Diversity of Biodeteriorative Bacterial and Fungal Consortia in Winter and Summer on Historical Sandstone of the Northern Pergola, Museum of King John III’s Palace at Wilanow, Poland
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristic of Samples and Sampling Location
2.2. Mosses and Lichens Identification Methodology
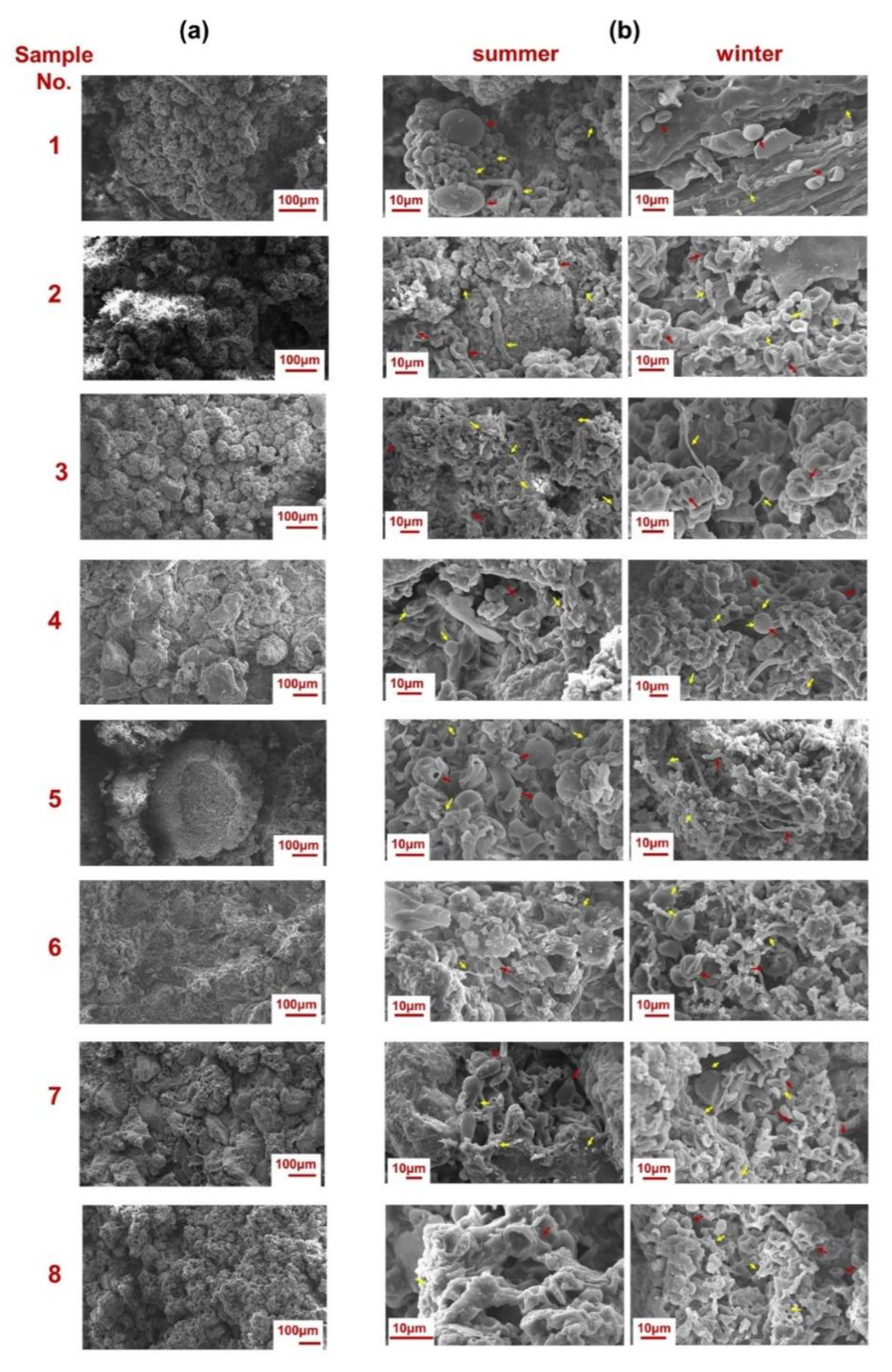
2.3. Microscopic Observations of the Biofilm Surfaces through Scanning Electron Microscopy (SEM)
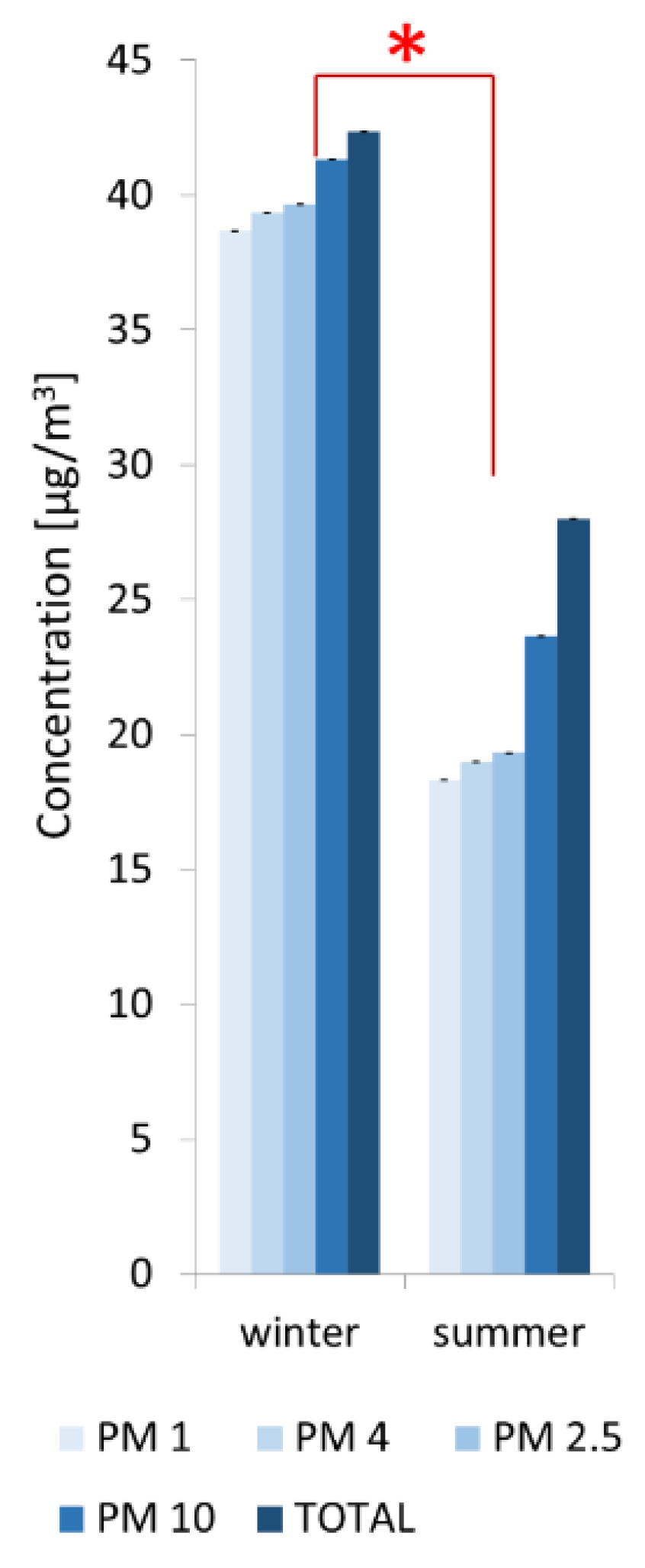
2.4. Quantitative Analysis of Particulate Matter (PM) Concentrations in Air
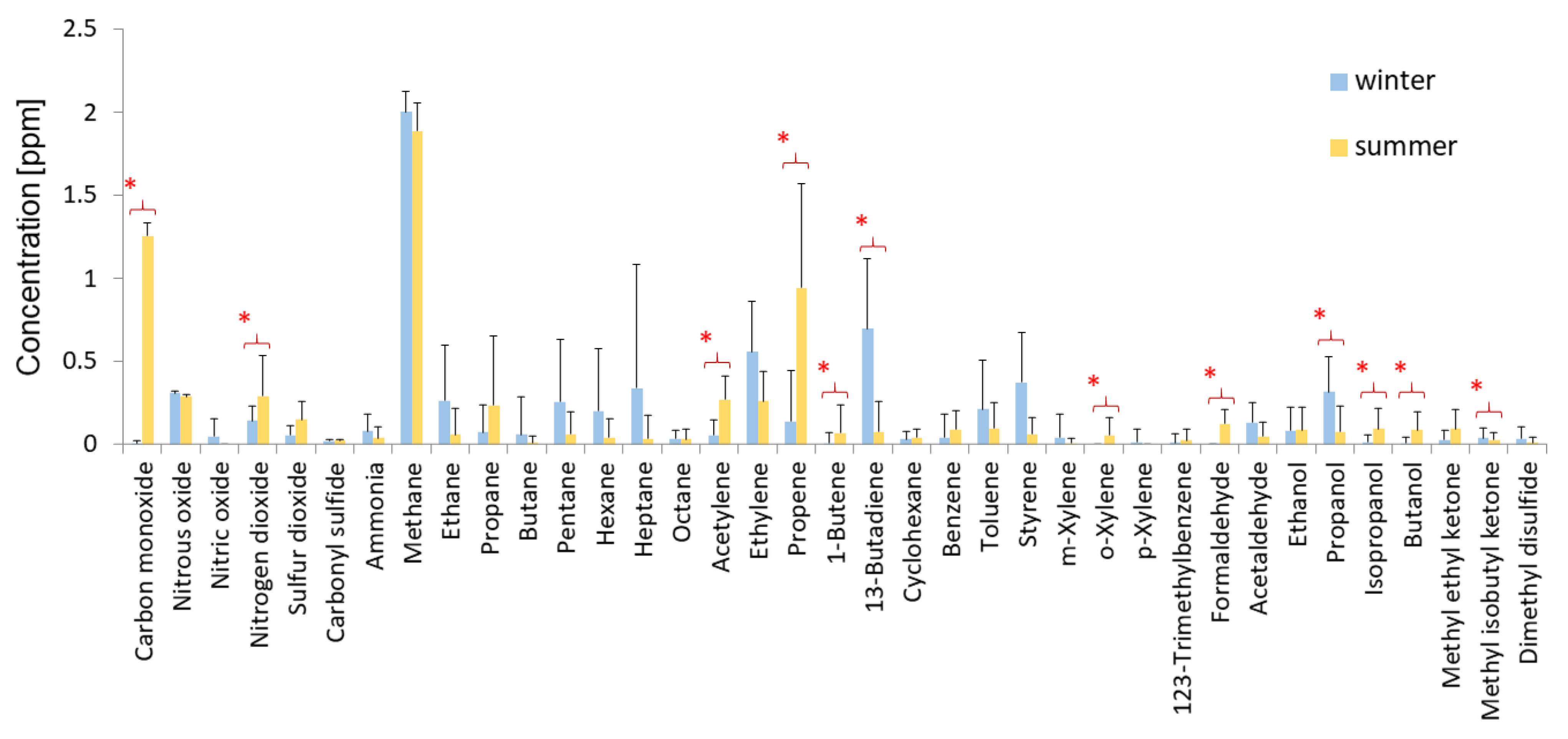
2.5. Measurements of the Chemical Composition of Air
2.6. DNA Isolation and Purification
2.7. Amplicon Preparation
2.8. DNA Sequencing
2.9. Processing of Raw Amplicon Reads
2.10. Accession Numbers of Nucleotide Sequences
2.11. Isolation and Analysis of Acidification Properties of Bacterial Strains
3. Results
3.1. Biodeterioration Patterns
- -
- green biofilms (GB) in sites 6 and 7.
- -
- yellow biofilms (YB) with lichen crusts (LC) and mosses (M) in site no. 8.
- -
