Pathologic Complete Response Prediction after Neoadjuvant Chemoradiation Therapy for Rectal Cancer Using Radiomics and Deep Embedding Network of MRI
Abstract
1. Introduction
- We propose a method for encapsulating multiple MRI sequences into an MSFI and generating MSFI embedding using 3D-CNN to extract novel volumetric features of tumors.
- We introduce multi-parametric MRI embedding that contains diverse discriminative features of tumors by incorporating MSFI embedding and radiomics.
- We show the superiority of the proposed method through extensive experiments using the pre-operative MRI sequences of 912 rectal cancer patients.
2. Related Works
2.1. Qualitative Evaluation of Rectal Cancer Using MRI
2.2. Quantitative Evaluation of Rectal Cancer using Radiomics Features
2.3. Quantitative Evaluation of Rectal Cancer using Deep Learning
3. Method
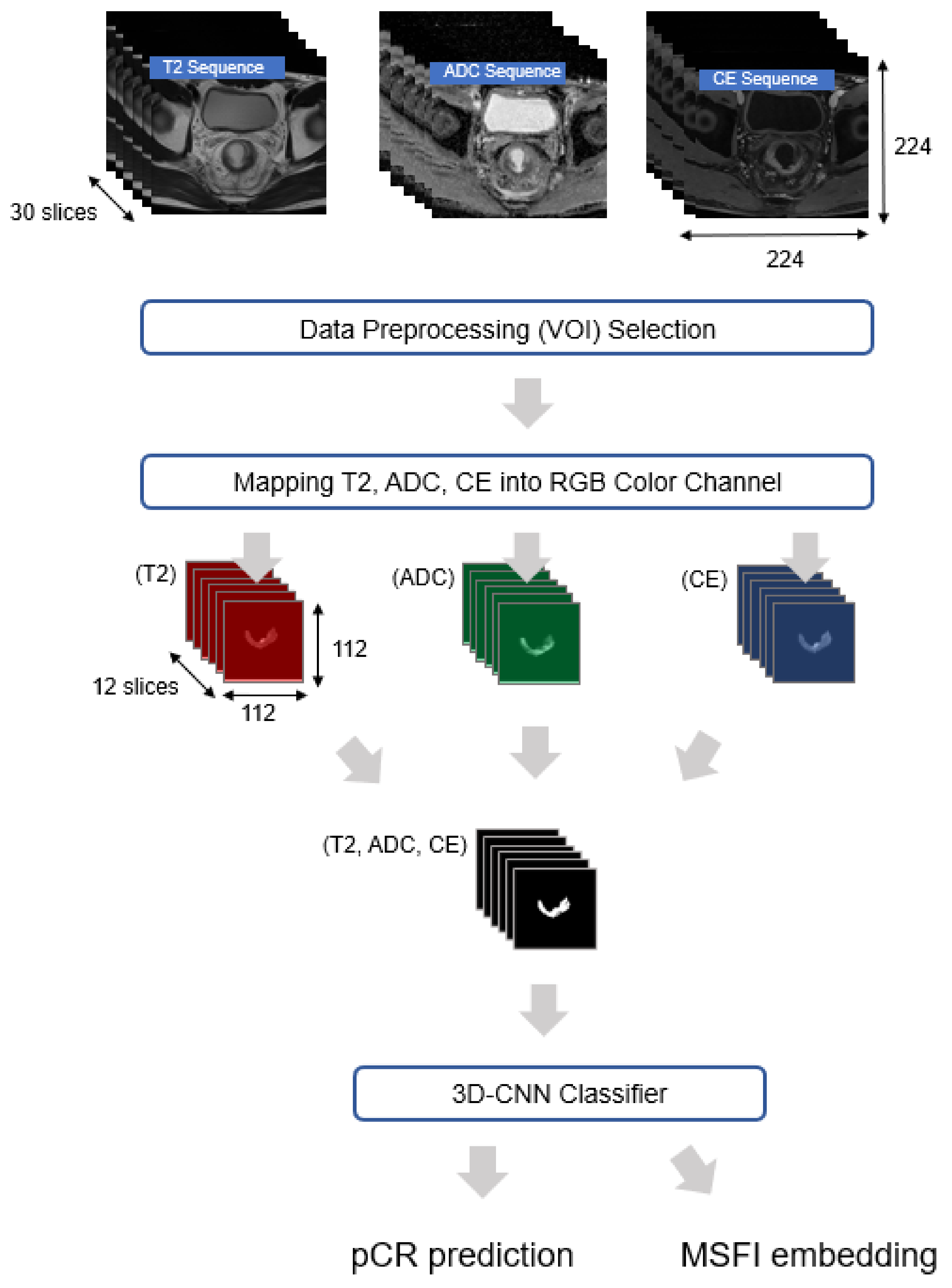
3.1. Data Preprocessing
3.2. Suggested Method
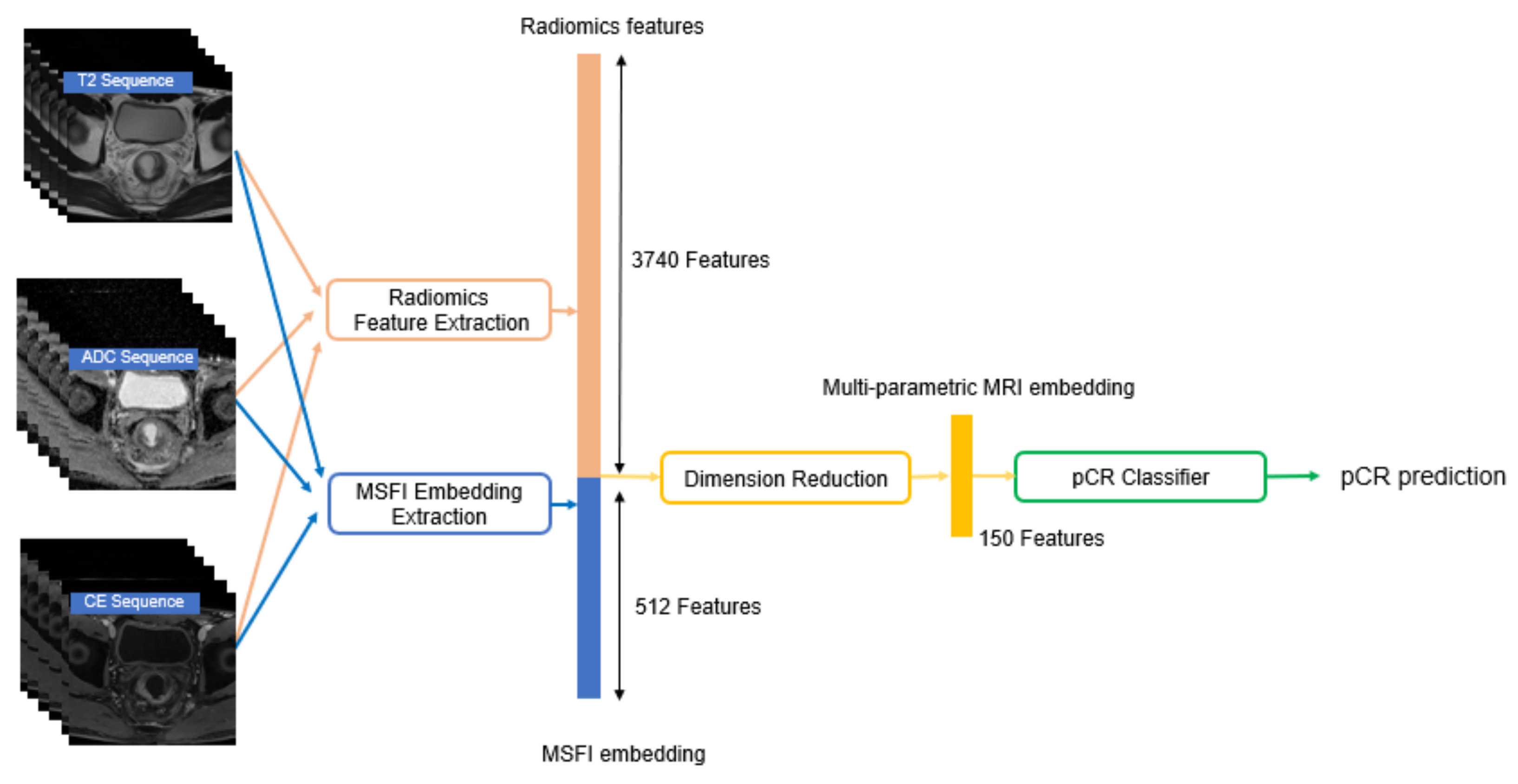
3.2.1. Representing Multiple MRI Sequences as MSFI Embedding
3.2.2. Extracting Radiomics Features
3.2.3. Predicting pCR using Both MSFI Embedding and Radiomics Features
4. Experiments
- Comparison of five types of input MRI sequences:
- (a)
- {T2}
- (b)
- {DWI/ADC}
- (c)
- {CE}
- (d)
- {T2, DWI/ADC} and
- (e)
- {T2, DWI/ADC, and CE} (ours).
