Consecutive Independence and Correlation Transform for Multimodal Data Fusion: Discovery of One-to-Many Associations in Structural and Functional Imaging Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Brain Data
2.1.1. Data Acquisition
2.1.2. Data Preprocessing and Feature Extraction
2.2. Background
2.2.1. ICA
2.2.2. IVA
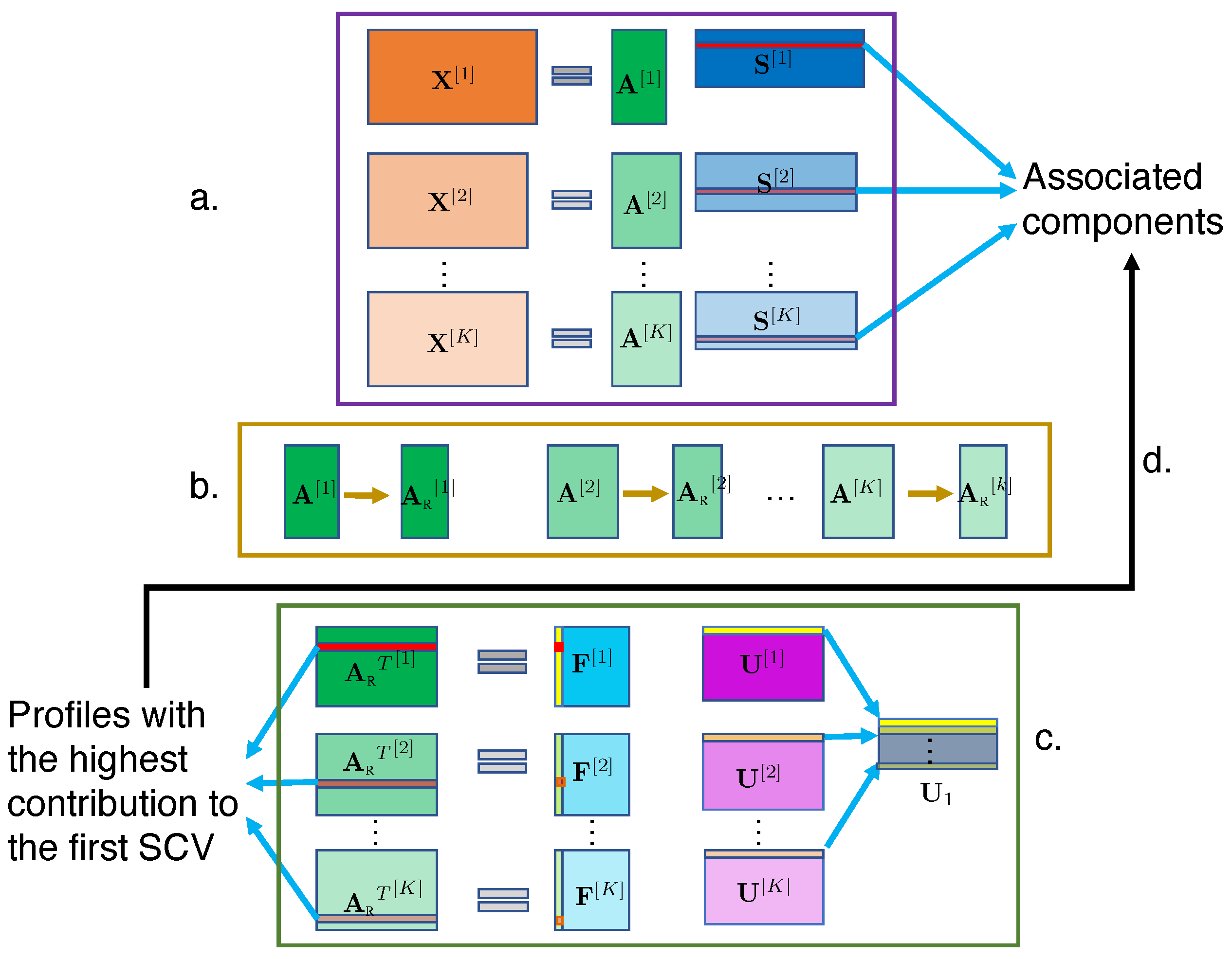
2.3. C-ICT Framework
2.3.1. C-ICT Step 1: ICA
2.3.2. C-ICT Step 2: Artifact Elimination
2.3.3. C-ICT Step 3: IVA
2.3.4. C-ICT Step 4: Tracing Back to Components
3. Implementation and Results
3.1. Implementation
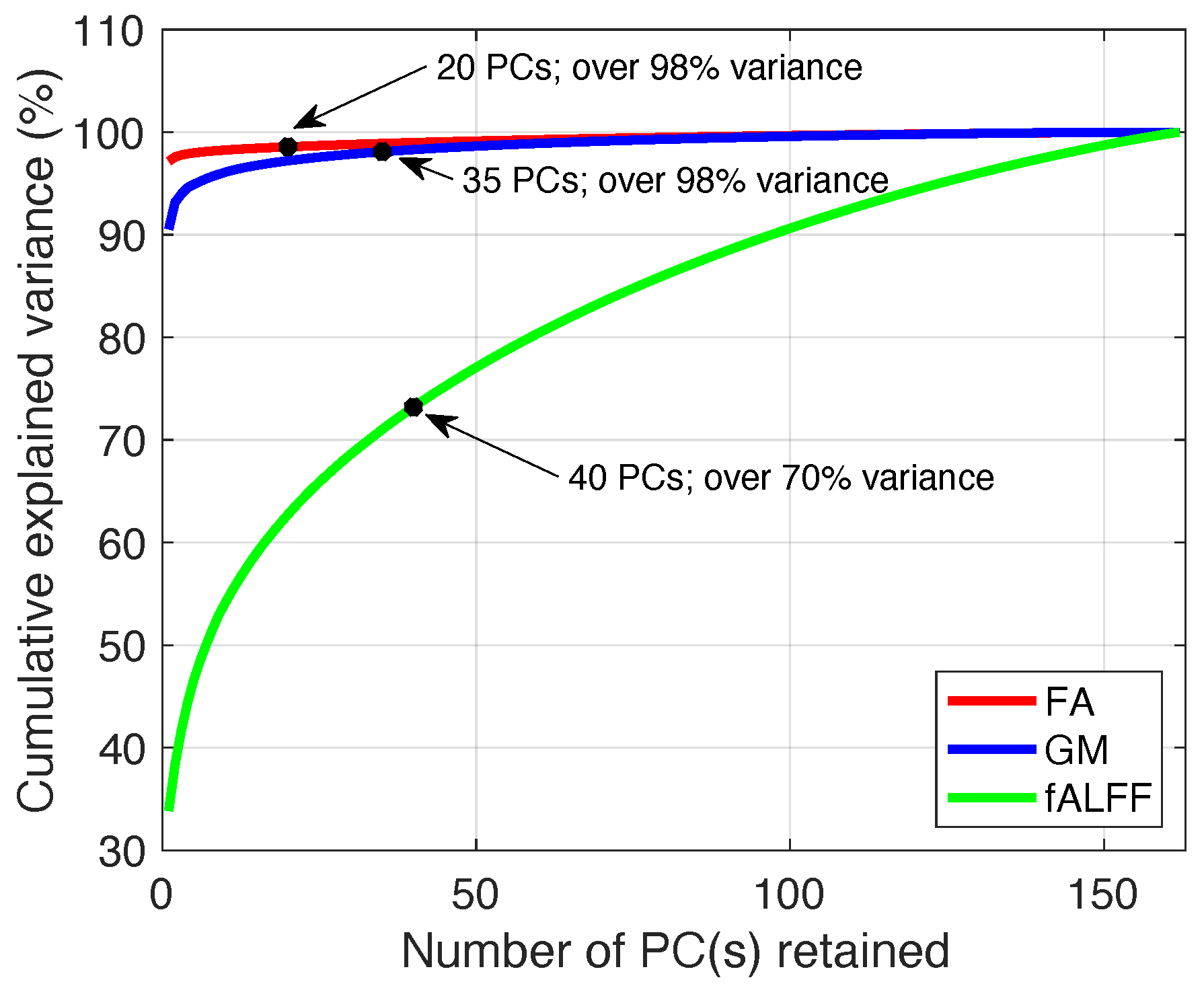
3.1.1. Order Selection
3.1.2. Algorithm Choice
3.1.3. Artifact Elimination
3.1.4. Group Differences
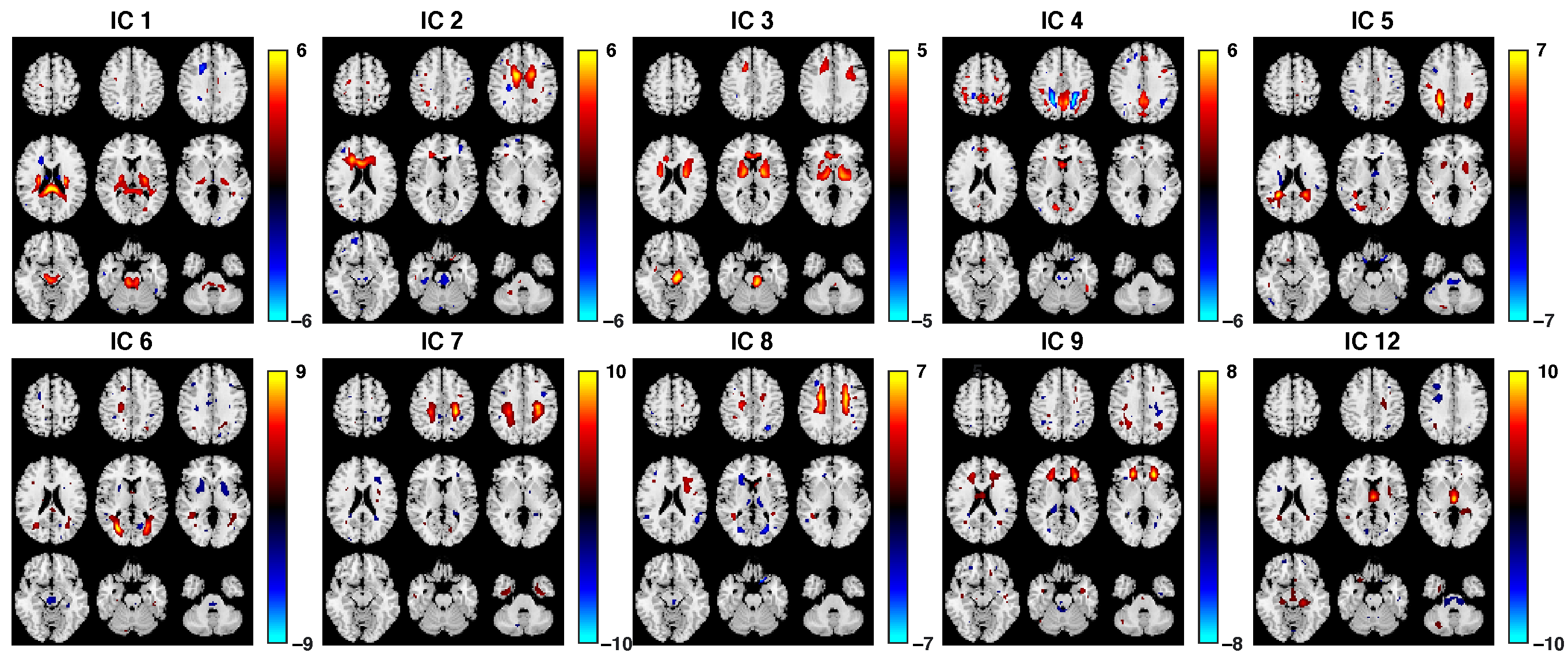
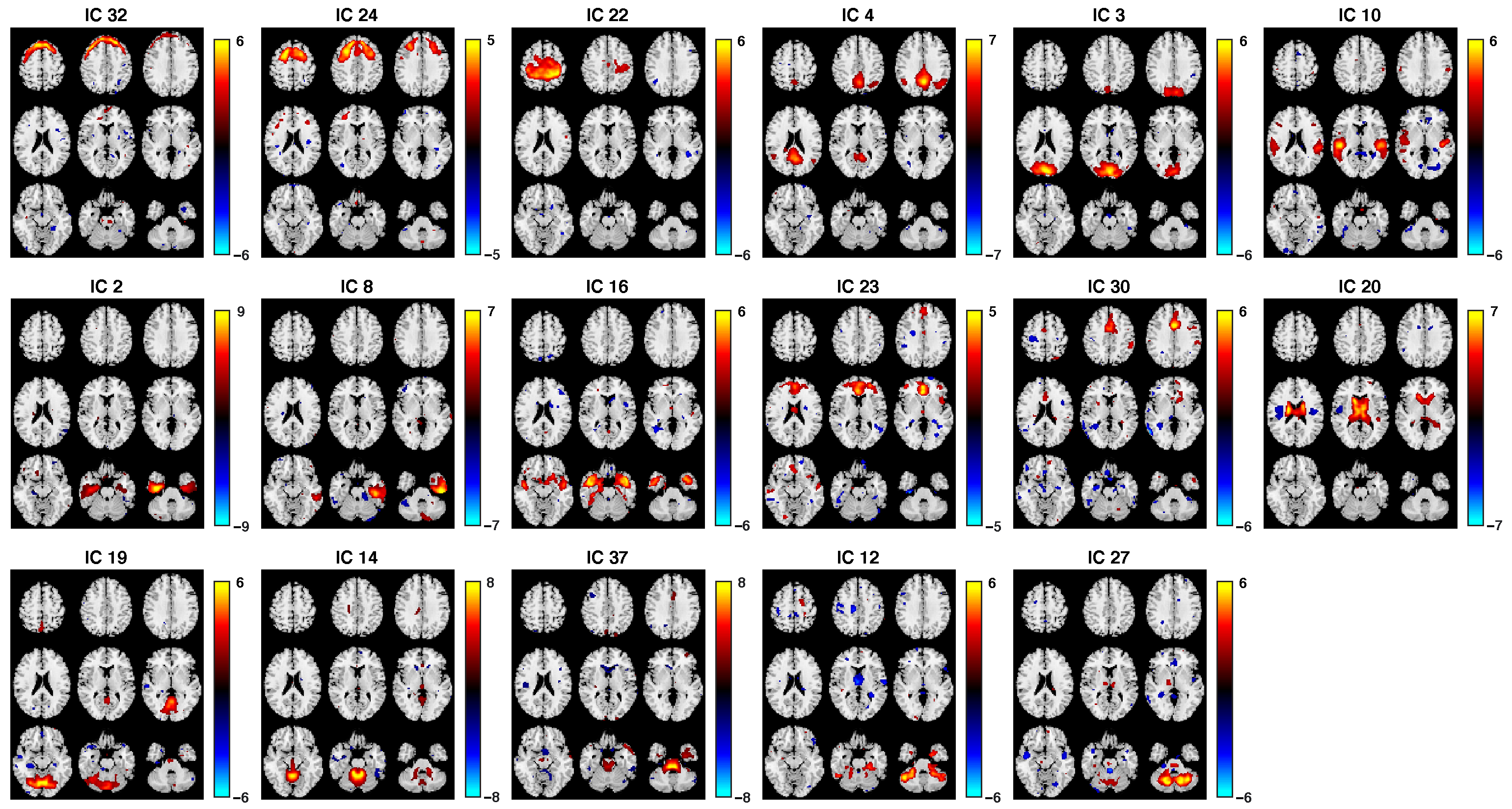
3.2. Fusion Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le Bihan, D.; Johansen-Berg, H. Diffusion MRI at 25: Exploring brain tissue structure and function. NeuroImage 2012, 61, 324–341. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Dai, Z.; Gong, G.; Zhou, C.; He, Y. Understanding structural-functional relationships in the human brain: A large-scale network perspective. Neuroscientist 2015, 21, 290–305. [Google Scholar] [CrossRef] [PubMed]
- Birur, B.; Kraguljac, N.V.; Shelton, R.C.; Lahti, A.C. Brain structure, function, and neurochemistry in schizophrenia and bipolar disorder—A systematic review of the magnetic resonance neuroimaging literature. NPJ Schizophr. 2017, 3, 1–15. [Google Scholar] [CrossRef]
- Dumoulin, S.O.; Fracasso, A.; van der Zwaag, W.; Siero, J.C.; Petridou, N. Ultra-high field MRI: Advancing systems neuroscience towards mesoscopic human brain function. NeuroImage 2018, 168, 345–357. [Google Scholar] [CrossRef]
- Le Bihan, D.; Mangin, J.F.; Poupon, C.; Clark, C.A.; Pappata, S.; Molko, N.; Chabriat, H. Diffusion tensor imaging: Concepts and applications. J. Magn. Reson. Imaging Off. J. Int. Soc. Magn. Reson. Med. 2001, 13, 534–546. [Google Scholar] [CrossRef]
- Coffman, J.A.; Bornstein, R.A.; Olson, S.C.; Schwarzkopf, S.B.; Nasrallah, H.A. Cognitive impairment and cerebral structure by MRI in bipolar disorder. Biol. Psychiatry 1990, 27, 1188–1196. [Google Scholar] [CrossRef]
- Biswal, B.; Zerrin Yetkin, F.; Haughton, V.M.; Hyde, J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 1995, 34, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fan, G. Alterations of structural and functional connectivity in profound sensorineural hearing loss infants within an early sensitive period: A combined DTI and fMRI study. Dev. Cogn. Neurosci. 2019, 38, 100654. [Google Scholar] [CrossRef]
