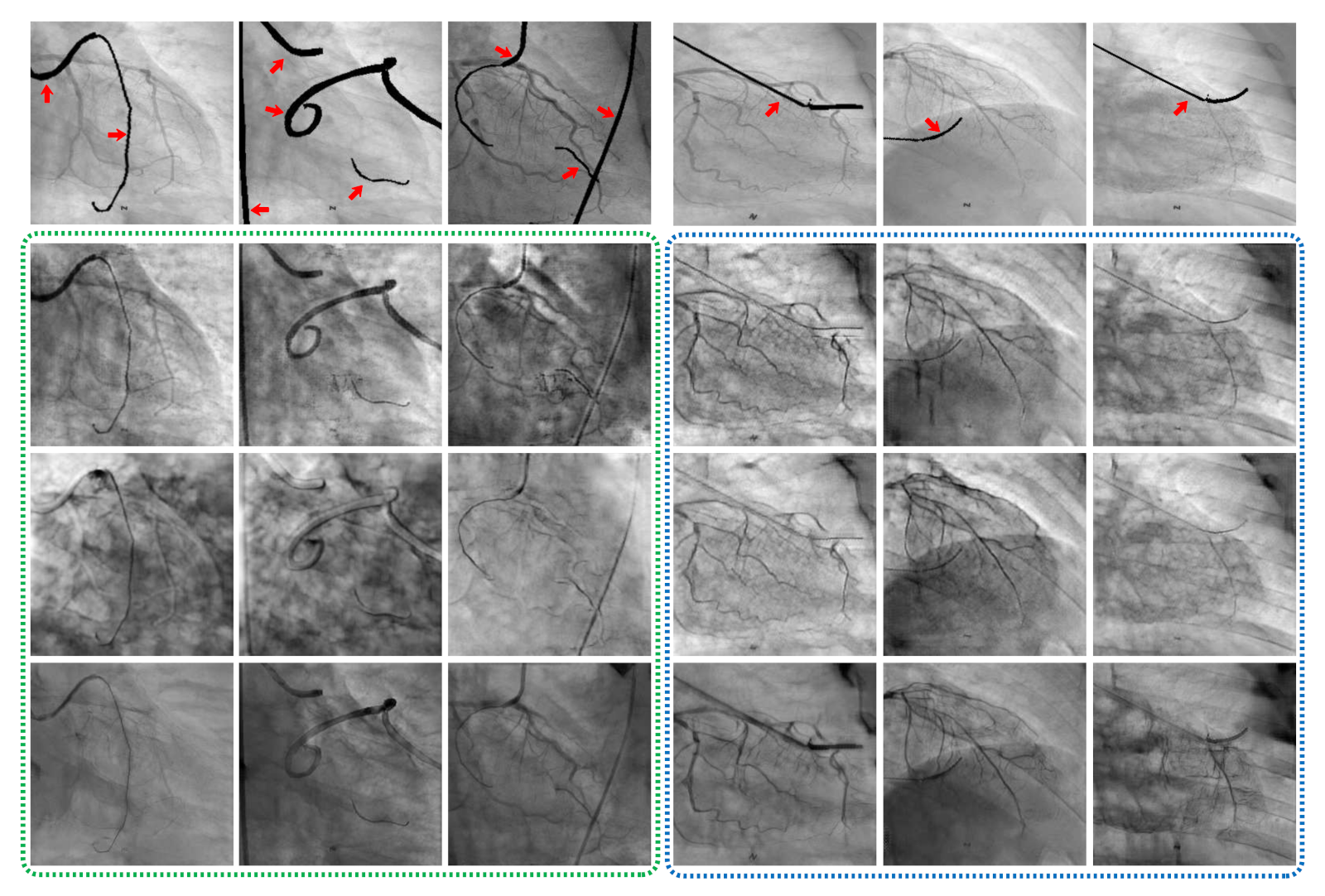
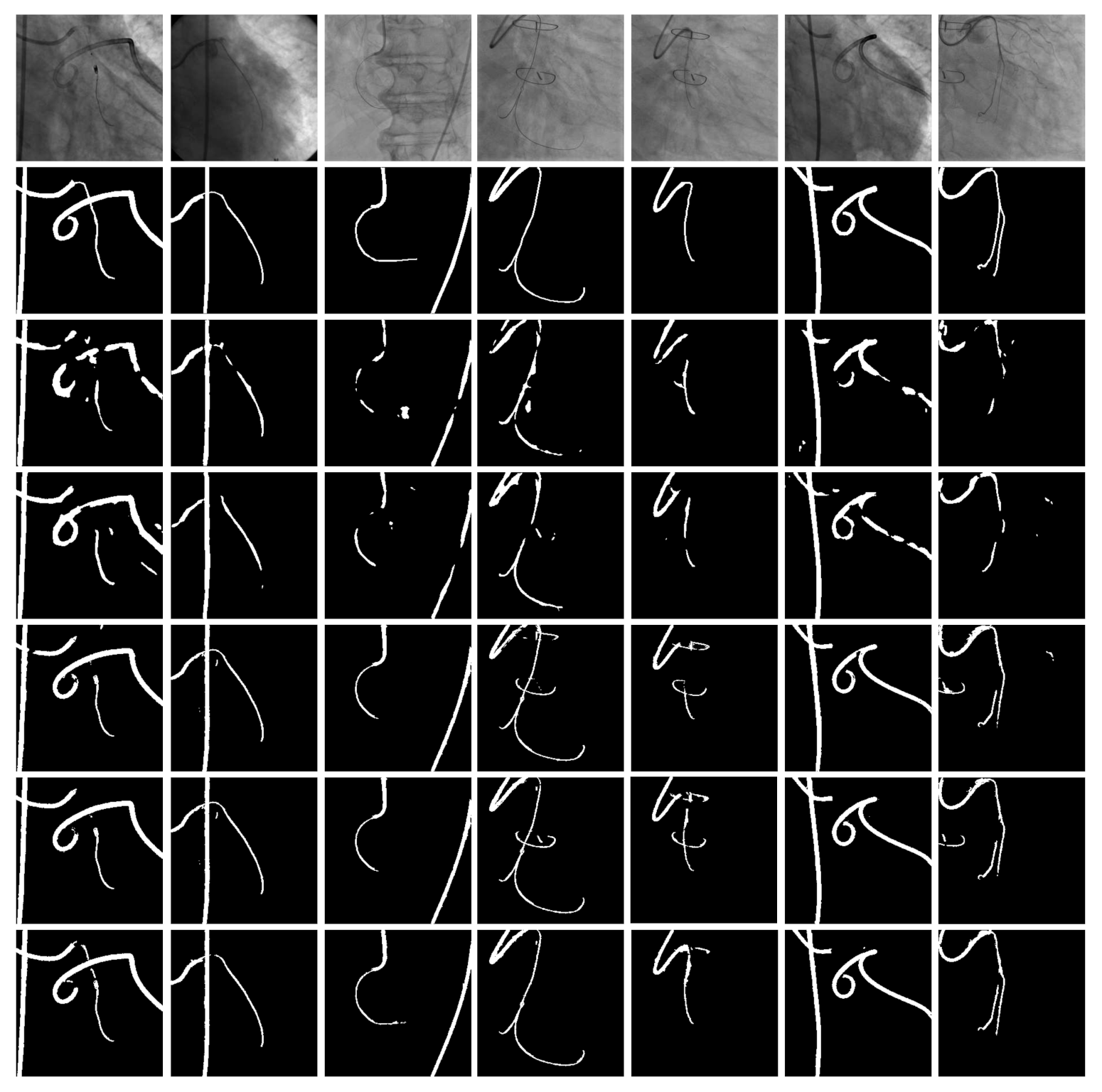
4.4. Qualitative Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kern, M.M.; Gustafson, L.; Kapur, R.; Wasek, S. Angiographic projections made simple: An easy guide to understanding oblique views. Cath Lab Digest 2011, 19. [Google Scholar]
- Zhou, Y.J.; Xie, X.L.; Bian, G.B.; Hou, Z.G.; Wu, Y.D.; Liu, S.Q.; Zhou, X.H.; Wang, J.X. Fully Automatic Dual-Guidewire Segmentation for Coronary Bifurcation Lesion. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–6. [Google Scholar]
- Guo, S.; Tang, S.; Zhu, J.; Fan, J.; Ai, D.; Song, H.; Liang, P.; Yang, J. Improved U-Net for Guidewire Tip Segmentation in X-ray Fluoroscopy Images. In Proceedings of the 2019 3rd International Conference on Advances in Image Processing, Chengdu, China, 8–10 November 2019; pp. 55–59. [Google Scholar]
- Kao, E.F.; Jaw, T.S.; Li, C.W.; Chou, M.C.; Liu, G.C. Automated detection of endotracheal tubes in paediatric chest radiographs. Comput. Methods Programs Biomed. 2015, 118, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, R.R. Image-Based Medical Device Localization. U.S. Patent 7,190,819, 13 March 2007. [Google Scholar]
- Uherčík, M.; Kybic, J.; Zhao, Y.; Cachard, C.; Liebgott, H. Line filtering for surgical tool localization in 3D ultrasound images. Comput. Biol. Med. 2013, 43, 2036–2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandini, A.; Glocker, B.; Hamady, M.; Yang, G.Z. Robust guidewire tracking under large deformations combining segment-like features (SEGlets). Med Image Anal. 2017, 38, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.G.; Laeseke, P.; Speidel, M.A. Deep learning based guidewire segmentation in x-ray images. In Proceedings of the Medical Imaging 2019: Physics of Medical Imaging. International Society for Optics and Photonics, San Diego, CA, USA, 16–21 February 2019; Volume 10948, p. 1094844. [Google Scholar]
- Subramanian, V.; Wang, H.; Wu, J.T.; Wong, K.C.; Sharma, A.; Syeda-Mahmood, T. Automated Detection and Type Classification of Central Venous Catheters in Chest X-Rays. arXiv 2019, arXiv:1907.01656. [Google Scholar]
- Breininger, K.; Würfl, T.; Kurzendorfer, T.; Albarqouni, S.; Pfister, M.; Kowarschik, M.; Navab, N.; Maier, A. Multiple device segmentation for fluoroscopic imaging using multi-task learning. In Intravascular Imaging and Computer Assisted Stenting and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis; Springer: Granada, Spain, 2018; pp. 19–27. [Google Scholar]