- mosses (M) in sites 1, 2, and 3.
- -
- mosses (M) with lichen crusts (LC) in site no. 4.
- -
- lichen crusts (LC) in site no. 5.
3.2. Abiotic Factors Affecting Biodeterioration
3.3. Seasonal Changes of Microbial Biodiversity
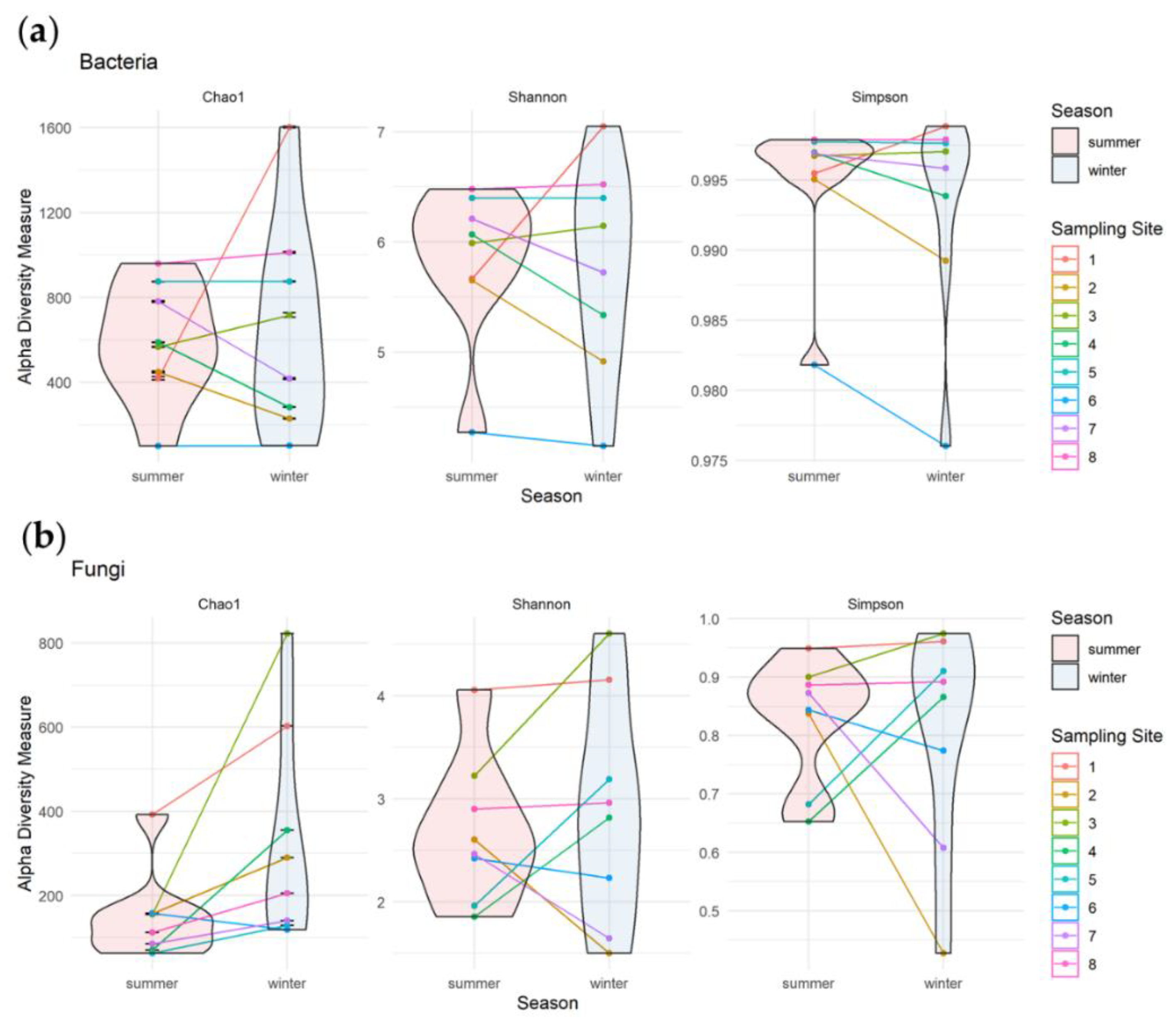
3.3.1. Alpha Diversity of Bacterial and Fungal Communities
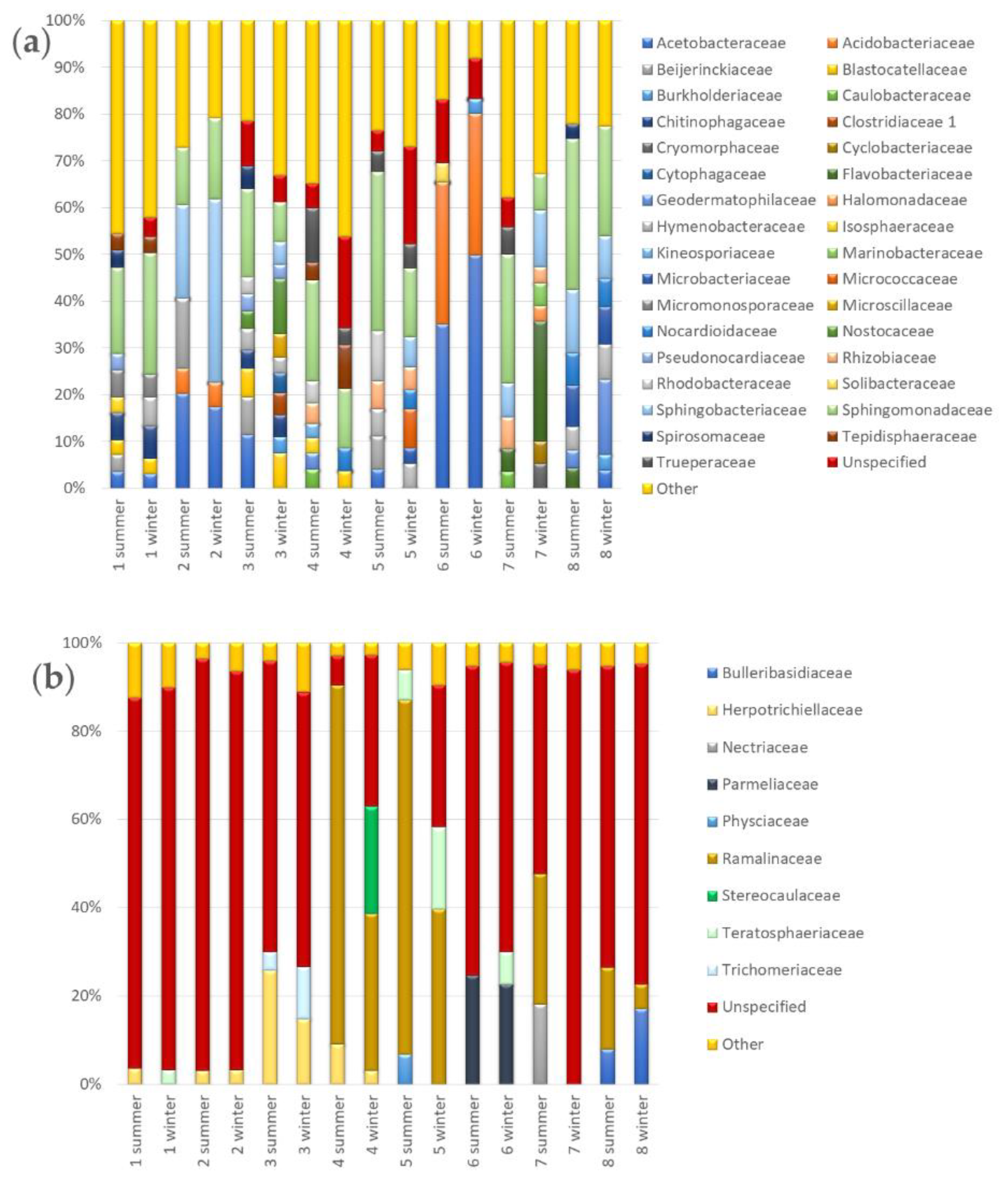
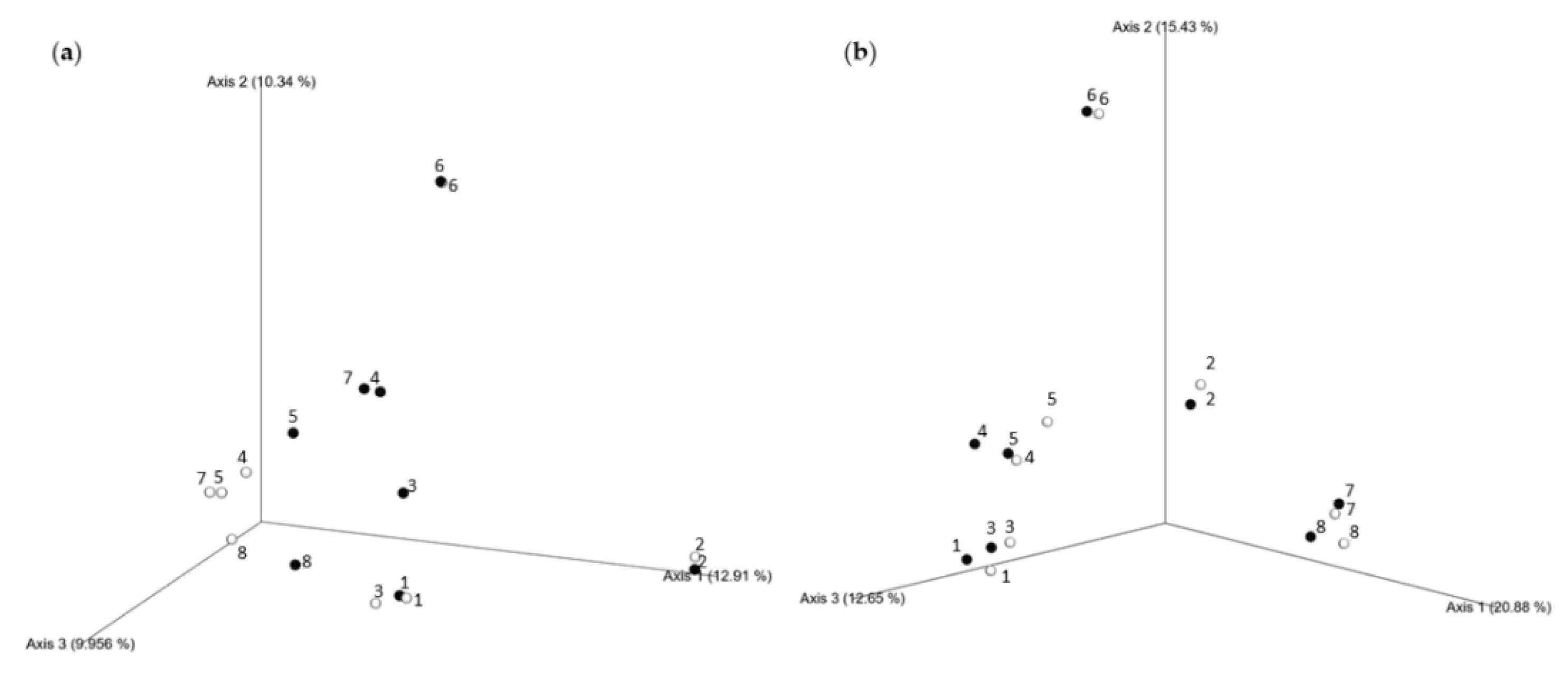
3.3.2. Seasonal Biodiversity of Bacterial and Fungal Communities
3.4. Acidifying Properties of Bacterial Strains Isolated within Subsequent Seasons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pangallo, D.; Bučková, M.; Kraková, L.; Puškárová, A.; Šaková, N.; Grivalský, T.; Chovanová, K.; Zemánková, M. Biodeterioration of epoxy resin: A microbial survey through culture-independent and culture-dependent approaches. Environ. Microbiol. 2015, 17, 462–479. [Google Scholar] [CrossRef] [PubMed]
- Piñar, G.; Garcia-Valles, M.; Gimeno-Torrente, D.; Fernandez-Turiel, J.L.; Ettenauer, J.; Sterflinger, K. Microscopic, chemical, and molecular-biological investigation of the decayed medieval stained window glasses of two Catalonian churches. Int. Biodeter. Biodegr. 