Abstract
With the outbreak of COVID-19 that has prompted an increased focus on self-care, more and more people hope to obtain disease knowledge from the Internet. In response to this demand, medical question answering and question generation tasks have become an important part of natural language processing (NLP). However, there are limited samples of medical questions and answers, and the question generation systems cannot fully meet the needs of non-professionals for medical questions. In this research, we propose a BERT medical pretraining model, using GPT-2 for question augmentation and T5-Small for topic extraction, calculating the cosine similarity of the extracted topic and using XGBoost for prediction. With augmentation using GPT-2, the prediction accuracy of our model outperforms the state-of-the-art (SOTA) model performance. Our experiment results demonstrate the outstanding performance of our model in medical question answering and question generation tasks, and its great potential to solve other biomedical question answering challenges.
1. Introduction and Background
In recent years, human diseases and healthcare have received extensive attention. With the outbreak of COVID-19 at the beginning of 2020, huge efforts have been dedicated to the prevention, treatment, diagnosis, and rehabilitation of COVID-19 [1]. Many studies on COVID-19 have been published in major medical research communities (such as PubMed [2,3], bioRxiv [4], WHO [5], and medRxiv [2]). However, medical terminologies in academic research and publications hinder the majority of non-professionals from better acquiring the urgently needed medical knowledge. This has become a deep and insurmountable gap between advanced research and public demand [6]. Since medical question answering (QA) systems based on natural language processing (NLP) play a critical role to improve the quality of current health care systems, developing accurate and robust medical QA models has become a research priority [7].
Disease knowledge includes various information about a disease, such as signs, symptoms, diagnosis, and treatment [8,9,10], which are essential for NLP in healthcare and biomedicine. NLP in healthcare addresses at least three issues: (1) medical question answering (QA) system, ranking candidate paragraphs to answer the question; (2) medical terminology inference [11], predicting whether a given hypothesis (a description of the patient) could be inferred from a given premise (another description of the patient); (3) disease name recognition [12], detecting disease information from the text. To address the above issues, it is crucial for NLP models to capture disease knowledge, that is, to detect the semantic relationship between the description text and the disease.
Due to the development of deep learning, NLP has made unprecedented progress. The NLP community attaches great importance to automated question answering (QA). In contrast, the reverse research—automatic question generation—has received significantly less attention. However, question generation (QG) [13,14,15,16] has a large number of potential applications, such as improving the training of QA systems [17,18] and helping create new resources in the field of medical question answering. Automatic question generation and personalized questions can help identify relevant medical questions and retrieve the most desired answers. It helps with the insufficiency in the medical question database and alleviates the burden on healthcare providers to manually answer questions, which further benefit the prevention and treatment of diseases, such as COVID-19. Consumers can also get the most understandable answers with a fast response, through simple and straightforward medical questions.
In the research field of medical question answering and question generation, there are still lots of challenges. First of all, key problems in medical question answering include: (1) the number of samples is insufficient, thus, deep learning models cannot learn well; (2) the manual labeling of medical professional terminologies is challenging and time consuming. Meanwhile, in medical automatic question generation (AQG) systems there are also challenges: (1) similar to question answering systems, the samples of questions and medical terminologies are limited; (2) the universality of problem generation is challenging. The generated problems may be too professional, which does not match the simple questions the public demands. In order to overcome the above challenges, data augmentation together with transfer learning becomes a better option.
In this paper, we propose a model for medical question answering based on BERT, GPT-2 [19], and T5-Small [20], three latest variants of the transformer architecture [21]. Our study aims to improve the performance of medical question answering. The main idea is that since question answering and question generation are naturally related tasks, some work, such as training a QA system with limited samples, would be beneficial to both tasks. Essentially, the transformer architecture consists of two main blocks, namely stacked encoders and decoders, which are connected in cascade. The encoder includes two parts: self-attention sublayer and feedforward neural network. Self-attention allows the current node not only to focus on a word, but also to obtain the semantics of the context. The decoder also contains the two layers of the encoder, as well as an encoder-decoder attention layer between the two, which helps process specific parts that need attention. Compared with the original transformer architecture, GPT-2 discards the encoder block and keeps the decoder stack. It provides the functions of a conventional language model and is powerful at predicting the next token in a sequence. Therefore, it is suitable for the question generation task. However, it cannot guarantee that the generated questions are valid and answerable because it is not optimized.
In contrast, BERT is a masked language model. It establishes word embeddings in a context-specific and bidirectional manner. Furthermore, by applying a specific regression head, BERT is trained for discriminative QA. Specifically, it predicts answers in the given paragraph for a given question. Beyond QA, BERT has demonstrated extreme versatility in many downstream tasks. Compared with conventional transformer and in contrast to GPT-2, BERT discards the decoder block and keeps the encoder stack. As BERT is a general purpose language model, most disease names and medical terminologies are not included in BERT’s vocabulary. We pretrained BERT on a large scale biomedical corpus and it demonstrated much better performance in biomedical text mining tasks.