- Hirjak, D.; Rashidi, M.; Kubera, K.M.; Northoff, G.; Fritze, S.; Schmitgen, M.M.; Sambataro, F.; Calhoun, V.D.; Wolf, R.C. Multimodal magnetic resonance imaging data fusion reveals distinct patterns of abnormal brain structure and function in catatonia. Schizophr. Bull. 2020, 46, 202–210. [Google Scholar] [CrossRef] [Green Version]
- Sui, J.; Yu, Q.; He, H.; Pearlson, G.; Calhoun, V.D. A selective review of multimodal fusion methods in schizophrenia. Front. Hum. Neurosci. 2012, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahat, D.; Adali, T.; Jutten, C. Multimodal data fusion: An overview of methods, challenges, and prospects. Proc. IEEE 2015, 103, 1449–1477. [Google Scholar] [CrossRef] [Green Version]
- Adali, T.; Levin-Schwartz, Y.; Calhoun, V.D. Multimodal data fusion using source separation: Application to medical imaging. Proc. IEEE 2015, 103, 1494–1506. [Google Scholar] [CrossRef]
- Calhoun, V.D.; Adali, T. Feature-based fusion of medical imaging data. IEEE Trans. Inf. Technol. Biomed. 2008, 13, 711–720. [Google Scholar] [CrossRef] [Green Version]
- James, A.P.; Dasarathy, B.V. Medical image fusion: A survey of the state of the art. Inf. Fusion 2014, 19, 4–19. [Google Scholar] [CrossRef] [Green Version]
- Adali, T.; Levin-Schwartz, Y.; Calhoun, V.D. Multimodal data fusion using source separation: Two effective models based on ICA and IVA and their properties. Proc. IEEE 2015, 103, 1478–1493. [Google Scholar] [CrossRef] [Green Version]
- Adali, T.; Akhonda, M.; Calhoun, V.D. ICA and IVA for data fusion: An overview and a new approach based on disjoint subspaces. IEEE Sens. Lett. 2018, 3, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Comon, P.; Jutten, C. Handbook of Blind Source Separation: Independent Component Analysis and Applications; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Harold, H. Relations between two sets of variates. Biometrika 1936, 28, 321–377. [Google Scholar]
- Correa, N.M.; Li, Y.O.; Adali, T.; Calhoun, V.D. Canonical correlation analysis for feature-based fusion of biomedical imaging modalities and its application to detection of associative networks in schizophrenia. IEEE J. Sel. Top. Signal Process. 2008, 2, 998–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.; Eltoft, T.; Lee, T.W. Independent vector analysis: An extension of ICA to multivariate components. In Proceedings of the International Conference on Independent Component Analysis and Signal Separation, Charleston, SC, USA, 5–8 March 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 165–172. [Google Scholar]