- Gozes, O.; Greenspan, H. Bone Structures Extraction and Enhancement in Chest Radiographs via CNN Trained on Synthetic Data. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 858–861. [Google Scholar]
- Vlontzos, A.; Mikolajczyk, K. Deep segmentation and registration in X-ray angiography video. arXiv 2018, arXiv:1805.06406. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 2672–2680. [Google Scholar]
- Tmenova, O.; Martin, R.; Duong, L. CycleGAN for style transfer in X-ray angiography. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Lee, D.H.; Li, Y.; Shin, B.S. Generalization of intensity distribution of medical images using GANs. Hum. Centric Comput. Inf. Sci. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Ullah, I.; Chikontwe, P.; Park, S.H. Catheter Synthesis in X-Ray Fluoroscopy with Generative Adversarial Networks. In International Workshop on Predictive Intelligence In Medicine; Springer: Shenzhen, China, 2019; pp. 125–133. [Google Scholar]
- Mercan, C.A.; Celebi, M.S. An approach for chest tube detection in chest radiographs. IET Image Process. 2013, 8, 122–129. [Google Scholar] [CrossRef]
- Nguyen, A.; Kundrat, D.; Dagnino, G.; Chi, W.; Abdelaziz, M.E.; Guo, Y.; Ma, Y.; Kwok, T.M.; Riga, C.; Yang, G.Z. End-to-End Real-time Catheter Segmentation with Optical Flow-Guided Warping during Endovascular Intervention. In Proceedings of the 2020 IEEE International Conference on Robotics and Automation (ICRA), Paris, France, 31 May–31 August 2020; pp. 9967–9973. [Google Scholar]
- Mountney, P.; Maier, A.; Ionasec, R.I.; Boese, J.; Comaniciu, D. Method and System for Obtaining a Sequence of X-ray Images Using a Reduced Dose of Ionizing Radiation. US Patent 9,259,200, 16 February 2016. [Google Scholar]
- Wang, L.; Xie, X.L.; Bian, G.B.; Hou, Z.G.; Cheng, X.R.; Prasong, P. Guide-wire detection using region proposal network for X-ray image-guided navigation. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 3169–3175. [Google Scholar]
- Ullah, I.; Chikontwe, P.; Park, S.H. Real-time tracking of guidewire robot tips using deep convolutional neural networks on successive localized frames. IEEE Access 2019, 7, 159743–159753. [Google Scholar] [CrossRef]