In our proposed model, we first use BERT to build word vectors for medical samples and obtain medical word-embedding vectors. After that, we use GPT-2 to augment the question (Q). After the augmentation, we use T5-Small from Google to extract predictions and using answer (A) and question (Q), respectively. Finally, we calculate the cosine similarity of and , and evaluate the accuracy with XGBoost. The main contributions of this research are as follows:
- We use GPT-2 to augment the question (Q) samples and use S (meaning) and A (answer) as keywords to generate complete sentences as data augmentation for a problem;
- We use T5-Small, a transfer learning model, to perform the extraction on answer (A) and the extraction on question (Q), which is based on a larger corpus. We put more biomedical corpus into our proposed augmentation model for learning, which improves the prediction accuracy of the model in medical question answering tasks.
Experiments on the benchmark datasets for medical question and answer tasks show that, compared with state-of-the-art (SOTA) performance that integrates BERT with disease knowledge, our proposed model is more effective and more competitive. Specifically, we introduce GPT-2 for data augmentation and use T5-Small for extraction, which improves the overall performance of the system.
The rest of this paper is organized as follows. Section 2 reviews research work related to QA and QG systems in the biomedical field. Section 3 explains our proposed model. In Section 4, several comprehensive experiments are conducted to evaluate the effectiveness of the proposed system. Finally in Section 5 we conclude the paper and provide future work.
2. Related Work
2.1. Medical Question Answering and Data Augmentation
The main issues faced by medical question answering tasks are focused on insufficient samples [22] and medical terminologies [23]. To address those issues, various transfer learning and augmentation models have been proposed to improve the prediction performance of the system [24,25]. Semantic biomedical question answering is an important task in the application of biomedical question answering [26,27]. Due to the reliability of answers it can provide, it has attracted widespread attention. In a question answering system, a better word representation is very important, and proper word embeddings can usually considerably improve the performance of the system. With the successful application of pretraining models in general natural language processing tasks, pretraining models have also been widely used in the field of biomedicine. Pretraining models have demonstrated their effectiveness in biomedical question answering tasks [28]. In addition to proper word embedding, named entity recognition is also important in biomedical question answering. Inspired by transfer learning, Peng et al. developed a model to fine-tune BioBERT with a named entity dataset to improve the performance of question answering. In addition, the model uses BiLSTM to encode the question text to obtain sentence-level information. The model also uses bagging to further improve its overall performance, which better combines question-level and token-level information. The model has been evaluated on BioASQ 6b and 7b datasets, and the results demonstrate its advantages and promising potentials [29]. When adapting to specialized domains, such as the COVID-19 literature, model fine-tuning and pretraining can be costly. In order to improve their domain adaptation, Pergola et al. proposed an approach called biomedical entity-aware masking (BEM), which allowed masked language models to learn entity-centric knowledge based on pivotal entities at hand, and used these entities to drive the fine-tuning of the language model (LM). The performance of this model on several biomedical quality assurance datasets was comparable to state-of-the-art models [30].
Healthcare has attracted significant attention, especially during the pandemic. In order to seek healthcare information, tons of questions have appeared on the Internet, which makes it even more urgent to develop an efficient and reliable question answering system. However, people often provide unnecessary information in their questions, such as a patient’s medical history, demographic information, etc. It adds the challenges of understanding natural language questions. In addition, it is crucial to provide accurate and relevant answers instead of paragraphs or even documents. To achieve a reliable medical question answering system, the main tasks include question summarization, as well as multi-answer summarization. The MEDIQA 2021 challenge tackled three summarization tasks in the medical domain: consumer health question summarization, multi-answer summarization, and radiology report summarization. Yadav et al. (NLM at MEDIQA 2021) proposed an approach that first pretrained transformer models on a task-specific summarization dataset and introduced a transfer learning method for question summarization and multi-answer summarization tasks, and then fine-tuned the model for these two tasks by incorporating medical entities. Their approach won the second, sixth and fourth place for the question summarization task in ROUGE-1 (the overlap of unigram (each word) between the system and the reference summaries), ROUGE-2 (the overlap of bigrams between the system and the reference summaries) and ROUGE-L (the longest common subsequence (LCS)-based statistics) scores, respectively, ref. [31].
Question generation (QG) can automatically generate questions from a given context [13], which can be used to build QA datasets. Yue et al. (2020) applied the QG method to synthesize QA pairs on new clinical contexts without requiring manual annotation, and showed that the generated datasets can be used to improve QA models on new contexts [32]. Suwarningsih [33] has created an educational electronic health system in the form of a health question and answer system. It uses a dynamic neural network to validate information with answers, which can effectively provide answers that focus on valid information. The model provides a corpus in the form of a QA pair, which can be automatically generated and provide accurate information for users with upper respiratory tract infections. The accuracy rate of the model is 71.6%. Based on all above research on medical question answering and question generation, transfer learning can effectively improve the accuracy of model prediction. This paper adopts T5-Small, a transfer learning model of text-to-text transformer, to improve the accuracy of the system.