- Adali, T.; Anderson, M.; Fu, G.S. Diversity in independent component and vector analyses: Identifiability, algorithms, and applications in medical imaging. IEEE Signal Process. Mag. 2014, 31, 18–33. [Google Scholar] [CrossRef]
- Kettenring, J.R. Canonical analysis of several sets of variables. Biometrika 1971, 58, 433–451. [Google Scholar] [CrossRef]
- Correa, N.M.; Eichele, T.; Adali, T.; Li, Y.; Calhoun, V.D. Multi-set canonical correlation analysis for the fusion of concurrent single trial ERP and functional MRI. NeuroImage 2010, 50, 1438–1445. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.; Fu, G.S.; Phlypo, R.; Adali, T. Independent vector analysis: Identification conditions and performance bounds. IEEE Trans. Signal Process. 2014, 62, 4399–4410. [Google Scholar] [CrossRef] [Green Version]
- Calhoun, V.D.; Adali, T.; Pearlson, G.; Kiehl, K.A. Neuronal chronometry of target detection: Fusion of hemodynamic and event-related potential data. NeuroImage 2006, 30, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; He, H.; Pearlson, G.D.; Adali, T.; Kiehl, K.A.; Yu, Q.; Clark, V.P.; Castro, E.; White, T.; Mueller, B.A.; et al. Three-way (N-way) fusion of brain imaging data based on mCCA+ jICA and its application to discriminating schizophrenia. NeuroImage 2013, 66, 119–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin-Schwartz, Y.; Calhoun, V.D.; Adali, T. Data-driven fusion of EEG, functional and structural MRI: A comparison of two models. In Proceedings of the 2014 48th Annual Conference on Information Sciences and Systems (CISS), Princeton, NJ, USA, 19–21 March 2014; pp. 1–6. [Google Scholar]
- Akhonda, M.A.B.S.; Levin-Schwartz, Y.; Bhinge, S.; Calhoun, V.D.; Adali, T. Consecutive independence and correlation transform for multimodal fusion: Application to EEG and fMRI data. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 2311–2315. [Google Scholar]
- Jia, C.; Akhonda, M.A.B.S.; Long, Q.; Calhoun, V.D.; Waldstein, S.; Adali, T. C-ICT for Discovery of Multiple Associations in Multimodal Imaging Data: Application to Fusion of fMRI and DTI Data. In Proceedings of the 2019 53rd Annual Conference on Information Sciences and Systems (CISS), Baltimore, MD, USA, 20–22 March 2019; pp. 1–5. [Google Scholar]