- Comparison of the proposed method with its variants that differ in two factors, MRI feature vector extraction and pCR classification:
- Three MRI feature vectors: radiomics features, MSFI embedding, and multi-parametric MRI embedding (ours).
- Six classification models: logistic regression, xgboost, lightgbm, random forest (ours), MLP, and ensemble of the five classifiers.
- Comparative evaluation of the proposed method with four competing baselines:
4.1. Experimental Setup
4.2. Impact of Input MRI Sequences
4.3. Analysis of Our pCR Prediction Model
4.4. Comparison with Existing pCR Prediction Methods
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. MRI Protocol
| 1.5 T | 3.0 T | |||||
|---|---|---|---|---|---|---|
| Fast Spin-Echo T2-Weighted Image (T2) | Diffusion-Weighted Image (DWI) | 3D T1-Weighted Gradient Echo (CE) | Fast Spin-Echo T2-Weighted Image (T2) | Diffusion-Weighted Image (DWI) | 3D T1-Weighted Gradient Echo (CE) | |
| Plane | Axial, Sagittal, Oblique axial, Oblique coronal | Axial | Axial | Axial, Sagittal, Oblique axial, Oblique coronal | Axial | Axial |
| Repetition time(ms) | 2740–4200 | 6900–9100 | 3.51 | 3800–5500 | 9500–12,000 | 3.51 |
| Echo time(ms) | 80 | 64–90 | 1.44 | 80–120 | 62–95 | 1.44 |
| Flip angle(degrees) | 137 | 90 | 90–150 | 90 | ||
| B factor(s/mm) | 0, 300, 1000 | 0, 300, 1000 | ||||
| Field of view(mm) | 180 or 240 | 220 | 240 | 180 or 240 | 220 | 240 |
| Matrix without interpolation | 304 | 128 or 150 | 240 | 320–448 | 126 or 153 | 240 |
| Slice thickness (mm) | 3 | 3 | 3 | 3 | 3 | 3 |
| Slice gap (mm) | 0 | 0 | 0 | 0 | ||
| Echo train length | 16 | 17 or 35 | ||||
References
- Jung, K.W.; Won, Y.J.; Kong, H.J.; Lee, E.S. Prediction of cancer incidence and mortality in Korea, 2019. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2019, 51, 431. [Google Scholar] [CrossRef] [PubMed]
- Renehan, A.G.; Malcomson, L.; Emsley, R.; Gollins, S.; Maw, A.; Myint, A.S.; Rooney, P.S.; Susnerwala, S.; Blower, A.; Saunders, M.P.; et al. Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): A propensity-score matched cohort analysis. Lancet Oncol. 2016, 17, 174–183. [Google Scholar] [CrossRef]
- Maas, M.; Lambregts, D.M.; Nelemans, P.J.; Heijnen, L.A.; Martens, M.H.; Leijtens, J.W.; Sosef, M.; Hulsewé, K.W.; Hoff, C.; Breukink, S.O.; et al. Assessment of clinical complete response after chemoradiation for rectal cancer with digital rectal examination, endoscopy, and MRI: Selection for organ-saving treatment. Ann. Surg. Oncol. 2015, 22, 3873–3880. [Google Scholar] [CrossRef] [PubMed]
- Patel, U.B.; Brown, G.; Rutten, H.; West, N.; Sebag-Montefiore, D.; Glynne-Jones, R.; Rullier, E.; Peeters, M.; Van Cutsem, E.; Ricci, S.; et al. Comparison of magnetic resonance imaging and histopathological response to chemoradiotherapy in locally advanced rectal cancer. Ann. Surg. Oncol. 2012, 19, 2842–2852. [Google Scholar] [CrossRef] [PubMed]
