Interactive Effects of Scion and Rootstock Genotypes on the Root Microbiome of Grapevines (Vitis spp. L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Soil Sampling
2.2. DNA Extraction and Sequencing
2.3. Sequence Quality Filtering and Data Analysis
3. Results
3.1. Bacterial α-Diversity across Different Cultivar-Rootstock Combinations
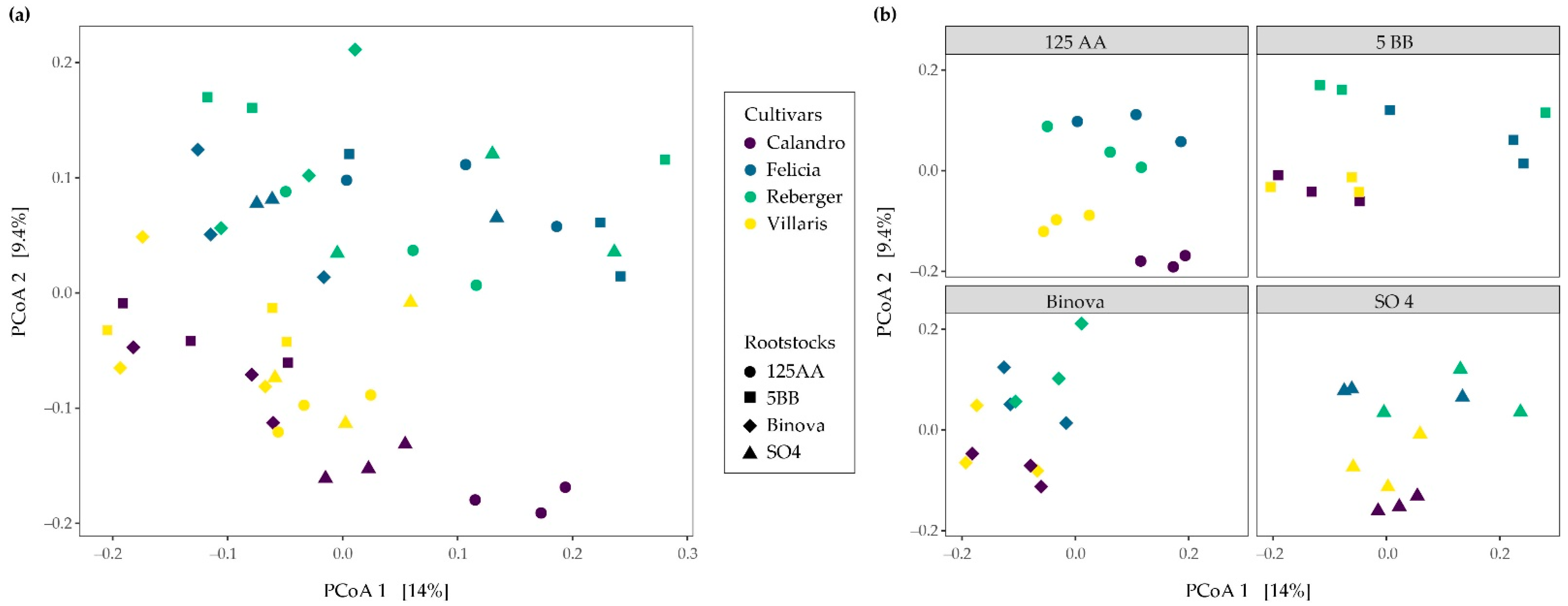
3.2. Bacterial β-Diversity across Different Cultivar-Rootstock Combinations
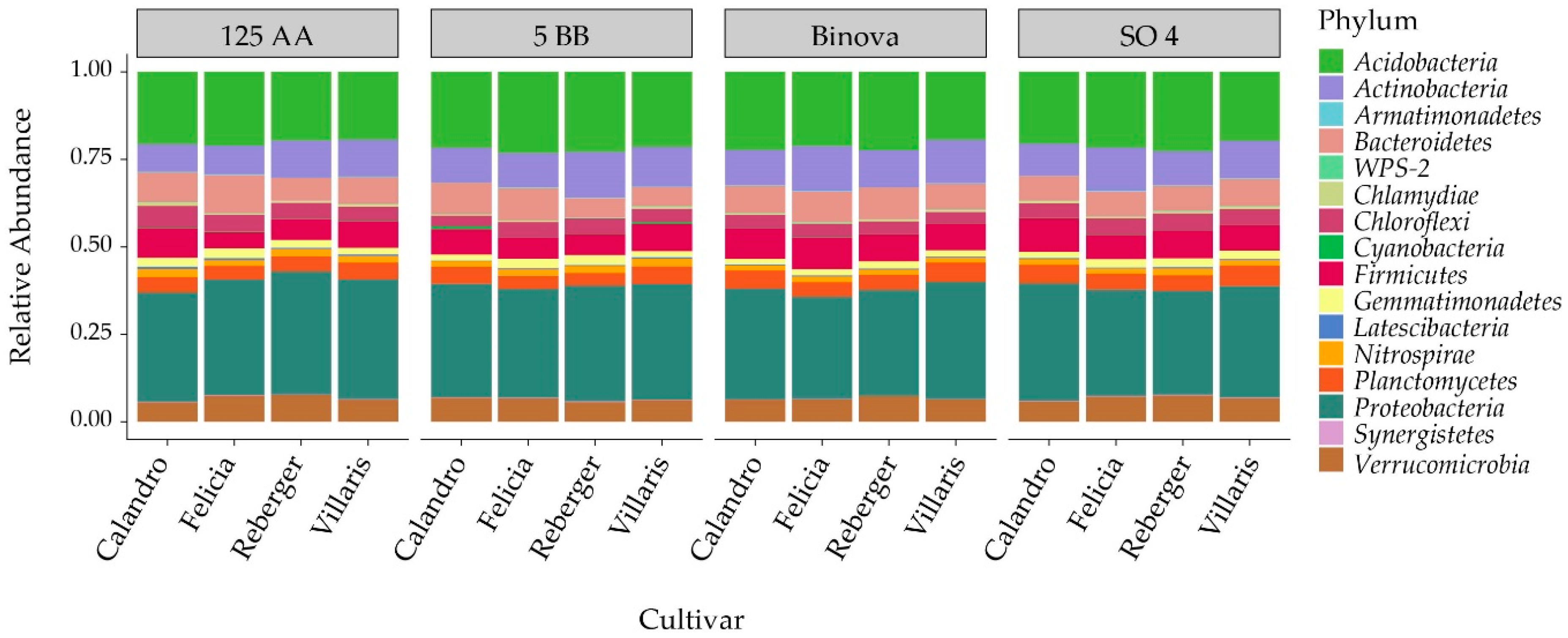
3.3. Taxonomic Level Analysis
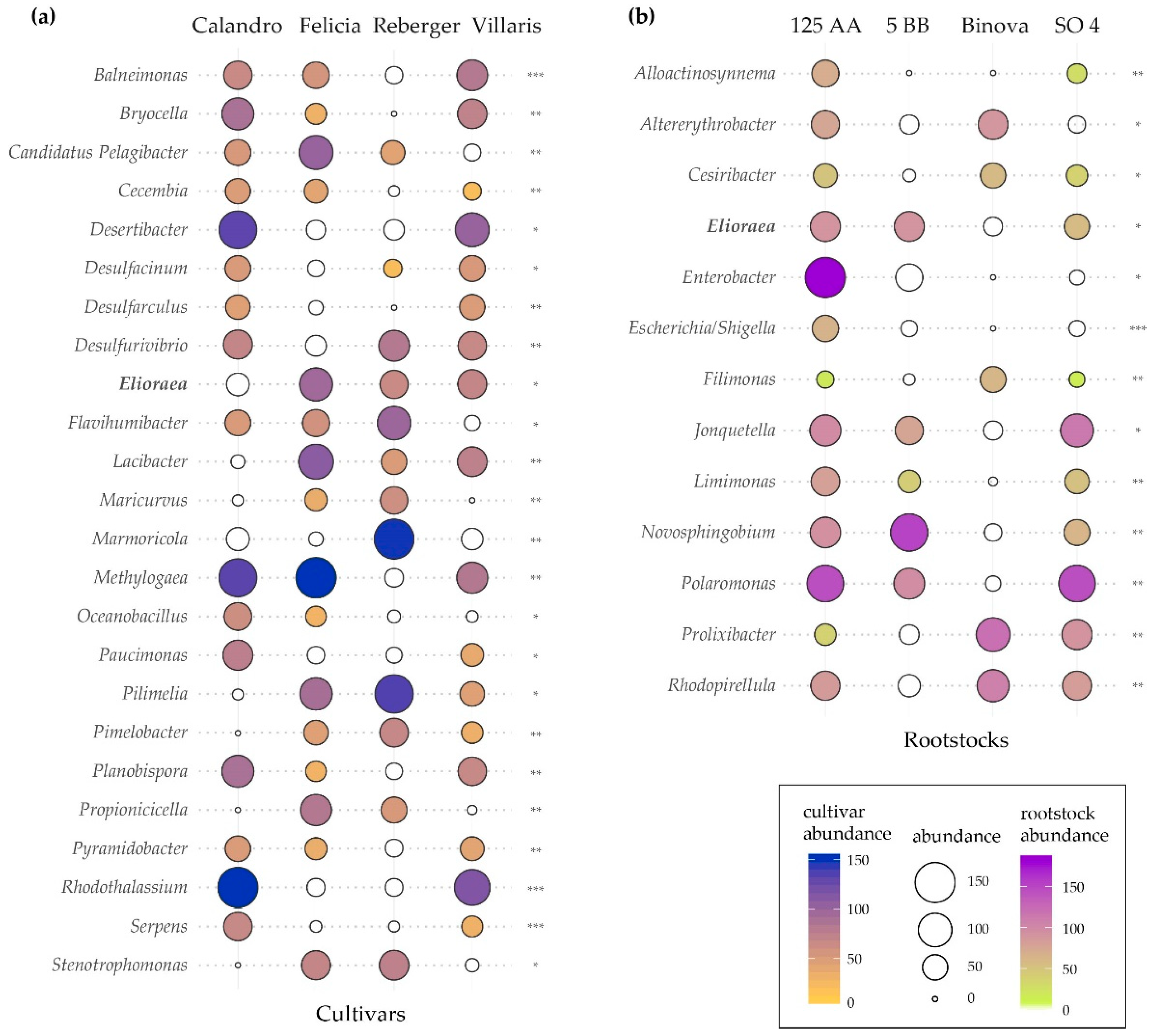
3.4. Bacterial Indicator Species Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottova, D.; Kristufek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef]
- Mallon, C.A.; van Elsas, J.D.; Salles, J.F. Microbial Invasions: The Process, Patterns, and Mechanisms. Trends Microbiol. 2015, 23, 719–729. [Google Scholar] [CrossRef]
- Kurm, V.; van der Putten, W.H.; Pineda, A.; Hol, W.H.G. Soil microbial species loss affects plant biomass and survival of an introduced bacterial strain, but not inducible plant defences. Ann. Bot. 2018, 121, 311–319. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Goh, C.-H.; Veliz Vallejos, D.F.; Nicotra, A.B.; Mathesius, U. The Impact of Beneficial Plant-Associated Microbes on Plant Phenotypic Plasticity. J. Chem. Ecol. 2013, 39, 826–839. [Google Scholar] [CrossRef]