2013, 84, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K.; Piñar, G. Microbial deterioration of cultural heritage and works of art—Tilting at windmills? Appl. Microbiol. Biotechnol. 2013, 97, 963–9646. [Google Scholar] [CrossRef] [PubMed]
- Zanardini, E.; May, E.; Inkpen, R.; Cappitelli, F.; Murrell, J.C.; Purdy, K.J. Diversity of archaeal and bacterial communities on exfoliated sandstone from Portchester Castle (UK). Int. Biodeter. Biodegr. 2016, 109, 78–87. [Google Scholar] [CrossRef]
- Pinheiro, A.C.; Mesquita, N.; Trovão, J.; Soaresa, F.; Tiagoa, I.; Coelhoa, C.; Paiva de Carvalhoa, H.; Gil, F.; Catarino, L.; Piñar, G.; et al. Limestone biodeterioration: A review on the Portuguese cultural heritage scenario. J. Cult. Herit. 2019, 36, 275–285. [Google Scholar] [CrossRef]
- Gadd, G.M. Geomycology: Biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 2007, 111, 3–49. [Google Scholar] [CrossRef]
- Gaylarde, C.C.; Rodríguez, C.H.; Navarro-Noya, Y.E.; Ortega-Morales, B.O. Microbial Biofilms on the Sandstone Monuments of the Angkor Wat Complex, Cambodia. Curr. Microbiol. 2012, 64, 85–92. [Google Scholar] [CrossRef]