- Dzik-Jurasz, A.; Domenig, C.; George, M.; Wolber, J.; Padhani, A.; Brown, G.; Doran, S. Diffusion MRI for prediction of response of rectal cancer to chemoradiation. Lancet 2002, 360, 307–308. [Google Scholar] [CrossRef]
- Villers, A.; Puech, P.; Mouton, D.; Leroy, X.; Ballereau, C.; Lemaitre, L. Dynamic contrast enhanced, pelvic phased array magnetic resonance imaging of localized prostate cancer for predicting tumor volume: Correlation with radical prostatectomy findings. J. Urol. 2006, 176, 2432–2437. [Google Scholar] [CrossRef] [PubMed]
- Weiser, M.R.; Gollub, M.J.; Saltz, L.B. Assessment of clinical complete response after chemoradiation for rectal cancer with digital rectal examination, endoscopy, and MRI. Ann. Surg. Oncol. 2015, 22, 3769–3771. [Google Scholar] [CrossRef][Green Version]
- Horvat, N.; Veeraraghavan, H.; Khan, M.; Blazic, I.; Zheng, J.; Capanu, M.; Sala, E.; Garcia-Aguilar, J.; Gollub, M.J.; Petkovska, I. MR imaging of rectal cancer: Radiomics analysis to assess treatment response after neoadjuvant therapy. Radiology 2018, 287, 833–843. [Google Scholar] [CrossRef]
- Lambregts, D.M.; Maas, M.; Riedl, R.G.; Bakers, F.C.; Verwoerd, J.L.; Kessels, A.G.; Lammering, G.; Boetes, C.; Beets, G.L.; Beets-Tan, R.G. Value of ADC measurements for nodal staging after chemoradiation in locally advanced rectal cancer—A per lesion validation study. Eur. Radiol. 2011, 21, 265–273. [Google Scholar] [CrossRef]
- Nie, K.; Shi, L.; Chen, Q.; Hu, X.; Jabbour, S.K.; Yue, N.; Niu, T.; Sun, X. Rectal cancer: Assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin. Cancer Res. 2016, 22, 5256–5264. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, X.Y.; Shi, Y.J.; Wang, L.; Zhu, H.T.; Tang, Z.; Wang, S.; Li, X.T.; Tian, J.; Sun, Y.S. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin. Cancer Res. 2017, 23, 7253–7262. [Google Scholar] [CrossRef]
- Cui, Y.; Yang, X.; Shi, Z.; Yang, Z.; Du, X.; Zhao, Z.; Cheng, X. Radiomics analysis of multiparametric MRI for prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Eur. Radiol. 2019, 29, 1211–1220. [Google Scholar] [CrossRef]
- Huynh, B.Q.; Antropova, N.; Giger, M.L. Comparison of breast DCE-MRI contrast time points for predicting response to neoadjuvant chemotherapy using deep convolutional neural network features with transfer learning. In Proceedings of the Medical imaging 2017: Computer-Aided Diagnosis. International Society for Optics and Photonics, Orlando, FL, USA, 11–16 February 2017; Volume 10134, p. 101340. [Google Scholar]
- Ravichandran, K.; Braman, N.; Janowczyk, A.; Madabhushi, A. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI. In Proceedings of the Medical Imaging 2018: Computer-Aided Diagnosis, International Society for Optics and Photonics, Houston, TX, USA, 10–15 February 2018; Volume 10575, p. 105750C. [Google Scholar]