- Anderson, M.; Adali, T.; Li, X.L. Joint blind source separation with multivariate Gaussian model: Algorithms and performance analysis. IEEE Trans. Signal Process. 2011, 60, 1672–1683. [Google Scholar] [CrossRef]
- Scott, A.; Courtney, W.; Wood, D.; De la Garza, R.; Lane, S.; Wang, R.; King, M.; Roberts, J.; Turner, J.A.; Calhoun, V.D. COINS: An innovative informatics and neuroimaging tool suite built for large heterogeneous datasets. Front. Neuroinform. 2011, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.; King, M.; Landis, D.; Courtney, W.; Wang, R.; Kelly, R.; Turner, J.A.; Calhoun, V.D. Harnessing modern web application technology to create intuitive and efficient data visualization and sharing tools. Front. Neuroinform. 2014, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.C. DSM-IV: Diagnostic and statistical manual of mental disorders. JAMA 1994, 272, 828–829. [Google Scholar] [CrossRef]
- Aine, C.; Bockholt, H.J.; Bustillo, J.R.; Cañive, J.M.; Caprihan, A.; Gasparovic, C.; Hanlon, F.M.; Houck, J.M.; Jung, R.E.; Lauriello, J.; et al. Multimodal neuroimaging in schizophrenia: Description and dissemination. Neuroinformatics 2017, 15, 343–364. [Google Scholar] [CrossRef]
- Jones, D.K.; Horsfield, M.A.; Simmons, A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 1999, 42, 515–525. [Google Scholar] [CrossRef]
- Deichmann, R.; Gottfried, J.A.; Hutton, C.; Turner, R. Optimized EPI for fMRI studies of the orbitofrontal cortex. NeuroImage 2003, 19, 430–441. [Google Scholar] [CrossRef]
- Shin, J.; Ahn, S.; Hu, X. Correction for the T1 effect incorporating flip angle estimated by Kalman filter in cardiac-gated functional MRI. Magn. Reson. Med. 2013, 70, 1626–1633. [Google Scholar] [CrossRef] [Green Version]
- Assaf, Y.; Pasternak, O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: A review. J. Mol. Neurosci. 2008, 34, 51–61. [Google Scholar] [CrossRef]
- Van Erp, T.G.; Greve, D.N.; Rasmussen, J.; Turner, J.; Calhoun, V.D.; Young, S.; Mueller, B.; Brown, G.G.; McCarthy, G.; Glover, G.H.; et al. A multi-scanner study of subcortical brain volume abnormalities in schizophrenia. Psychiatry Res. Neuroimaging 2014, 222, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Bockholt, H.J.; Scully, M.; Courtney, W.; Rachakonda, S.; Scott, A.; Caprihan, A.; Fries, J.; Kalyanam, R.; Segall, J.; De La Garza, R.; et al. Mining the mind research network: A novel framework for exploring large scale, heterogeneous translational neuroscience research data sources. Front. Neuroinform. 