- Lee, H.; Mansouri, M.; Tajmir, S.; Lev, M.H.; Do, S. A deep-learning system for fully-automated peripherally inserted central catheter (PICC) tip detection. J. Digit. Imaging 2018, 31, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, S. Deep learning based non-rigid device tracking in ultrasound image. In Proceedings of the 2018 2nd International Conference on Computer Science and Artificial Intelligence, Shenzhen, China, 8–10 December 2018; pp. 354–358. [Google Scholar]
- Ambrosini, P.; Ruijters, D.; Niessen, W.J.; Moelker, A.; van Walsum, T. Fully automatic and real-time catheter segmentation in X-ray fluoroscopy. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Quebec City, QC, Canada, 2017; pp. 577–585. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Munich, Germany, 2015; pp. 234–241. [Google Scholar]
- Wu, Y.D.; Xie, X.L.; Bian, G.B.; Hou, Z.G.; Cheng, X.R.; Chen, S.; Liu, S.Q.; Wang, Q.L. Automatic guidewire tip segmentation in 2D X-ray fluoroscopy using convolution neural networks. In Proceedings of the 2018 International Joint Conference on Neural Networks (IJCNN), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–7. [Google Scholar]
- Breininger, K.; Albarqouni, S.; Kurzendorfer, T.; Pfister, M.; Kowarschik, M.; Maier, A. Intraoperative stent segmentation in X-ray fluoroscopy for endovascular aortic repair. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Zaman, A.; Park, S.H.; Bang, H.; Park, C.; Park, I.; Joung, S. Generative approach for data augmentation for deep learning-based bone surface segmentation from ultrasound images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Wolterink, J.M.; Dinkla, A.M.; Savenije, M.H.; Seevinck, P.R.; van den Berg, C.A.; Išgum, I. Deep MR to CT synthesis using unpaired data. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Québec City, QC, Canada, 2017; pp. 14–23. [Google Scholar]
- Dar, S.U.; Yurt, M.; Karacan, L.; Erdem, A.; Erdem, E.; Çukur, T. Image synthesis in multi-contrast MRI with conditional generative adversarial networks. IEEE Trans. Med Imaging 2019, 38, 2375–2388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiasa, Y.; Otake, Y.; Takao, M.; Matsuoka, T.; Takashima, K.; Carass, A.; Prince, J.L.; Sugano, N.; Sato, Y. Cross-modality image synthesis from unpaired data using CycleGAN. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Granada, Spain, 2018; pp. 31–41. [Google Scholar]
- Chartsias, A.; Joyce, T.; Dharmakumar, R.; Tsaftaris, S.A. Adversarial image synthesis for unpaired multi-modal cardiac data. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Québec City, QC, Canada, 2017; pp. 3–13. [Google Scholar]