- Hu, Q.; Whitney, H.M.; Giger, M.L. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Yun, J.; Park, J.E.; Lee, H.; Ham, S.; Kim, N.; Kim, H.S. Radiomic features and multilayer perceptron network classifier: A robust MRI classification strategy for distinguishing glioblastoma from primary central nervous system lymphoma. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Fu, J.; Zhong, X.; Li, N.; Van Dams, R.; Lewis, J.; Sung, K.; Raldow, A.C.; Jin, J.; Qi, X.S. Deep learning-based radiomic features for improving neoadjuvant chemoradiation response prediction in locally advanced rectal cancer. Phys. Med. Biol. 2020, 65, 075001. [Google Scholar] [CrossRef]
- Li, H.; Boimel, P.; Janopaul-Naylor, J.; Zhong, H.; Xiao, Y.; Ben-Josef, E.; Fan, Y. Deep convolutional neural networks for imaging data based survival analysis of rectal cancer. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 846–849. [Google Scholar]
- Kornblith, S.; Shlens, J.; Le, Q.V. Do better imagenet models transfer better? In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2661–2671. [Google Scholar]
- Lubner, M.G.; Stabo, N.; Abel, E.J.; del Rio, A.M.; Pickhardt, P.J. CT textural analysis of large primary renal cell carcinomas: Pretreatment tumor heterogeneity correlates with histologic findings and clinical outcomes. Am. J. Roentgenol. 2016, 207, 96–105. [Google Scholar] [CrossRef]
- Park, J.H.; Seo, N.; Lim, J.S.; Hahm, J.; Kim, M.J. Feasibility of Simultaneous Multislice Acceleration Technique in Diffusion-Weighted Magnetic Resonance Imaging of the Rectum. Korean J. Radiol. 2020, 21, 77–87. [Google Scholar] [CrossRef]
- Gollub, M.; Gultekin, D.; Akin, O.; Do, R.; Fuqua, J.; Gonen, M.; Kuk, D.; Weiser, M.; Saltz, L.; Schrag, D.; et al. Dynamic contrast enhanced-MRI for the detection of pathological complete response to neoadjuvant chemotherapy for locally advanced rectal cancer. Eur. Radiol. 2012, 22, 821–831. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Bibault, J.E.; Giraud, P.; Housset, M.; Durdux, C.; Taieb, J.; Berger, A.; Coriat, R.; Chaussade, S.; Dousset, B.; Nordlinger, B.; et al. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci. Rep. 2018, 8, 1–8. [Google Scholar]
- Cox, D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. (Methodol.) 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Pieper, S.; Halle, M.; Kikinis, R. 3D Slicer. In Proceedings of the 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821), Arlington, VA, USA, 15–18 April 2004; pp. 632–635. [Google Scholar]
- Gosain, A.; Sardana, S. Handling class imbalance problem using oversampling techniques: A review. In Proceedings of the 2017 International Conference on Advances in Computing, Communications and Informatics (ICACCI), Udupi, India, 13–16 September 2017; pp. 79–85. [Google Scholar]
- Unser, M.; Aldroubi, A.; Eden, M. B-spline signal processing. I. Theory. IEEE Trans. Signal Process. 1993, 41, 821–833. [Google Scholar] [CrossRef]