- Rolli, E.; Marasco, R.; Vigani, G.; Ettoumi, B.; Mapelli, F.; Deangelis, M.L.; Gandolfi, C.; Casati, E.; Previtali, F.; Gerbino, R.; et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait: Root bacteria protect plants from drought. Environ. Microbiol. 2015, 17, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Friesen, M.L.; Porter, S.S.; Stark, S.C.; von Wettberg, E.J.; Sachs, J.L.; Martinez-Romero, E. Microbially Mediated Plant Functional Traits. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 23–46. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere: Plant species, soil type and rhizosphere communities. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A.H.M. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- İnceoğlu, Ö.; Al-Soud, W.A.; Salles, J.F.; Semenov, A.V.; van Elsas, J.D. Comparative Analysis of Bacterial Communities in a Potato Field as Determined by Pyrosequencing. PLoS ONE 2011, 6, e23321. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, S.; Li, R.; Zhang, J.; Liu, Y.; Lv, L.; Zhu, H.; Wu, W.; Li, W. Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 2017, 109, 145–155. [Google Scholar] [CrossRef]
- Oger, P.M.; Mansouri, H.; Nesme, X.; Dessaux, Y. Engineering Root Exudation of Lotus toward the Production of Two Novel Carbon Compounds Leads to the Selection of Distinct Microbial Populations in the Rhizosphere. Microb. Ecol. 2004, 47, 96–103. [Google Scholar] [CrossRef]
- Wang, D.; Yang, S.; Tang, F.; Zhu, H. Symbiosis specificity in the legume—Rhizobial mutualism: Host specificity in root nodule symbiosis. Cell. Microbiol. 2012, 14, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.R. Host Specificity in the Genus Agrobacterium. Phytopathology 1979, 69, 320. [Google Scholar] [CrossRef]