- Cuzman, O.A.; Tiano, P.; Ventura, S.; Frediani, P. Biodiversity on Stone Artifacts. In The Importance of Biological Interactions in the Study of Biodiversity, 1st ed.; Lõpez-Pujol, J., Ed.; InTech: Rijeka, Croatia, 2011; pp. 367–390. Available online: http://www.intechopen.com/books/the-importance-ofbiological-interactions-in-the-study-of-biodiversity/biodiversity-on-stone-artifacts (accessed on 8 September 2020). [CrossRef]
- Cutler, N.A.; Chaput, D.L.; Oliver, A.E.; Viles, H.A. The spatial organization and microbial community structure of an epilithic biofilm. FEMS Microbiol. Ecol. 2015, 91, fiu027. [Google Scholar] [CrossRef]
- Hueck, H.J. The biodeterioration of materials—An appraisal. Int. Biodeter. Biodegr. 2001, 48, 5–11. [Google Scholar] [CrossRef]
- Perito, B.; Cavalieri, D. Innovative metagenomic approaches for detection of microbial communities involved in biodeteriorattion of cultural heritage. In Proceedings of the Florence Heri-Tech—The Future of Heritage Science and Technologies, Florence, Italy, 16–18 May 2018. IOP Conf. Ser. Mater. Sci. Eng. 2018, 364, 012074. [Google Scholar] [CrossRef]