- Gherardini, M.; Mazomenos, E.; Menciassi, A.; Stoyanov, D. Catheter segmentation in X-ray fluoroscopy using synthetic data and transfer learning with light U-nets. In Computer Methods and Programs in Biomedicine; Elsevier: Amsterdam, The Netherlands, 2020; p. 105420. [Google Scholar]
- Yi, X.; Adams, S.; Babyn, P.; Elnajmi, A. Automatic Catheter and Tube Detection in Pediatric X-ray Images Using a Scale-Recurrent Network and Synthetic Data. J. Digit. Imaging 2019, 33, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frid-Adar, M.; Amer, R.; Greenspan, H. Endotracheal Tube Detection and Segmentation in Chest Radiographs using Synthetic Data. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Shenzhen, China, 2019; pp. 784–792. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Zhao, H.; Gallo, O.; Frosio, I.; Kautz, J. Loss functions for image restoration with neural networks. IEEE Trans. Comput. Imaging 2016, 3, 47–57. [Google Scholar] [CrossRef]
- Johnson, J.; Alahi, A.; Fei-Fei, L. Perceptual losses for real-time style transfer and super-resolution. In European Conference on Computer Vision; Springer: Amsterdam, The Netherlands, 2016; pp. 694–711. [Google Scholar]
- Chaurasia, A.; Culurciello, E. Linknet: Exploiting encoder representations for efficient semantic segmentation. In Proceedings of the 2017 IEEE Visual Communications and Image Processing (VCIP), St. Petersburg, FL, USA, 10–13 December 2017; pp. 1–4. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Li, H.; Xiong, P.; An, J.; Wang, L. Pyramid attention network for semantic segmentation. arXiv 2018, arXiv:1805.10180. [Google Scholar]
- Frangi, A.F.; Niessen, W.J.; Vincken, K.L.; Viergever, M.A. Multiscale vessel enhancement filtering. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 1998; pp. 130–137. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and flexible image augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef] [Green Version]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best Practices for Convolutional Neural Networks Applied to Visual Document Analysis. In Proceedings of the ICDAR, Edinburgh, UK, 3–6 August 2003; Volume 3. [Google Scholar]


| Model | Training Images | Dice Score |
|---|---|---|
| 140 Labeled Images (Baseline) | 0.8156 | |
| UNet [26] | 30,000 Synthetic catheter (Generated Images) | 0.8023 |
| 140 Labeled Images + 30,000 Synthetic catheter (Generated Images) | 0.8595 | |