- Jacques, M.-A.; Arlat, M.; Boulanger, A.; Boureau, T.; Carrère, S.; Cesbron, S.; Chen, N.W.G.; Cociancich, S.; Darrasse, A.; Denancé, N.; et al. Using Ecology, Physiology, and Genomics to Understand Host Specificity in Xanthomonas. Annu. Rev. Phytopathol. 2016, 54, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Mendes, R.; Tsai, S.M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018, 12, 212–224. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [PubMed]
- International Organisation of Vine and Wine. 2019 Statistical Report on World Vitiviniculture; OIV: Paris, France, 2019. [Google Scholar]
- Mudge, K.; Janick, J.; Scofield, S.; Goldschmidt, E.E. A History of Grafting. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 437–493. ISBN 978-0-470-59377-6. [Google Scholar]
- Alsina, M.M.; Smart, D.R.; Bauerle, T.; de Herralde, F.; Biel, C.; Stockert, C.; Negron, C.; Save, R. Seasonal changes of whole root system conductance by a drought-tolerant grape root system. J. Exp. Bot. 2011, 62, 99–109. [Google Scholar] [CrossRef]
- Serra, I.; Strever, A.; Myburgh, P.A.; Deloire, A. Review: The interaction between rootstocks and cultivars (Vitis vinifera L.) to enhance drought tolerance in grapevine: Rootstocks to enhance drought tolerance in grapevine. Aust. J. Grape Wine Res. 2014, 20, 1–14. [Google Scholar] [CrossRef]
- Peccoux, A.; Loveys, B.; Zhu, J.; Gambetta, G.A.; Delrot, S.; Vivin, P.; Schultz, H.R.; Ollat, N.; Dai, Z. Dissecting the rootstock control of scion transpiration using model-assisted analyses in grapevine. Tree Physiol. 2018, 38, 1026–1040. [Google Scholar] [CrossRef] [PubMed]