- Korkanç, A.; Savran, A. Impact of the surface roughness of stones used in historical buildings on biodeterioration. Construct. Build. Mater. 2015, 80, 279–294. [Google Scholar] [CrossRef]
- Warscheid, T.; Braams, J. Biodeterioration of stone: A review. Int. Biodeter. Biodegr. 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Kusumi, A.; Li, X.S.; Katayama, Y. Mycobacteria isolated from Angkor monument sandstones grow chemolithoautotrophically by oxidizing elemental sulfur. Front. Microbiol. 2011, 2, 104. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M.; Bahri-Esfahani, J.; Li, Q.; Rhee, Y.J.; Wei, Z.; Fomina, M.; Liang, X. Oxalate production by fungi: Significance in geomycology, biodeterioration and bioremediation. Fungal Biol. Rev. 2014, 28, 36–55. [Google Scholar] [CrossRef]
- Lisci, M.; Monte, M.; Pacini, E. Lichens and higher plants on stone: A review. Int. Biodeter. Biodegr. 2003, 51, 1–17. [Google Scholar] [CrossRef]
- Hoppert, M.; Flies, C.; Pohl, W.; Gunzl, B.; Schneider, J. Colonization strategies of lithobiontic microorganisms on carbonate rocks. Environ. Geol. 2004, 46, 421–428. [Google Scholar] [CrossRef]
- McNamara, C.J.; Perry, T.D., 4th; Bearce, K.A.; Hernandez-Duque, G.; Mitchell, R. Epilithic and Endolithic Bacterial Communities in Limestone from a Maya Archaeological Site. Microb. Ecol. 2006, 51, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Gorbushina, A.A.; Heyrman, J.; Dornieden, T.; Gonzalez-Delvalle, M.; Krumbein, W.E.; Laiz, L.; Petersen, K.; Saiz-Jimenez, C.; Swings, J. Bacterial and fungal diversity and biodeterioration problems in mural painting environments of St. Martins church (Greene-Kreiensen, Germany). Int. Biodeter. Biodegr. 2004, 53, 13–24. [Google Scholar] [CrossRef]
- Gorbushina, A.A. Life on the rocks. Environ. Microbiol. 2007, 9, 1613–1631. [Google Scholar] [CrossRef]
- Mohammadi, P.; Maghboli-Balasjin, N. Isolation and molecular identification of deteriorating fungi from Cyrus the Great tomb Stones. Iran. J. Microbiol. 2014, 6, 361–370. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4385579/ (accessed on 11 September 2020).