2010, 3, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freire, L.; Roche, A.; Mangin, J.F. What is the best similarity measure for motion correction in fMRI time series? IEEE Trans. Med. Imaging 2002, 21, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Zhu, C.; Yang, Y.; Zuo, X.; Long, X.; Cao, Q.; Wang, Y.; Zang, Y. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. J. Neurosci. Methods 2008, 172, 137–141. [Google Scholar] [CrossRef] [Green Version]
- Heni, M.; Kullmann, S.; Ketterer, C.; Guthoff, M.; Linder, K.; Wagner, R.; Stingl, K.; Veit, R.; Staiger, H.; Häring, H.U.; et al. Nasal insulin changes peripheral insulin sensitivity simultaneously with altered activity in homeostatic and reward-related human brain regions. Diabetologia 2012, 55, 1773–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.Y.; Tang, R.; Posner, M.I. Brief meditation training induces smoking reduction. Proc. Natl. Acad. Sci. USA 2013, 110, 13971–13975. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.C.; Oathes, D.J.; Chang, C.; Bradley, T.; Zhou, Z.W.; Williams, L.M.; Glover, G.H.; Deisseroth, K.; Etkin, A. Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 19944–19949. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.Z.; Belgard, T.G.; Mao, D.; Chen, L.; Berto, S.; Preuss, T.M.; Lu, H.; Geschwind, D.H.; Konopka, G. Correspondence between resting-state activity and brain gene expression. Neuron 2015, 88, 659–666. [Google Scholar] [CrossRef] [Green Version]
- Kong, F.; Hu, S.; Wang, X.; Song, Y.; Liu, J. Neural correlates of the happy life: The amplitude of spontaneous low frequency fluctuations predicts subjective well-being. Neuroimage 2015, 107, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.J.; Hollinshead, M.O.; O’keefe, T.M.; Petrov, V.I.; Fariello, G.R.; Wald, L.L.; Fischl, B.; Rosen, B.R.; Mair, R.W.; Roffman, J.L.; et al. Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci. Data 2015, 2, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.A.; Damaraju, E.; Van ERP, T.G.; Mathalon, D.H.; Ford, J.M.; Voyvodic, J.; Mueller, B.A.; Belger, A.; Bustillo, J.; McEwen, S.C.; et al. A multi-site resting state fMRI study on the amplitude of low frequency fluctuations in schizophrenia. Front. Neurosci. 2013, 7, 137. [Google Scholar] [CrossRef] [Green Version]
- Comon, P. Independent component analysis, a new concept? Signal Process. 1994, 36, 287–314. [Google Scholar] [CrossRef]