2.2. GPT-2 and Question Answering System
Created by OpenAI in 2019, Generative Pre-trained Transformer 2 (GPT-2) is an unsupervised deep learning transformer-based language model. It is widely used in text translation, question answering, summarization, etc. Esteva et al. used Wikipedia to train a multi-hop question answering model, which treats a query as a question and generates answers from the retrieved documents. In the same way, they trained an abstractive summarizer to generate a summary. The summarizer consists of a BERT encoder and a modified GPT-2 decoder. The model was tested on COVID-19-related datasets and achieved top performance based on key metrics: normalized discounted cumulative gain, precision, mean average precision, and binary preference [34]. To address the challenge of real-world relation extraction (RE) tasks, Papanikolaou proposed the GPT-1-based Data Augmented Relation Extraction (DARE). DARE is designed to augment training data by appropriately fine-tuning GPT-2 to generate examples. Combined with a gold standard dataset (a set of data that has been manually prepared or verified and considered to represent “the objective truth” as closely as possible), the generated training data is used to train a BERT-based RE classifier. A series of experimental results demonstrates the advantage of the proposed method, which improves up to 11 F1 scores. Furthermore, DARE reaches a new level in three widely used biomedical RE datasets [35]. Oniani et al. used the GPT-2 language model to automatically answer questions related to COVID-19. They applied transfer learning to retrain it on the COVID-19 open research dataset corpus. They used four different approaches to improve the quality of the generated responses. Performance evaluation showed that the work achieved significant results in designing a chatbot to produce high-quality COVID-19-related question answering [36].
2.3. T5 and Question Answering
The Text-To-Text Transfer Transformer (T5) model is a modern, large-scale multi-tasking model that is trained by multiple NLP tasks in a unified text-to-text framework [37]. Through extensive pretraining and transfer learning, it has achieved tremendous success in various NLP benchmark tasks, including the GLUE benchmark [38]. This text-to-text framework can be conveniently adapted to any NLP task, including machine translation, document summarization, question answering, and classification tasks. T5 is flexible enough and successful for many tasks. In [39], a generative closed-book question-answering task is studied. In [40], a QA system based on the T5 model with 770M parameters is provided, which explored the efficacy of generating COVID-19 answers from an input question. The advantage of this framework is that it is context-free. However, the results in [40] are not directly comparable to other frameworks.
3. Materials and Method
3.1. Dataset
Pretraining Dataset. In this paper, we used the disease knowledge dataset used in DiseaseBERT [41] for pretraining. The Medical Subject Headings (MeSH) disease and mental disorder branch was selected as the disease vocabulary. A total of 5853 target disease terms from MeSH were searched through Wikipedia articles and 14,617 paragraphs of disease knowledge were collected. The code and data for the dataset are provided in [17]. We used the training procedure proposed in DiseaseBERT [41]: disease knowledge infusion training, which augments BERT-like pretrained models with disease knowledge to achieve better performance in answering medical questions, medical inference, and disease name recognition. In the DiseaseBERT pretraining dataset, the extraction of S is as follows. Taking the first item in the first column of Table 1 as an example: “hemorrhagic septicemia” (disease) is the name of the disease, which is extracted from an article title on Wikipedia and “diagnosis” (aspect) is from a section title in that same Wikipedia article. Q is constructed by the disease and aspect in S. A is the answer to Q. Table 1 shows some examples of the disease knowledge dataset.

Table 1.
Disease knowledge dataset examples.
Validation Dataset. The following two datasets are used as the validation dataset: MEDIQA-2019 [42] and TRECQA-2017 [43], as shown in Table 2, Table 3 and Table 4.

Table 2.
Basics about MEDIQA-2019 and TRECQA-2017 [41]: number of questions (outside parenthesis) and number of associated answers (inside parenthesis).

Table 3.
MEDIQA-RQE test set examples: premise–hypothesis pair [42].

Table 4.
TRECQA-2017 dataset examples.
The MEDIQA-2019 or MEDIQA 2019 challenge was based on questions submitted to the medical QA system CHiQA14. There were mainly three tasks in the MEDIQA 2019 challenge: natural language inference (NLI), recognizing question entailment (RQE), and question answering (QA). The objective was to filter and improve the ranking of automatically retrieved answers. Medical experts manually re-ranked the retrieved answers and provided reference ranks and scores. In the MEDIQA-QA validation dataset, there are 25 consumer health questions and 234 associated answers returned by CHiQA and judged manually. In the MEDIQA-QA test set, there are 150 consumer health questions and 1107 related answers. All QA training, validation, and testing sets are published on its website. The purpose of the test set is for the official and final challenge evaluation.
The Text Retrieval Conference 2017 Live QA (TRECQA-2017) organized a medical question answering task. The medical task was organized in the scope of the CHQA project. The task aimed to address the automatic question answering of consumer health questions submitted to the National Library of Medicine (NLM). The NLM is on the NIH campus and it is the world’s largest biomedical library, leading the research, development, and training in biomedical informatics and health information technology. There are more than 100,000 requests submitted to the NLM every year, including over 10,000 consumer health questions (CHQs). CHQs cover a broad range of questions related to diseases, medications, or medical procedures. The NLM also constructs relevant resources by manually annotating relevant question elements.