- Tandonnet, J.-P.; Cookson, S.J.; Vivin, P.; Ollat, N. Scion genotype controls biomass allocation and root development in grafted grapevine: Scion/rootstock interactions in grapevine. Aust. J. Grape Wine Res. 2009, 16, 290–300. [Google Scholar] [CrossRef]
- Clingeleffer, P.; Morales, N.; Davis, H.; Smith, H. The significance of scion × rootstock interactions. OENO ONE 2019, 53. [Google Scholar] [CrossRef]
- Ferlito, F.; Distefano, G.; Gentile, A.; Allegra, M.; Lakso, A.N.; Nicolosi, E. Scion–rootstock interactions influence the growth and behaviour of the grapevine root system in a heavy clay soil. Aust. J. Grape Wine Res. 2020, 26, 68–78. [Google Scholar] [CrossRef]
- Ling, N.; Song, Y.; Raza, W.; Huang, Q.; Guo, S.; Shen, Q. The response of root-associated bacterial community to the grafting of watermelon. Plant Soil 2015, 391, 253–264. [Google Scholar] [CrossRef]
- Poudel, R.; Jumpponen, A.; Kennelly, M.M.; Rivard, C.L.; Gomez-Montano, L.; Garrett, K.A. Rootstocks Shape the Rhizobiome: Rhizosphere and Endosphere Bacterial Communities in the Grafted Tomato System. Appl. Environ. Microbiol. 2018, 85, e01765-18. [Google Scholar] [CrossRef] [PubMed]
- Ruehl, E.; Schmid, J.; Eibach, R.; Töpfer, R. Grapevine breeding programmes in Germany. In Grapevine Breeding Programs for the Wine Industry; Elsevier: Amsterdam, The Netherlands, 2015; pp. 77–101. ISBN 978-1-78242-075-0. [Google Scholar]
- Maul, E.; Toepfer, R. Vitis International Variety Catalogue. 2020. Available online: www.vivc.de (accessed on 29 November 2020).