- Schwier, M.; van Griethuysen, J.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of multiparametric prostate MRI radiomics features. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Lim, H.; Bae, B.K.; Hahm, M.H.; Chong, G.O.; Jeong, S.Y.; Kim, J.C. Robustness of magnetic resonance radiomic features to pixel size resampling and interpolation in patients with cervical cancer. Cancer Imaging 2021, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Duron, L.; Balvay, D.; Vande Perre, S.; Bouchouicha, A.; Savatovsky, J.; Sadik, J.C.; Thomassin-Naggara, I.; Fournier, L.; Lecler, A. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS ONE 2019, 14, e0213459. [Google Scholar] [CrossRef] [PubMed]
- Hussain, Z.; Gimenez, F.; Yi, D.; Rubin, D. Differential data augmentation techniques for medical imaging classification tasks. In Proceedings of the AMIA Annual Symposium Proceedings, American Medical Informatics Association, Washington, DC, USA, 6–8 November 2017; Volume 2017, p. 979. [Google Scholar]
- Perez, F.; Vasconcelos, C.; Avila, S.; Valle, E. Data augmentation for skin lesion analysis. In OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis; Springer: Berlin, Germany, 2018; pp. 303–311. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Almourish, M.H.; Saif, A.A.; Radman, B.M.; Saeed, A.Y. Covid-19 Diagnosis Based on CT Images Using Pre-Trained Models. In Proceedings of the 2021 International Conference of Technology, Science and Administration (ICTSA), Taiz, Yemen, 22–24 March 2021; pp. 1–5. [Google Scholar]
- Tran, D.; Wang, H.; Torresani, L.; Ray, J.; LeCun, Y.; Paluri, M. A closer look at spatiotemporal convolutions for action recognition. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6450–6459. [Google Scholar]
- Kay, W.; Carreira, J.; Simonyan, K.; Zhang, B.; Hillier, C.; Vijayanarasimhan, S.; Viola, F.; Green, T.; Back, T.; Natsev, P.; et al. The kinetics human action video dataset. arXiv 2017, arXiv:1705.06950. [Google Scholar]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 1–8. [Google Scholar] [CrossRef]
- Sarveniazi, A. An actual survey of dimensionality reduction. Am. J. Comput. Math. 2014, 4, 55–72. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.; Müller, K.R. Kernel principal component analysis. In International Conference on Artificial Neural Networks; Springer: Berlin, Germany, 1997; pp. 583–588. [Google Scholar]
- Breiman, L. Random forests. Machine Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ospina, J.D.; Zhu, J.; Chira, C.; Bossi, A.; Delobel, J.B.; Beckendorf, V.; Dubray, B.; Lagrange, J.L.; Correa, J.C.; Simon, A.; et al. Random forests to predict rectal toxicity following prostate cancer radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 1024–1031. [Google Scholar] [CrossRef]
- Petkovska, I.; Tixier, F.; Ortiz, E.J.; Pernicka, J.S.G.; Paroder, V.; Bates, D.D.; Horvat, N.; Fuqua, J.; Schilsky, J.; Gollub, M.J.; et al. Clinical utility of radiomics at baseline rectal MRI to predict complete response of rectal cancer after chemoradiation therapy. Abdom. Radiol. 2020, 45, 3608–3617. [Google Scholar] [CrossRef]