- Beckmann, C.F.; DeLuca, M.; Devlin, J.T.; Smith, S.M. Investigations into resting-state connectivity using independent component analysis. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1001–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pasquale, F.; Della Penna, S.; Snyder, A.Z.; Lewis, C.; Mantini, D.; Marzetti, L.; Belardinelli, P.; Ciancetta, L.; Pizzella, V.; Romani, G.L.; et al. Temporal dynamics of spontaneous MEG activity in brain networks. Proc. Natl. Acad. Sci. USA 2010, 107, 6040–6045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calhoun, V.D.; Adali, T. Multisubject independent component analysis of fMRI: A decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev. Biomed. Eng. 2012, 5, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.O.; Yang, F.G.; Nguyen, C.T.; Cooper, S.R.; LaHue, S.C.; Venugopal, S.; Mukherjee, P. Independent component analysis of DTI reveals multivariate microstructural correlations of white matter in the human brain. Hum. Brain Mapp. 2012, 33, 1431–1451. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.; Bhinge, S.; Calhoun, V.D.; Adali, T. Independent vector analysis for common subspace analysis: Application to multi-subject fMRI data yields meaningful subgroups of schizophrenia. NeuroImage 2020, 216, 116872. [Google Scholar] [CrossRef] [PubMed]
- McKeown, M.J.; Makeig, S.; Brown, G.G.; Jung, T.P.; Kindermann, S.S.; Bell, A.J.; Sejnowski, T.J. Analysis of fMRI data by blind separation into independent spatial components. Hum. Brain Mapp. 1998, 6, 160–188. [Google Scholar] [CrossRef]
- Beckmann, C.F. Modelling with independent components. NeuroImage 2012, 62, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Calhoun, V.D.; Adali, T. Group ICA of fMRI Toolbox (GIFT). 2004. Available online: https://trendscenter.org/software/ (accessed on 23 February 2019).
- Abou-Elseoud, A.; Starck, T.; Remes, J.; Nikkinen, J.; Tervonen, O.; Kiviniemi, V. The effect of model order selection in group PICA. Hum. Brain Mapp. 2010, 31, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Correa, N.M.; Adali, T.; Li, Y.; Calhoun, V.D. Canonical correlation analysis for data fusion and group inferences. IEEE Signal Process. Mag. 2010, 27, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Calhoun, V.D.; Liu, J. ICA order selection based on consistency: Application to genotype data. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 360–363. [Google Scholar]