Table 4 shows several examples of consumer health questions. The first CHQ asks about the treatment of a disease (retinitis pigmentosa) and includes a lot of descriptive and personal information. The second CHQ is about a problem (“abetalipoproteinemia”) and multiple questions. The third CHQ asks about ingredients in a medicine (Kapvay).
One approach to question answering is question analysis, which retrieves relevant question elements that lead to correct answers. Another approach is to retrieve similar or equivalent questions from history questions. CHQ may include a lot of irrelevant information, for example some background or descriptive information, which introduce more challenges, as shown in Table 4. If there are multiple subquestions in one question, the answer should cover each subquestion. We recommend trusted medical website for relevant answers, such as NIH and PubMed abstracts.
3.2. System Architecture
Our system is based on BERT, GPT-2, and T5-Small. We first pretrain BERT with disease knowledge, in order to obtain medical-word-embedding vectors. Next, we apply GPT-2 for sample augmentation to generate complete sentences, using S and Q (in Table 1) as input data. After that, we use T5-Small (the T5 model with 60 million parameters) to extract and from A and Q, respectively. Because and are text, we use word2vec to convert them to vectors. We then calculate the cosine similarity between vector and vector, which is used as input for XGBoost to find the optimal answer. We also use the last hidden state (LHS) from T5-Small, which is the sequence of hidden states at the output of the last layer of the decoder of the model. We calculate the cosine similarity between and , which is also used as input information for XGBoost. The system architecture is shown in Figure 1.
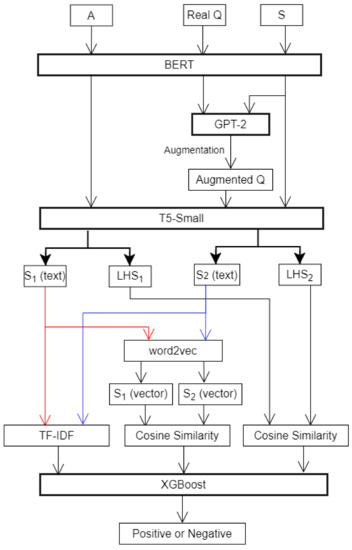
Figure 1.
System architecture.
3.2.1. Fine-Tuning GPT-2
Transformer-based models [21] have become the most advanced technology for several NLP tasks. One of the tasks is language generation. It requires the generated text to be grammatically correct, cohesive, and meaningful. GPT-2 model released by OpenAI [44] is a transformer-based language model. It can generate remarkably fluent sentences, even paragraphs, for a given topic. In addition, GPT-2 can also perform various NLP tasks, such as classification.
GPT-2 is trained to predict the next word, given all the previous words in some text. Its architecture implements a deep neural network, specifically a transformer model, and uses an attention mechanism. The language model outperforms RNN/CNN/LSTM-based models. GPT-2 is pretrained on a very large corpus of English data, only on raw text without labeling. It is trained to predict the next word in sentences only using words before it but no future ones, that is, using k previously seen words to predict the next word. This is achieved by maximizing the following possibilities as shown in Equation (1):
where is the neural network parameter and the objective is to maximize the probability of token being at position i based on k previously seen tokens: .
In this paper, we employed the GPT-2 model to generate a complete sentence, taking S and answer A as the inputs. The generation algorithm works as follows: keyword x is the starting word, model G is the GPT-2 model, and length is the set length of the result sentence. Question Q is the sentence input to the model, and finally the result sentence is generated. Here, we define the input as {x, G, length}. At each timestep, the model G is initialized, and based on the input sentence, the most likely token is predicted as the next token. Then, the token together with the input sentence Q becomes the input to G for the next timestep. When the result sentence reaches the set length, the generation is complete and the result sentence is the final output.
3.2.2. Extraction of and Using T5-Small
In this paper, we chose the T5 model with 60 M parameters, known as T5-Small. T5-Small is convenient for fine-tuning and pretraining. Compared with other variants of the T5 architecture, the training speed of T5-Small is faster. We use the T5-Small model to extract and from answer (A) and question (Q) in the dataset. Specifically, we use A as the input and as the output. Similarly, we use Q as the input and as the output. Real Q is all the unprocessed and unextracted relevant texts in the DiseaseBERT pretraining dataset. We use Equation (2) to represent how we get using T5-Small as the model and A as the input:
Similarly, we use Equation (3) to represent how we get using the T5-Small as the model and Q as the input:
Here, [POS] and [SEP] are special symbols. Taking as an example, it can be inferred from Equation (3) that the trained generator will directly generate for the target domain document Q, where Q is considered as a relevant (positive) document of . Irrelevant (negative) documents can be sampled from the target corpus.