- Caporaso, J.G.; Ackermann, G.; Apprill, A.; Bauer, M.; Berg, D.; Betley, J.; Fierer, N.; Fraser, L.; Fuhrman, A.J.; Gilbert, A.J.; et al. EMP 16S Illumina Amplicon Protocol. Available online: https://www.protocols.io/view/emp-16s-illumina-amplicon-protocol-nuudeww (accessed on 3 February 2018).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. AEM 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, T. Btools: A Suite of R Function for All Types of Microbial Diversity Analyses. R Package Version 0.0.1. Available online: https://rdrr.io/github/twbattaglia/btools (accessed on 10 July 2019).
- Kay, M.; Wobbrock, J. ARTool: Aligned Rank Transform for Nonparametric Factorial ANOVAs. R Package Version 0.10.7. Available online: https://github.com/mjskay/ARTool (accessed on 10 July 2019).
- Lenth, R. Emmeans: Estimated Marginal Means, aka Least-Squares Means. R Package Version 1.4.5. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 10 July 2019).
- Oksanen, J.; Blanche, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-6. Available online: https://github.com/vegandevs/vegan (accessed on 10 July 2019).
- Cáceres, M.D.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.; Ramaley, R.F.; Meyer, T.; Kyndt, J.A. Whole-Genome Sequence of a Unique Elioraea Species Strain Isolated from a Yellowstone National Park Hot Spring. Microbiol. Resour. Announc. 2019, 8, e00907-19. [Google Scholar] [CrossRef] [PubMed]
- Yurkov, V.V.; Beatty, J.T. Aerobic Anoxygenic Phototrophic Bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 695–724. [Google Scholar] [CrossRef] [PubMed]
- Evtushenko, L.; Ariskina, E. Part B. Order XII. Propionibacteriales ord. nov., Family II. Nocardioidaceae. In Bergey’s Manual® of Systematic Bacteriology; Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; pp. 1189–1197. ISBN 978-0-387-95043-3. [Google Scholar]
- Octavia, S.; Lan, R. The Family Enterobacteriaceae. In The Prokaryotes: Gammaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 225–286. ISBN 978-3-642-38921-4. [Google Scholar]
- The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; ISBN 978-3-642-39043-2. [Google Scholar]
- Marasco, R.; Rolli, E.; Fusi, M.; Michoud, G.; Daffonchio, D. Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 2018, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Vivanco, J.M. Regulation and Function of Root Exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Mönchgesang, S.; Strehmel, N.; Schmidt, S.; Westphal, L.; Taruttis, F.; Müller, E.; Herklotz, S.; Neumann, S.; Scheel, D. Natural variation of root exudates in Arabidopsis thaliana -linking metabolomic and genomic data. Sci. Rep. 2016, 6, 29033. [Google Scholar] [CrossRef] [PubMed]