- Laney, J.; Adali, T.; Waller, S.M.; Westlake, K.P. Quantifying motor recovery after stroke using independent vector analysis and graph-theoretical analysis. NeuroImage Clin. 2015, 8, 298–304. [Google Scholar] [CrossRef] [Green Version]
- Bell, A.J.; Sejnowski, T.J. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995, 7, 1129–1159. [Google Scholar] [CrossRef]
- Correa, N.M.; Adali, T.; Li, Y.; Calhoun, V.D. Comparison of blind source separation algorithms for fMRI using a new Matlab toolbox: GIFT. In Proceedings of the (ICASSP ’05), IEEE International Conference on Acoustics, Speech, and Signal Processing, Philadelphia, PA, USA, 23 March 2005; Volume 5, p. v-401. [Google Scholar]
- Long, Q.; Jia, C.; Boukouvalas, Z.; Gabrielson, B.; Emge, D.; Adali, T. Consistent run selection for independent component analysis: Application to fMRI analysis. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 2581–2585. [Google Scholar]
- Mori, S.; Wakana, S.; Van Zijl, P.C.; Nagae-Poetscher, L. MRI Atlas of Human White Matter; Elsevier: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Wakana, S.; Caprihan, A.; Panzenboeck, M.M.; Fallon, J.H.; Perry, M.; Gollub, R.L.; Hua, K.; Zhang, J.; Jiang, H.; Dubey, P.; et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage 2007, 36, 630–644. [Google Scholar] [CrossRef] [Green Version]
- Hua, K.; Zhang, J.; Wakana, S.; Jiang, H.; Li, X.; Reich, D.S.; Calabresi, P.A.; Pekar, J.J.; van Zijl, P.C.; Mori, S. Tract probability maps in stereotaxic spaces: Analyses of white matter anatomy and tract-specific quantification. NeuroImage 2008, 39, 336–347. [Google Scholar] [CrossRef] [Green Version]
- Kelly, R.E., Jr.; Alexopoulos, G.S.; Wang, Z.; Gunning, F.M.; Murphy, C.F.; Morimoto, S.S.; Kanellopoulos, D.; Jia, Z.; Lim, K.O.; Hoptman, M.J. Visual inspection of independent components: Defining a procedure for artifact removal from fMRI data. J. Neurosci. Methods 2010, 189, 233–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calhoun, V.D.; Adali, T. Unmixing fMRI with independent component analysis. IEEE Eng. Med. Biol. Mag. 2006, 25, 79–90. [Google Scholar] [CrossRef]
- Ray, K.L.; McKay, D.R.; Fox, P.M.; Riedel, M.C.; Uecker, A.M.; Beckmann, C.F.; Smith, S.M.; Fox, P.T.; Laird, A. ICA model order selection of task co-activation networks. Front. Neurosci. 2013, 7, 237. [Google Scholar] [CrossRef] [Green Version]