3.2.3. , and XGBoost Prediction
We used the same dataset to train and cross-validate the models of different classifiers. Based on the average of the training accuracy and the test accuracy, and the standard deviation of the test accuracy, we finally chose XGBoost which was the most accurate and robust classifier. We used the XGBoost model to predict the answer. Based on the test results, considering the feature range, feature correlation, data distribution (such as the ratio of positive and negative examples) and model parameters, we iteratively improved the results of the model.
Cosine similarity is one of the most commonly used text analysis methods to measure text similarity. Therefore, it is popular in NLP tasks. Many NLP applications need to calculate the semantic similarity between two short texts. It is flexible enough to be applied in almost any setting, as long as the document can be represented as a vector. Meanwhile, calculating cosine similarity is not a time-consuming task [45].
Cosine similarity observes the angle between vectors without considering weight and magnitude. Equation (4) calculates cosine similarity, where and are vectors.
The term frequency-inverse document frequency (TF-IDF) technology is used to find relevant words in files or documents. It measures the frequency of any word in a given document or dataset. TF-IDF is mainly used for text mining and increasingly used for natural language processing. In this paper, we used the TF-IDF of and as one of the three-dimensional input of XGBoost. Equations (5) and (6) shows the calculation:
The training process of the sentence similarity model is as follows: we calculate the cosine similarity between and , the cosine similarity between and , and the TF-IDF between and . These results are used as the three-dimensional input to the XGBoost classifier. After testing, we iteratively adjust previous modules to maximize the overall performance of the model.
4. Experiments and Results
4.1. Model and Experiment Design
Large-scale LMs, such as BERT and its variants, can capture real-world knowledge (collected from its massive encyclopedic training corpus) and can be directly applied for tasks, such as QA. RoBERTa, BlueBERT, BioBERT, ClinicalBERT, SCiBERT are all variants of BERT, with information from knowledge bases, such as WikiData and WordNet, injected into BERT. We finally chose the best performance model in [41] as SOTA and compared its performance to that of the model in this paper.
OpenAI has released four GPT-2 models: 124 million (124 M), 355 million (355 M), 774 million (774 M), and 1.5 billion (1.5 B) parameters models. The 1.5 billion model is ten times larger than the original GPT-2 model. The 1.5 B model outperforms all other models in the original paper, however, it is hard to fine-tune and to use for transfer learning. It’s very time-consuming to train the model even on the Tensor Processing Units (TPUs) provided by Google Colaboratory. In this paper, we chose the original GPT-2, the TPU-trainable version of GPT-2 [46].
With a batch size of eight, after the first 2000 iterations the loss did not decrease, so we continued for an additional 500 iterations, and then stopped training. In terms of hardware, we used the cloud TPU provided by Google Colaboratory. Due to the memory limit of Google Colaboratory, we chose eight batches. With 25 GB RAM and taking advantage of Google drive [35], we had plenty of storage for transfer learning. For the optimizer, we used Adam [36] and set the learning rate to 0.0001 (1 × 10).
For the QA task, we used T5-Small as the encoder-decoder model. For the perturbation function q, we added two feedforward layers with ReLU on the encoder. For the T5-Small model, we trained it using three epochs with batch size of 20 for extracting and batch size of 64 for extracting , and used the Adam optimizer with a learning rate of 0.0001. We used a beam search with a width of four to generate answers to generative questions. The probability of dropout was 0.1, which was used for regularization. In terms of hardware, we used a GPU Tesla V100 16G and a CPU i7-10875h.
The parameters that XGBoost needed to adjust were max_depth, learning_rate, n_estimators, reg_lambda, and reg_alpha. Through experiments, we found that when other parameters stayed unchanged, parameters reg_lambda and reg_alpha did not change the performance of the XGBoost model. On the other hand, any change in n_estimators, max_depth, and learning_rate changed the performance of the model, which had a negative correlation with the evaluation results. A smaller value of those parameters was associated with a higher accuracy. We finally chose the parameters as: “, “, “(1 × 10).
4.2. Evaluation Metrics
Evaluation methods play an important role in assessing and measuring the performance of a QA system. The main metrics we used were accuracy (Acc), mean reciprocal rank (MRR), and precision. We used these metrics to evaluate our model before and after augmentation as well as to compare with the SOTA model. We used Equation (7) to calculate the accuracy.
In Equation (7), TP is the number of true positives, which means a segment is correctly selected. TN is the number of true negatives, which means a segment is correctly not selected. FP represents the number of false positives, which means a segment is incorrectly selected. FN is the number of false negatives, which means a segment is incorrectly not selected.
It can be observed from Equation (7) that a system with high computational accuracy may be found to have a high TN rate. To solve this issue, precision can be used as a second metric, which is calculated as the number of true positives divided by the total number of true positives and false positives. It can be calculated through Equation (8).
Finally, we used mean reciprocal rank (MRR) as shown in Equation (9) to calculate the answer relevance.