- Caneva, G.; Bartoli, F.; Ceschin, S.; Salvadori, O.; Futagami, Y.; Salvati, L. Exploring ecological relationships in the biodeterioration patterns of Angkor temples (Cambodia) along a forest canopy gradient. J. Cult. Herit. 2015, 16, 728–735. [Google Scholar] [CrossRef]
- Cennamo, P.; Montuori, N.; Trojsi, G.; Fatigati, G.; Moretti, A. Biofilms in churches built in grottoes. Sci. Total Environ. 2016, 543, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, A.; Li, X.S.; Osuga, Y.; Kawashima, A.; Gu, J.-D.; Nasu, M.; Katayama, Y. Bacterial Communities in Pigmented Biofilms Formed on the Sandstone Bas-Relief Walls of the Bayon Temple, Angkor Thom, Cambodia. Microbes Environ. 2013, 28, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Kremer, B.P.; Muhle, H. Porosty, Mszaki, Paprotniki (Polish Edition); Swiat Ksiazki Press: Warsaw, Poland, 1998; ISBN 8372270619. [Google Scholar]
- Wójciak, H. Flora Polski. Porosty, Mszaki, Paprotniki, 2nd ed.; MULTICO Oficyna Wydawnicza: Warsaw, Poland, 2007; ISBN 9788370735524. (In Polish) [Google Scholar]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- BBolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Herrero, A.; Flores, E. (Eds.) The Cyanobacteria: Molecular Biology, Genomics and Evolution, 1st ed.; Caister Academic Press: Sevilla, Spain, 2008; ISBN 978-1-904455-15-8. [Google Scholar]
- Shi, X.L.; Lepère, C.; Scanlan, D.J.; Vaulot, D. Plastid 16S rRNA Gene Diversity among Eukaryotic Picophytoplankton Sorted by Flow Cytometry from the South Pacific Ocean. PLoS ONE 2011, 6, e18979. [Google Scholar] [CrossRef]
- Caneva, G.; Bartoli, F.; Savo, V.; Futagami, Y.; Strona, G. Combining Statistical Tools and Ecological Assessments in the Study of Biodeterioration Patterns of Stone Temples in Angkor (Cambodia). Sci. Rep. 2016, 6, 32601. [Google Scholar] [CrossRef]
- Caneva, G.; Fidanza, M.R.; Tonon, C.; Favero-Longo, S.E. Biodeterioration Patterns and Their Interpretation for Potential Applications to Stone Conservation: A Hypothesis from Allelopathic Inhibitory Effects of Lichens on the Caestia Pyramid (Rome). Sustainability 2020, 12, 1132. [Google Scholar] [CrossRef]
- Ramesh, C.; Vinithkumar, N.V.; Kirubagaran, R.; Venil, C.K.; Dufossé, L. Multifaceted Applications of Microbial Pigments: Current Knowledge, Challenges and Future Directions for Public Health Implications. Microorganisms 2019, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Panzer, K.; Yilmaz, P.; Weiß, M.; Reich, L.; Richter, M.; Wiese, J.; Schmaljohann, R.; Labes, A.; Imhoff, J.F.; Glöckner, F.O.; et al. Identification of Habitat-Specific Biomes of Aquatic Fungal Communities Using a Comprehensive Nearly Full-Length 18S rRNA Dataset Enriched with Contextual Data. PLoS ONE 2015, 10, e0134377. [Google Scholar] [CrossRef] [PubMed]
- Stielow, J.B.; Lévesque, C.A.; Seifert, K.A.; Meyer, W.; Iriny, L.; Smits, D.; Renfurm, R.; Verkley, G.J.M.; Groenewald, M.; Chaduli, D.; et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 2015, 35, 242–263. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, F.; Casanova Municchia, A.; Futagami, Y.; Kashiwadani, H.; Moon, K.H.; Caneva, G. Biological colonization patterns on the ruins of Angkor temples (Cambodia) in the biodeterioration vs bioprotection debate. Int. Biodeter. Biodegr. 2014, 96, 157–165. [Google Scholar] [CrossRef]
- Matwiejuk, A. Matuzalem Genus of Lichens—Rhizocarpon geographiucm (L.) dc.—Its Properties and Application. Kosmos 2007, 56, 175–180. Available online: https://kosmos.ptpk.org/index.php/Kosmos/article/view/1322/1301 (accessed on 15 October 2020). (In Polish).
- Gehrmann, C.; Krumbein, W.E.; Petersen, K. Lichen weathering activities on mineral and rock surfaces. Stud. Geobot. 1988, 8, 33–45. [Google Scholar]
- Seaward, M.R.D. Lichens, agents of monumental destruction. Microbiol. Today 2003, 30, 110–112. Available online: https://socgenmicrobiol.org.uk/pubs/micro_today/pdf/080303.pdf (accessed on 15 October 2020).
- Studzińska, E.; Witkowska-Banaszczak, E.; Bylka, W. Bioactive compounds of lichen. Herb. Polon. 2008, 54, 79–88. Available online: http://herbapolonica.pl/magazines-files/9811528-Zwi%C4%85zki%20biologicznie.pdf (accessed on 16 October 2020). (In Polish).