- Calders, T.; Jaroszewicz, S. Efficient AUC optimization for classification. In European Conference on Principles of Data Mining and Knowledge Discovery; Springer: Berlin, Germany, 2007; pp. 42–53. [Google Scholar]
- Liu, L.; Jiang, H.; He, P.; Chen, W.; Liu, X.; Gao, J.; Han, J. On the variance of the adaptive learning rate and beyond. arXiv 2019, arXiv:1908.03265. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Comelli, A.; Stefano, A.; Coronnello, C.; Russo, G.; Vernuccio, F.; Cannella, R.; Salvaggio, G.; Lagalla, R.; Barone, S. Radiomics: A new biomedical workflow to create a predictive model. In Annual Conference on Medical Image Understanding and Analysis; Springer: Berlin, Germany, 2020; pp. 280–293. [Google Scholar]
- Marini, N.; Otálora, S.; Müller, H.; Atzori, M. Semi-supervised training of deep convolutional neural networks with heterogeneous data and few local annotations: An experiment on prostate histopathology image classification. Med. Image Anal. 2021, 73, 102165. [Google Scholar] [CrossRef]
- Granzier, R.; Verbakel, N.; Ibrahim, A.; van Timmeren, J.; van Nijnatten, T.; Leijenaar, R.; Lobbes, M.; Smidt, M.; Woodruff, H. MRI-based radiomics in breast cancer: Feature robustness with respect to inter-observer segmentation variability. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 2015; pp. 234–241. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Comelli, A.; Dahiya, N.; Stefano, A.; Vernuccio, F.; Portoghese, M.; Cutaia, G.; Bruno, A.; Salvaggio, G.; Yezzi, A. Deep Learning-Based Methods for Prostate Segmentation in Magnetic Resonance Imaging. Appl. Sci. 2021, 11, 782. [Google Scholar] [CrossRef]
- Habr-Gama, A.; Perez, R.O.; Nadalin, W.; Sabbaga, J.; Ribeiro, U., Jr.; Silva e Sousa, A.H., Jr.; Campos, F.G.; Kiss, D.R.; Gama-Rodrigues, J. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: Long-term results. Ann. Surg. 2004, 240, 711. [Google Scholar] [CrossRef]
- Habr-Gama, A.; Perez, R.O.; Proscurshim, I.; Campos, F.G.; Nadalin, W.; Kiss, D.; Gama-Rodrigues, J. Patterns of failure and survival for nonoperative treatment of stage c0 distal rectal cancer following neoadjuvant chemoradiation therapy. J. Gastrointest. Surg. 2006, 10, 1319–1329. [Google Scholar] [CrossRef]
- Horvat, N.; Carlos Tavares Rocha, C.; Clemente Oliveira, B.; Petkovska, I.; Gollub, M.J. MRI of rectal cancer: Tumor staging, imaging techniques, and management. Radiographics 2019, 39, 367–387. [Google Scholar] [CrossRef]
- Beets-Tan, R.G.; Lambregts, D.M.; Maas, M.; Bipat, S.; Barbaro, B.; Curvo-Semedo, L.; Fenlon, H.M.; Gollub, M.J.; Gourtsoyianni, S.; Halligan, S.; et al. Magnetic resonance imaging for clinical management of rectal cancer: Updated recommendations from the 2016 European Society of Gastrointestinal and Abdominal Radiology (ESGAR) consensus meeting. Eur. Radiol. 2018, 28, 1465–1475. [Google Scholar] [CrossRef]