4.3. Performance Evaluation
In this section, we present the experimental results and performance evaluation of our system. We especially evaluate the effectiveness of the T5-Small model and data augmentation using the GPT-2 model. The performance of our model running on two test sets (MEDIQA-2019 and TRCEQA-2017) is shown in Table 5. The ALBERT + disease model is the best performance model as studied in [41] and we take it as the SOTA model. Without GPT-2 augmentation, our system can achieve a performance very close to the SOTA model, while after GPT-2 augmentation our system outperforms the SOTA model. For example, our system achieves 80.23% in terms of accuracy and 84.31% in terms of precision, which are superior to 79.49% in terms of accuracy and 84.02% in terms of precision obtained by the ALBERT + disease (SOTA) model on MEDIQA-2019. Similarly, our proposed system shows better performance on TRCEQA-2017 than the SOTA model in terms of accuracy, precision and MRR. The results demonstrate the advantage of using the GPT-2 model and the T5-Small model for medical question answering and question generation tasks.

Table 5.
Experimental results: comparison with SOTA.
Table 6 and Table 7 present the results of getting from Q before and after data augmentation. The test set is MEDIQA-2019. As shown in Table 7, after augmentation using GPT-2, the training loss and validation loss at epoch 3 are significantly reduced, while Rouge-1 (the overlap of unigram (each word) between the system and the reference summaries), Rouge-2 (the overlap of bigrams between the system and the reference summaries), and Rouge-L (the longest common subsequence (LCS)-based statistics) are significantly increased. The results indicate the improvement of generating S with the augmentation using GPT-2.

Table 6.
Training process and results of Q2S before augmentation (on MEDIQA-2019).

Table 7.
Training process and results of Q2S after augmentation (on MEDIQA-2019).
We also provide the results of getting from Q on the test set TRECQA-2017, as shown in Table 8 and Table 9. Table 8 presents the results before augmentation and Table 9 presents the results after augmentation. Similar to the above results, as shown in Table 9, after augmentation using GPT-2, the training loss and validation loss at epoch 3 are significantly reduced, while all other results are also improved.

Table 8.
Training process and results of Q2S before augmentation (on TRECQA-2017).

Table 9.
Training process and results of Q2S after augmentation (on TRECQA-2017).
In Table 10 and Table 11, we demonstrate some full negative and full positive results. A full positive means the prediction is completely in line with the real answer, and a full negative means that the answer obtained by our prediction result is completely inconsistent with the real answer. After augmenting Q with the GPT-2 model and extracting Q2S with the T5-Small model, we are able to get a more accurate and a larger number of full positive predictions. Q2S-Prediction is the result achieved by augmenting Q (question) with GPT-2 and then extracting it using the T5-Small model. A2S-Prediction is the result of extracting A (answer) using T5-Small. The column cos_sim is the cosine similarity between Q2S-Prediction and A2S-Prediction. Table 10 demonstrates some full negatives while Table 11 demonstrates some full positives. From the results of the two tables, we can learn that we should not count only on cosine similarity to make decisions. That is also why we chose XGBoost, which takes a three-dimensional input and cosine similarity is part of it.

Table 10.
Prediction results of our system: full negatives.

Table 11.
Prediction results of our system: full positives.
5. Conclusions
Medical question answering and question generation systems are facing limitations in existing research, especially the lack of samples. In this paper, we designed a model for medical question answering based on BERT, GPT-2, and T5-Small. We pretrained BERT on medical samples for disease knowledge infusion, and used the GPT-2 model to augment questions, and then used T5-Small to do the extraction. We also used XGBoost to predict the answer and iteratively improve the results. Through extensive experiments, our system demonstrated better performance compared with current medical question answering and question generation system (SOTA method). Our study demonstrates the effectiveness of question augmentation and transfer learning. Overall, our system shows great potential to be applied to health question answering systems, especially COVID-19 question answering. It also helps solve the challenge to retrieve accurate answers for medical recommendation systems.