- Boggs, J.R. Principles of Sedimentology and Stratigraphy, 3rd ed.; Pentice Hall: New York, NY, USA, 2000; ISBN 0-13-099696-3. [Google Scholar]
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Microbial deterioration of stone monuments-an updated overview. Adv. Appl. Microbiol. 2009, 66, 97–139. [Google Scholar] [CrossRef]
- Cutler, N.A.; Oliver, A.E.; Viles, H.A.; Ahmad, S.; Whiteley, A.S. The characterisation of eukaryotic microbial communities on sandstone buildings in Belfast, UK, using TRFLP and 454 pyrosequencing. Int. Biodeter. Biodegr. 2013, 82, 124–133. [Google Scholar] [CrossRef]
- Rajkowska, K.; Otlewska, A.; Koziróg, A.; Piotrowska, M.; Nowicka-Krawczyk, P.; Hachułka, M.; Wolski, G.J.; Kunicka-Styczyńska, A.; Gutarowska, B.; Żydzik-Białek, A. Assessment of biological colonization of historic buildings in the former Auschwitz II-Birkenau concentration camp. Ann. Microbiol. 2014, 64, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ge, Q.; Zhu, Z.; Deng, Y.; Gu, J.-D. Microbiological community of the Royal Palace in Angkor Thom and Beng Mealea of Cambodia by Illumina sequencing based on 16S rRNA gene. Int. Biodeter. Biodegr. 2018, 134, 127–135. [Google Scholar] [CrossRef]
- Brewer, T.A.; Fierer, N. Tales from the tomb: The microbial ecology of exposed rock surfaces. Environ. Microbiol. 2018, 20, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Dyda, M.; Pyzik, A.; Wilkojc, E.; Kwiatkowska-Kopka, B.; Sklodowska, A. Bacterial and Fungal Diversity Inside the Medieval Building Constructed with Sandstone Plates and Lime Mortar as an Example of the Microbial Colonization of a Nutrient-Limited Extreme Environment (Wawel Royal Castle, Krakow, Poland). Microorganisms 2019, 7, 416. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, B.; He, Z.; Yang, X. Distribution and Diversity of Bacteria and Fungi Colonization in Stone Monuments Analyzed by High-Throughput Sequencing. PLoS ONE 2016, 11, e0163287. [Google Scholar] [CrossRef] [PubMed]
- Chimienti, G.; Piredda, R.; Pepe, G.; van der Werf, I.D.; Sabbatini, L.; Crecchio, C.; Ricciuti, P.; D’Erchia, A.M.; Manzari, C.; Pesole, G. Profile of microbial communities on carbonate stones of the medieval church of San Leonardo di Siponto (Italy) by Illumina-based deep sequencing. Appl. Microbiol. Biotechnol. 2016, 100, 8537–8548. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; Wang, L.; Ge, Q. Distribution and diversity of bacteria and fungi colonizing ancient Buddhist statues analyzed by high-throughput sequencing. Int. Biodeter. Biodegr. 2017, 117, 245–254. [Google Scholar] [CrossRef]
- Gadd, G.M. Geomicrobiology of the built environment. Nat. Microbiol. 2017, 2, 16275. [Google Scholar] [CrossRef]
- Gutarowska, B.; Celikkol-Aydin, S.; Bonifay, V.; Otlewska, A.; Aydin, E.; Oldham, A.L.; Brauer, J.I.; Duncan, K.E.; Adamiak, J.; Sunner, J.A.; et al. Metabolomic and high-throughput sequencing analysis—modern approach for the assessment of biodeterioration of materials from historic buildings. Front. Microbiol. 2015, 6, 979. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Otlewska, A.; Gutarowska, B. Environmental parameters conditioning microbially induced mineralization under the experimental model conditions. Acta Biochim. Pol. 2016, 63, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Nuhoglu, Y.; Oguz, E.; Uslu, H.; Ozbek, A.; Ipekoglu, B.; Ocak, I.; Hasenekoglu, I. The accelerating effects of the microorganisms on biodeterioration of stone monuments under air pollution and continental-cold climatic conditions in Erzurum, Turkey. Sci. Total Environ. 2006, 364, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Viles, H.A.; Gorbushina, A.A. Soiling and microbial colonisation on urban roadside limestone: A three year study in Oxford, England. Build. Environ. 2003, 38, 1217–1224. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyda, M.; Laudy, A.; Decewicz, P.; Romaniuk, K.; Ciezkowska, M.; Szajewska, A.; Solecka, D.; Dziewit, L.; Drewniak, L.; Skłodowska, A. Diversity of Biodeteriorative Bacterial and Fungal Consortia in Winter and Summer on Historical Sandstone of the Northern Pergola, Museum of King John III’s Palace at Wilanow, Poland. Appl. Sci. 2021, 11, 620. https://doi.org/10.3390/app11020620
Dyda M, Laudy A, Decewicz P, Romaniuk K, Ciezkowska M, Szajewska A, Solecka D, Dziewit L, Drewniak L, Skłodowska A. Diversity of Biodeteriorative Bacterial and Fungal Consortia in Winter and Summer on Historical Sandstone of the Northern Pergola, Museum of King John III’s Palace at Wilanow, Poland. Applied Sciences. 2021; 11(2):620. https://doi.org/10.3390/app11020620
Chicago/Turabian StyleDyda, Magdalena, Agnieszka Laudy, Przemyslaw Decewicz, Krzysztof Romaniuk, Martyna Ciezkowska, Anna Szajewska, Danuta Solecka, Lukasz Dziewit, Lukasz Drewniak, and Aleksandra Skłodowska. 2021. "Diversity of Biodeteriorative Bacterial and Fungal Consortia in Winter and Summer on Historical Sandstone of the Northern Pergola, Museum of King John III’s Palace at Wilanow, Poland" Applied Sciences 11, no. 2: 620. https://doi.org/10.3390/app11020620
APA StyleDyda, M., Laudy, A., Decewicz, P., Romaniuk, K., Ciezkowska, M., Szajewska, A., Solecka, D., Dziewit, L., Drewniak, L., & Skłodowska, A. (2021). Diversity of Biodeteriorative Bacterial and Fungal Consortia in Winter and Summer on Historical Sandstone of the Northern Pergola, Museum of King John III’s Palace at Wilanow, Poland. Applied Sciences, 11(2), 620. https://doi.org/10.3390/app11020620