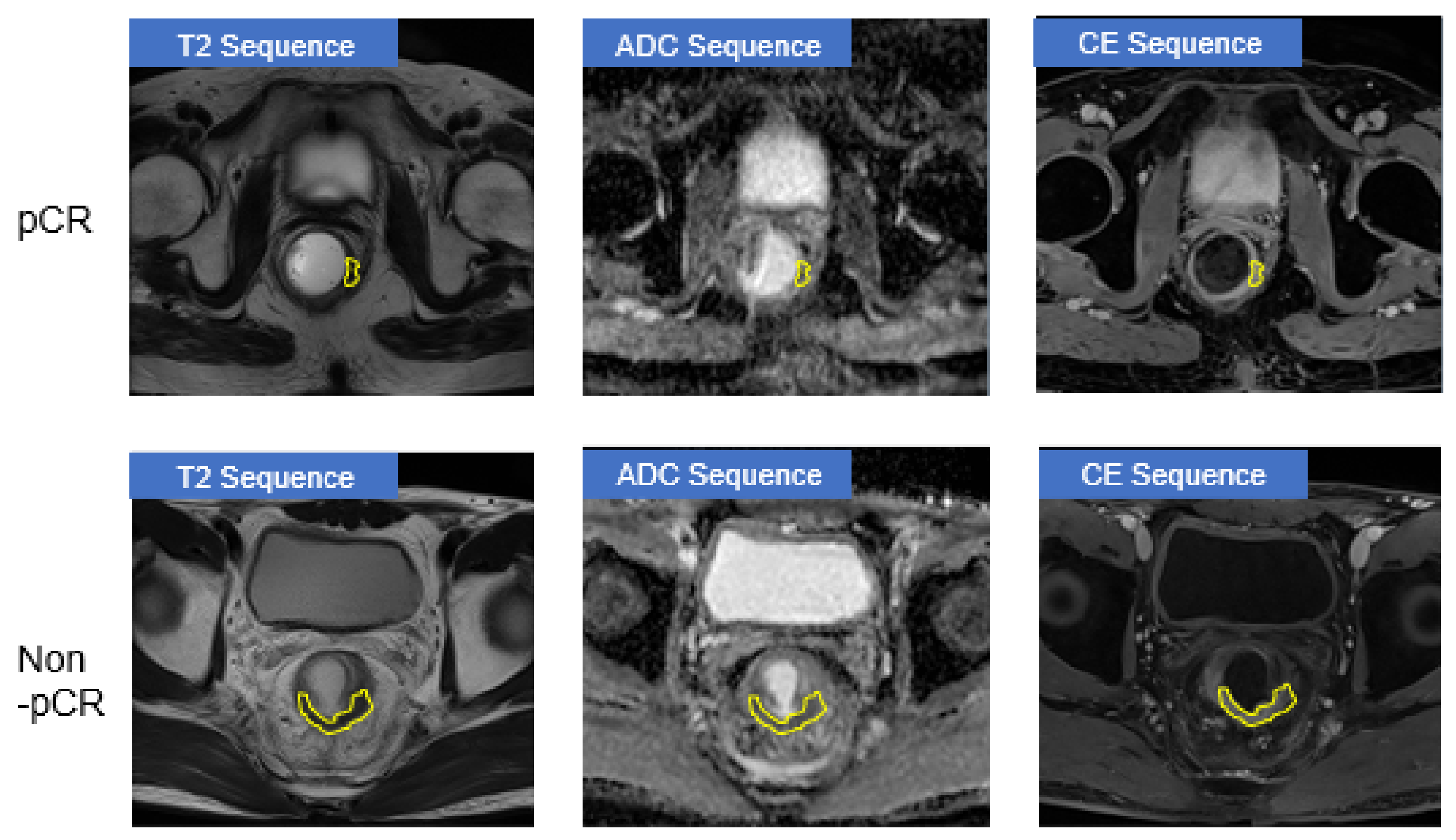


| Train (n = 592) | Validation (n = 320) | |
|---|---|---|
| Age (mean ± SD years) | 58.8 ± 12.1 | 59.5 ± 11.8 |
| Male (n (%))/Female (n (%)) | 388 (65.5)/204 (34.5) | 199 (62.2)/121 (37.8) |
| pCR (n (%)) | 114 (19.3) | 78 (24.4) |
| ypT stage (n (%)) | ||
| T0 | 114 (19.3) | 78 (24.4) |
| Tis | 6 (1.0) | 8 (2.5) |
| T1 | 36 (6.1) | 14 (4.4) |
| T2 | 145 (24.5) | 60 (18.8) |
| T3 | 285 (48.1) | 156 (48.8) |
| T4 | 6 (1.0) | 4 (1.3) |
| ypN stage (n (%)) | ||
| N0 | 409 (69.1) | 231 (72.2) |
| N1 | 139 (23.5) | 76 (23.8) |
| N2 | 44 (7.4) | 13 (4.1) |
| Input MRI Sequences | No Dimension Reduction | PCA | Kernel PCA |
|---|---|---|---|
| {T2} | 0.765 | 0.791 | 0.787 |
| {DWI/ADC} | 0.721 | 0.801 | 0.791 |
| {CE} | 0.716 | 0.800 | 0.801 |
| {T2, DWI/ADC} | 0.764 | 0.793 | 0.800 |
| {T2, DWI/ADC, and CE} | 0.811 | 0.804 | 0.819 |
| Feature Vector | No Dimension Reduction | PCA | Kernel PCA |
|---|---|---|---|
| MSFI embedding | 0.776 | 0.739 | 0.732 |
| Radiomics features | 0.811 | 0.754 | 0.819 |
| Concatenation of both | 0.796 | 0.746 | 0.837 |
| Classifier | Logistic Regression | Xgboost | Lightgbm | Random Forest | MLP | Ensemble |
|---|---|---|---|---|---|---|
| AUC | 0.804 | 0.783 | 0.792 | 0.837 | 0.798 | 0.819 |
| pCR Prediction Method | AUC | F1-Score | Specificity | Sensitivity |
|---|---|---|---|---|
| (B1) SVM classifier (input = radiomics features) [46] | 0.799 | 0.53 | 0.45 | 0.67 |
| (B2) RF classifier (input = radiomics features) [8] | 0.811 | 0.63 | 0.56 | 0.74 |
| (B3) MLP classifier (input = radiomics features) [25] | 0.763 | 0.54 | 0.49 | 0.62 |
| (B4) 3D-CNN classifier (input = MRI images) [18] | 0.807 | 0.63 | 0.59 | 0.68 |
| Proposed Method | 0.837 | 0.65 | 0.60 | 0.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Lim, J.; Shin, J.; Kim, S.; Hwang, H. Pathologic Complete Response Prediction after Neoadjuvant Chemoradiation Therapy for Rectal Cancer Using Radiomics and Deep Embedding Network of MRI. Appl. Sci. 2021, 11, 9494. https://doi.org/10.3390/app11209494
Lee S, Lim J, Shin J, Kim S, Hwang H. Pathologic Complete Response Prediction after Neoadjuvant Chemoradiation Therapy for Rectal Cancer Using Radiomics and Deep Embedding Network of MRI. Applied Sciences. 2021; 11(20):9494. https://doi.org/10.3390/app11209494
Chicago/Turabian StyleLee, Seunghyun, Joonseok Lim, Jaeseung Shin, Sungwon Kim, and Heasoo Hwang. 2021. "Pathologic Complete Response Prediction after Neoadjuvant Chemoradiation Therapy for Rectal Cancer Using Radiomics and Deep Embedding Network of MRI" Applied Sciences 11, no. 20: 9494. https://doi.org/10.3390/app11209494
APA StyleLee, S., Lim, J., Shin, J., Kim, S., & Hwang, H. (2021). Pathologic Complete Response Prediction after Neoadjuvant Chemoradiation Therapy for Rectal Cancer Using Radiomics and Deep Embedding Network of MRI. Applied Sciences, 11(20), 9494. https://doi.org/10.3390/app11209494