- Marastoni, L.; Lucini, L.; Miras-Moreno, B.; Trevisan, M.; Sega, D.; Zamboni, A.; Varanini, Z. Changes in Physiological Activities and Root Exudation Profile of Two Grapevine Rootstocks Reveal Common and Specific Strategies for Fe Acquisition. Sci. Rep. 2020, 10, 18839. [Google Scholar] [CrossRef]
- Berlanas, C.; Berbegal, M.; Elena, G.; Laidani, M.; Cibriain, J.F.; Sagües, A.; Gramaje, D. The Fungal and Bacterial Rhizosphere Microbiome Associated with Grapevine Rootstock Genotypes in Mature and Young Vineyards. Front. Microbiol. 2019, 10, 1142. [Google Scholar] [CrossRef]
- Wittebolle, L.; Marzorati, M.; Clement, L.; Balloi, A.; Daffonchio, D.; Heylen, K.; De Vos, P.; Verstraete, W.; Boon, N. Initial community evenness favours functionality under selective stress. Nature 2009, 458, 623–626. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, M.G.A.; Klironomos, J.N.; Ursic, M.; Moutoglis, P.; Streitwolf-Engel, R.; Boller, T.; Wiemken, A.; Sanders, I.R. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 1998, 396, 69–72. [Google Scholar] [CrossRef]
- Wagg, C.; Hautier, Y.; Pellkofer, S.; Banerjee, S.; Schmid, B.; van der Heijden, M.G.A. Diversity and asynchrony in soil microbial communities stabilizes ecosystem functioning. bioRxiv 2020. [Google Scholar] [CrossRef]
- Salles, J.F.; Le Roux, X.; Poly, F. Relating Phylogenetic and Functional Diversity among Denitrifiers and Quantifying their Capacity to Predict Community Functioning. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Jousset, A.; Schulz, W.; Scheu, S.; Eisenhauer, N. Intraspecific genotypic richness and relatedness predict the invasibility of microbial communities. ISME J. 2011, 5, 1108–1114. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed]
- Vink, S.N.; Chrysargyris, A.; Tzortzakis, N.; Salles, J.F. Bacterial Community Dynamics Varies with Soil Management and Irrigation Practices in Grapevines (Vitis Vinifera L.). Appl. Soil Ecol. 2021, 158, 103807. [Google Scholar] [CrossRef]
- Baldani, J.I.; Videira, S.S.; dos Santos Teixeira, K.R.; Reis, V.M.; de Oliveira, A.L.M.; Schwab, S.; de Souza, E.M.; Pedraza, R.O.; Baldani, V.L.D.; Hartmann, A. The Family Rhodospirillaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 533–618. ISBN 978-3-642-30197-1. [Google Scholar]