| 140 Labeled Images | 0.7589 | |
| PSPNet [43] | 30,000 Synthetic catheter (Generated Images) | 0.7455 |
| 140 Labeled Images + Synthetic catheter (Generated Images) | 0.8133 | |
| 140 Labeled Images | 0.8072 | |
| PAN [44] | 30,000 Synthetic catheter (Generated Images) | 0.7954 |
| 140 Labeled Images + 30,000 Synthetic catheter (Generated Images) | 0.8671 | |
| 140 Labeled Images | 0.8296 | |
| Linknet [42] | 30,000 Synthetic catheter (Generated Images) | 0.8044 |
| 140 Labeled Images + 30,000 Synthetic catheter (Generated Images) | 0.8806 |
| Model | Training Images | Dice Score |
|---|---|---|
| 5000 Camera catheter + 5000 Synthetic catheter | 0.8389 | |
| UNet [26] | 140 labeled images + 5000 Camera catheter + 5000 Synthetic catheter | 0.8974 |
| 140 labeled images + 10,000 Synthetic catheter | 0.8544 | |
| 5000 Camera catheter + 5000 Synthetic catheter | 0.7220 | |
| PSPNet [43] | 140 labeled images + 5000 Camera catheter + 5000 Synthetic catheter | 0.8112 |
| 140 labeled images + 10,000 Synthetic catheter | 0.8074 | |
| 5000 Camera catheter + 5000 Synthetic catheter | 0.8210 | |
| PAN [44] | 140 labeled images + 5000 Camera catheter + 5000 Synthetic catheter | 0.8764 |
| 140 labeled images + 10,000 Synthetic catheter | 0.8598 | |
| 5000 Camera catheter + 5000 Synthetic catheter) | 0.8273 | |
| Linknet [42] | 140 labeled images + 5000 Camera catheter + 5000 Synthetic catheter | 0.8894 |
| 140 labeled images + 10,000 Synthetic catheter | 0.8797 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, I.; Chikontwe, P.; Choi, H.; Yoon, C.H.; Park, S.H. Synthesize and Segment: Towards Improved Catheter Segmentation via Adversarial Augmentation. Appl. Sci. 2021, 11, 1638. https://doi.org/10.3390/app11041638
Ullah I, Chikontwe P, Choi H, Yoon CH, Park SH. Synthesize and Segment: Towards Improved Catheter Segmentation via Adversarial Augmentation. Applied Sciences. 2021; 11(4):1638. https://doi.org/10.3390/app11041638
Chicago/Turabian StyleUllah, Ihsan, Philip Chikontwe, Hongsoo Choi, Chang Hwan Yoon, and Sang Hyun Park. 2021. "Synthesize and Segment: Towards Improved Catheter Segmentation via Adversarial Augmentation" Applied Sciences 11, no. 4: 1638. https://doi.org/10.3390/app11041638
APA StyleUllah, I., Chikontwe, P., Choi, H., Yoon, C. H., & Park, S. H. (2021). Synthesize and Segment: Towards Improved Catheter Segmentation via Adversarial Augmentation. Applied Sciences, 11(4), 1638. https://doi.org/10.3390/app11041638