Author Contributions
Conceptualization and methodology, S.Z. and Y.Z.; software, S.Z.; validation, and original draft preparation, Y.Z.; review and editing, Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data and code supporting the conclusions of this article are available at https://github.com/ShuohuaZhou-NLPer/Question_Answering/, accessed on 17 November 2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhao, Y.; Cheng, S.; Yu, X.; Xu, H. Chinese Public’s Attention to the COVID-19 Epidemic on Social Media: Observational Descriptive Study. J. Med. Internet. Res. 2020, 22, e18825. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, Y.; Oide, S.; Arlie, T.; Tsujimoto, Y.; Furukawa, T. COVID-19 randomized controlled trials in medRxiv and PubMed. Eur. J. Int. Med. 2020, 81, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Dhingra, B.; Liu, Z.; Cohen, W.W.; Lu, X. PubMedQA: A Dataset for Biomedical Research Question Answering. arXiv 2019, arXiv:1909.06146v1. [Google Scholar]
- Ong, E.; Wong, M.U.; Huffman, A.; He, Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: WHO declares pandemic because of “alarming levels” of spread, severity, and inaction. BMJ 2020, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surita, G.; Nogueira, R.; Lotufo, R. Can questions summarize a corpus? Using question generation for characterizing COVID-19 research. arXiv 2020, arXiv:2009.092900. [Google Scholar]
- Yadav, S.; Gupta, D.; Abacha, A.; Demner-Fushman, D. Question-aware Transformer Models for Consumer Health Question Summarization. arXiv 2021, arXiv:2106.00219. [Google Scholar]
- He, Y.; Yu, H.; Ong, E.; Wang, Y.; Liu, Y.; Huffman, A.; Huang, H.H.; Beverley, J.; Hur, J.; Yang, X.; et al. CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci. Data 2020, 7, 181. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q. Social Media Use, eHealth Literacy, Disease Knowledge, and Preventive Behaviors in the COVID-19 Pandemic: Cross-Sectional Study on Chinese Netizens. J. Med. Internet Res. 2020, 22, e19684. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Du, L.; Wang, Y.; Wang, X.; Zhang, R. Disease knowledge and self-management behavior of COPD patients in China. Medicine 2019, 98, e14460. [Google Scholar] [CrossRef]
- Romanov, A.; Shivade, C.P. Lessons from Natural Language Inference in the Clinical Domain. arXiv 2018, arXiv:1808.06752. [Google Scholar]
- Doğan, R.I.; Leaman, R.; Lu, Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014, 47, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Shao, J.; Cardie, C. Learning to ask: Neural question generation for reading comprehension. arXiv 2017, arXiv:1705.00106. [Google Scholar]
- Tang, D.; Duan, N.; Qin, T.; Yan, Z.; Zhou, M. Question answering and question generation as dual tasks. arXiv 2017, arXiv:1706.02027. [Google Scholar]
- Kim, Y.; Lee, H.; Shin, J.; Jung, K. Improving neural question generation using answer separation. In Proceedings of the AAAI Conference on Artificial Intelligence. arXiv 2019, arXiv:1809.02393. [Google Scholar]
- Song, L.; Wang, Z.; Hamza, W.Z.; Zhang, Y.; Gildea, D. Leveraging context information for natural question generation. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 2 (Short Papers), New Orleans, LA, USA, 1–6 June 2018; Volume 2. Available online: https://aclanthology.org/N18-2090 (accessed on 22 November 2021).
- Lewis, P.; Denoyer, L.; Riedel, S. Unsupervised question answering by cloze translation. arXiv 2019, arXiv:1906.04980. [Google Scholar]
- Chen, Y.; Wu, L.; Zaki, M.J. Reinforcement learning based graph-to-sequence model for natural question generation. arXiv 2019, arXiv:1908.04942. [Google Scholar]
- Radford, A.; Wu, J.; Child, R.; Luan, D.; Amodei, D.; Sutskever, I. Language models are unsupervised multitask learners. OpenAI Blog 2019, 1, 9. [Google Scholar]
- Xue, L.; Constant, N.; Roberts, A.; Kale, M.; AI-Rfou, R.; Siddhant, A.; Barua, A.; Raffel, C. mT5: A massively multilingual pre-trained text-to-text transformer. arXiv 2020, arXiv:2010.11934. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gonez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Advances in neural information processing systems. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Jin, Q.; Yuan, Z.; Xiong, G.; Yu, Q. Biomedical question answering: A comprehensive review. arXiv 2021, arXiv:2102.05281. [Google Scholar]
- Xu, G.; Rong, W.; Wang, Y.; Ouyang, Y.; Xiong, Z. External features enriched model for biomedical question answering. BMC Bioinform. 2021, 22, 1–19. [Google Scholar] [CrossRef]
- Akdemir, A.; Shibuya, T. Transfer Learning for Biomedical Question Answering. In CLEF (Working Notes); 2020; Available online: http://ceur-ws.org/Vol-2696/paper_66.pdf (accessed on 22 November 2021).
- Jeong, M.; Sung, M.; Kim, G.; Kim, D.; Yoon, W.; Yoo, J.; Kang, J. Transferability of natural language inference to biomedical question answering. arXiv 2020, arXiv:2007.00217. [Google Scholar]
- Sarrouti, M.; Gupta, D.; Abacha, A.B.; Demner-Fushman, D. NLM at BioASQ Synergy 2021: Deep Learning-Based Methods for Biomedical Semantic Question Answering about COVID-19. CLEF 2021—Conference and Labs of the Evaluation Forum. Available online: http://ceur-ws.org/Vol-2936/paper-25.pdf (accessed on 22 November 2021).