- Pujalte, M.J.; Lucena, T.; Ruvira, M.A.; Arahal, D.R.; Macián, M.C. The Family Rhodobacteraceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 439–512. ISBN 978-3-642-30197-1. [Google Scholar]
- Sakarika, M.; Spanoghe, J.; Sui, Y.; Wambacq, E.; Grunert, O.; Haesaert, G.; Spiller, M.; Vlaeminck, S.E. Purple Non-sulphur Bacteria and Plant Production: Benefits for Fertilization, Stress Resistance and the Environment. Microb. Biotechnol. 2020, 13, 1336–1365. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; van der Lelie, D.; Dow, J.M. The Versatility and Adaptation of Bacteria from the Genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.; Chen, D.; Howell, K. From the Vineyard to the Winery: How Microbial Ecology Drives Regional Distinctiveness of Wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. mBio 2016, 7, e00631-16. [Google Scholar] [CrossRef]
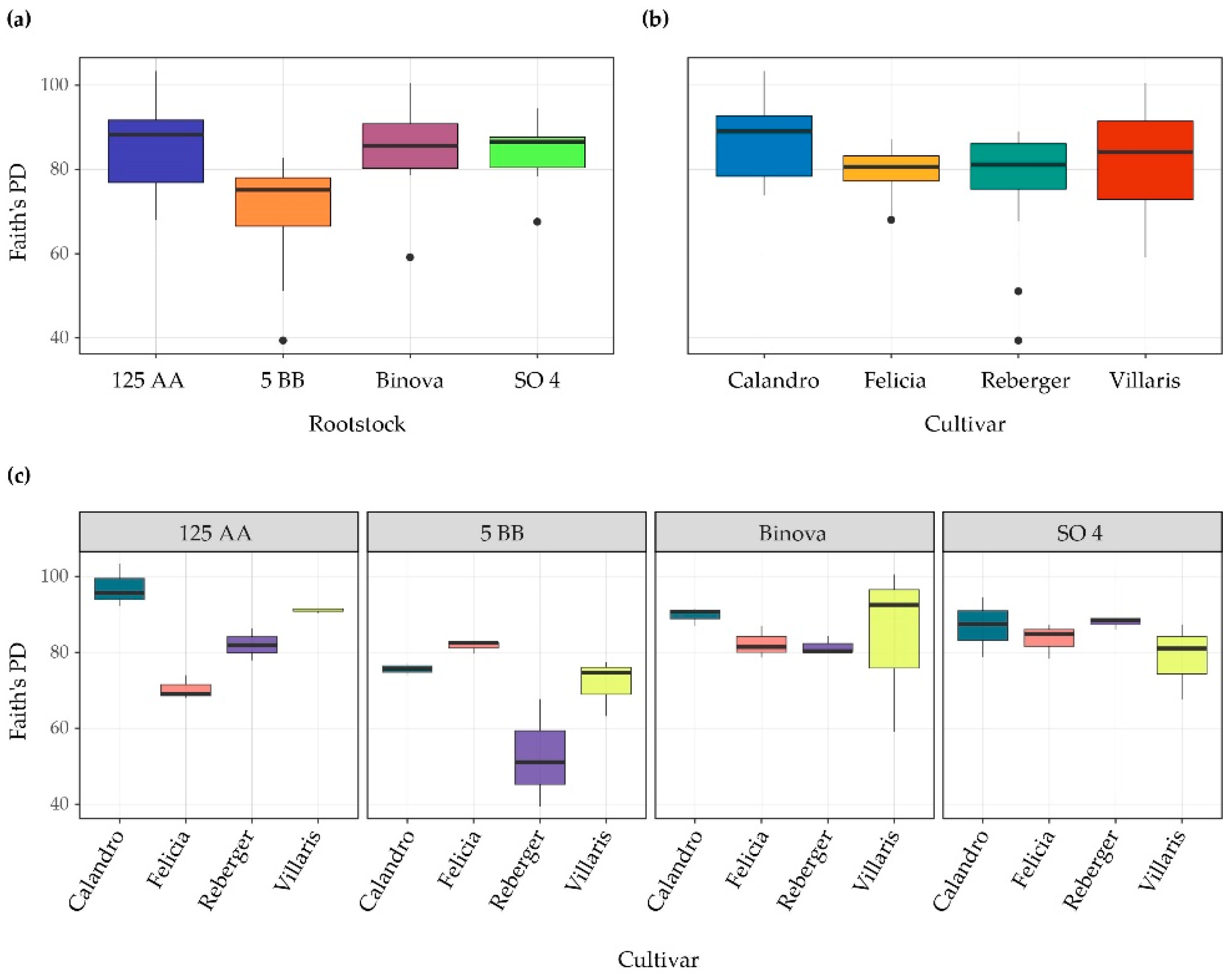
| Richness | Shannon | Simpson | Faith’s PD | |
|---|---|---|---|---|
| Rootstock | F3,32 = 4.6 ** | F3,32 = 5.2 ** | F3,32 = 3.2 * | F3,32 = 7.3 *** |
| Cultivar | F3,32 = 0.9 ns | F3,32 = 2.6 ns | F3,32 = 4.5 ** | F3,32 = 7.1 *** |
| Rootstock:Cultivars | F9,32 = 3.9 ** | F9,32 = 4.0 ** | F9,32 = 3.0 ** | F9,32 = 4.8 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vink, S.N.; Dini-Andreote, F.; Höfle, R.; Kicherer, A.; Salles, J.F. Interactive Effects of Scion and Rootstock Genotypes on the Root Microbiome of Grapevines (Vitis spp. L.). Appl. Sci. 2021, 11, 1615. https://doi.org/10.3390/app11041615
Vink SN, Dini-Andreote F, Höfle R, Kicherer A, Salles JF. Interactive Effects of Scion and Rootstock Genotypes on the Root Microbiome of Grapevines (Vitis spp. L.). Applied Sciences. 2021; 11(4):1615. https://doi.org/10.3390/app11041615
Chicago/Turabian StyleVink, Stefanie Nicoline, Francisco Dini-Andreote, Rebecca Höfle, Anna Kicherer, and Joana Falcão Salles. 2021. "Interactive Effects of Scion and Rootstock Genotypes on the Root Microbiome of Grapevines (Vitis spp. L.)" Applied Sciences 11, no. 4: 1615. https://doi.org/10.3390/app11041615
APA StyleVink, S. N., Dini-Andreote, F., Höfle, R., Kicherer, A., & Salles, J. F. (2021). Interactive Effects of Scion and Rootstock Genotypes on the Root Microbiome of Grapevines (Vitis spp. L.). Applied Sciences, 11(4), 1615. https://doi.org/10.3390/app11041615