- Sarrouti, M.; El Alaoui, S.O. SemBioNLQA: A semantic biomedical question answering system for retrieving exact and ideal answers to natural language questions. Artif. Intell. Med. 2020, 102, 101767. [Google Scholar] [CrossRef]
- Gouthaman, K.V.; Mittal, A. Reducing language biases in visual question answering with visually-grounded question encoder. Proceedings of Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020. Part XIII 16.s. [Google Scholar]
- Peng, K.; Yin, C.; Rong, W.; Lin, C.; Zhou, D.; Xiong, Z. Named Entity Aware Transfer Learning for Biomedical Factoid Question Answering. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021. [Google Scholar] [CrossRef]
- Pergola, G.; Kochkina, E.; Gui, L.; Liakata, M.; He, Y. Boosting Low-Resource Biomedical QA via Entity-Aware Masking Strategies. arXiv 2021, arXiv:2102.08366. [Google Scholar]
- Yadav, S.; Sarrouti, M.; Gupta, D. NLM at MEDIQA 2021: Transfer Learning-based Approaches for Consumer Question and Multi-Answer Summarization. In Proceedings of the 20th Workshop on Biomedical Language Processing (BIONLP 2021); Association for Computational Linguistics: Stroudsburg, PA, USA, 2021; pp. 291–301. [Google Scholar]
- Yue, X.; Zhang, X.; Yao, Z.; Lin, S.; Sun, H. CliniQG4QA: Generating Diverse Questions for Domain Adaptation of Clinical Question Answering. arXiv 2020, arXiv:2010.16021. [Google Scholar]
- Suwarningsih, W. e-Health Education Using Automatic Question Generation-Based Natural Language (Case Study: Respiratory Tract Infection). In Emerging Technologies in Biomedical Engineering and Sustainable TeleMedicine; Springer: Berlin/Heidelberg, Germany, 2021; pp. 69–79. [Google Scholar]
- Esteva, A.; Kale, A.; Paulu, S.R.; Hashimoto, K.; Yin, W.; Radev, D.; Socher, R. Co-search: COVID-19 information retrieval with semantic search, question answering, and abstractive summarization. arXiv 2020, arXiv:2006.09595. [Google Scholar]
- Papanikolaou, Y.; Pierleoni, A. DARE: Data Augmented Relation Extraction with GPT-2. arXiv 2020, arXiv:2004.13845. [Google Scholar]
- Oniani, D.; Wang, Y. A Qualitative Evaluation of Language Models on Automatic Question-Answering for COVID-19. In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. Association for Computing Machinery, Virtual Event, 21–24 September 2020; p. 33. [Google Scholar]
- Raffel, C.; Shazeer, N.; Roberts, A.; Lee, K.; Narang, S.; Matena, M.; Zhou, Y.; Li, W.; Liu, P. Exploring the limits of transfer learning with a unified text-to-text transformer. arXiv 2019, arXiv:1910.10683. [Google Scholar]
- Wang, A.; Singh, A.; Michael, J.; Hill, F.; Levy, O.; Bowman, S.R. Glue: A multi-task benchmark and analysis platform for natural language understanding. arXiv 2018, arXiv:1804.07461. [Google Scholar]
- Roberts, A.; Raffel, C.; Shazeer, N. How much knowledge can you pack into the parameters of a language model? arXiv 2020, arXiv:2002.08910. [Google Scholar]
- Ngai, H.; Park, Y.; Chen, J.; Parsa, M. Transfermer-Based Models for Question Answering on COVID19. arXiv 2021, arXiv:2101.11432v1. [Google Scholar]
- He, Y.; Zhu, Z.; Zhang, Y.; Chen, Q.; Caverlee, J. Infusing Disease Knowledge into BERT for Health Question Answering, Medical Inference and Disease Name Recognition. arXiv 2020, arXiv:2010.03746. [Google Scholar]
- Abacha, A.B.; Shivade, C.; Demner-Fushman, D. Overview of the mediqa 2019 shared task on textual inference, question entailment and question answering. In Proceedings of the 18th BioNLP Workshop and Shared Task, Florence, Italy, 1 August 2019; pp. 370–5039. Available online: https://aclanthology.org/W19-5039.pdf (accessed on 22 November 2021).
- Abacha, A.B.; Agichtein, E.; Pinter, Y.; Demner-Fushman, D. Overview of the Medical Question Answering Task at TREC 2017 LiveQA. In TREC; 2018. Available online: https://trec.nist.gov/pubs/trec26/papers/Overview-QA.pdf (accessed on 22 November 2021).
- Lee, J.-S.; Hsiang, J. Patent claim generation by fine-tuning OpenAI GPT-2. arXiv 2020, arXiv:1907.02052. [Google Scholar] [CrossRef]
- Prismana, I.; Prehanto, D.R.; Dermawan, D.A.; Herlingga, A.C.; Wibawa, S.C. Nazief & Adriani Stemming Algorithm With Cosine Similarity Method For Integrated Telegram Chatbots With Service. In IOP Conference Series: Materials Science and Engineering; Workshop on Environmental Science, Society, and Technology (WESTECH 2020); IOP: Makassar, Indonesia, 2021; Volume 1125. [Google Scholar]
- Cer, D.; Yang, Y.; Kong, S.Y.; Hua, N.; Limtiaco, N.; John, R.S.; Constant, N.; Guajardo-Céspedes, M.; Yuan, S.; Tar, C.; et al. Universal sentence encoder. arXiv 2018, arXiv:1803.11175. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).