- Feis, R.A.; Smith, S.M.; Filippini, N.; Douaud, G.; Dopper, E.G.; Heise, V.; Trachtenberg, A.J.; van Swieten, J.C.; van Buchem, M.A.; Rombouts, S.A.; et al. ICA-based artifact removal diminishes scan site differences in multi-center resting-state fMRI. Front. Neurosci. 2015, 9, 395. [Google Scholar] [CrossRef] [Green Version]
- Tohka, J.; Foerde, K.; Aron, A.R.; Tom, S.M.; Toga, A.W.; Poldrack, R.A. Automatic independent component labeling for artifact removal in fMRI. NeuroImage 2008, 39, 1227–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaganagarapu, K.; Jackson, G.D.; Abbott, D.F. An automated method for identifying artifact in independent component analysis of resting-state fMRI. Front. Hum. Neurosci. 2013, 7, 343. [Google Scholar] [CrossRef] [Green Version]
- Griffanti, L.; Douaud, G.; Bijsterbosch, J.; Evangelisti, S.; Alfaro-Almagro, F.; Glasser, M.F.; Duff, E.P.; Fitzgibbon, S.; Westphal, R.; Carone, D.; et al. Hand classification of fMRI ICA noise components. NeuroImage 2017, 154, 188–205. [Google Scholar] [CrossRef]
- Power, J.D.; Plitt, M.; Laumann, T.O.; Martin, A. Sources and implications of whole-brain fMRI signals in humans. NeuroImage 2017, 146, 609–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassinopoulos, M.; Mitsis, G.D. Identification of physiological response functions to correct for fluctuations in resting-state fMRI related to heart rate and respiration. NeuroImage 2019, 202, 116150. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pearlson, G.; Windemuth, A.; Ruano, G.; Perrone-Bizzozero, N.I.; Calhoun, V.D. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum. Brain Mapp. 2009, 30, 241–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bressler, S.L.; Menon, V. Large-scale brain networks in cognition: Emerging methods and principles. Trends Cogn. Sci. 2010, 14, 277–290. [Google Scholar] [CrossRef]
- Pessoa, L. Understanding brain networks and brain organization. Phys. Life Rev. 2014, 11, 400–435. [Google Scholar] [CrossRef] [Green Version]
- Hwang, K.; Bertolero, M.A.; Liu, W.B.; D’esposito, M. The human thalamus is an integrative hub for functional brain networks. J. Neurosci. 2017, 37, 5594–5607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, K.A.; Manaye, K.F.; Liang, C.L.; Hicks, P.B.; German, D.C. Reduced number of mediodorsal and anterior thalamic neurons in schizophrenia. Biol. Psychiatry 2000, 47, 944–953. [Google Scholar] [CrossRef]
- Sherman, S.M.; Guillery, R.W. Exploring the Thalamus and Its Role in Cortical Function; MIT Press: Cambridge, MA, USA, 2006. [Google Scholar]
- McIntosh, A.M.; Maniega, S.M.; Lymer, G.K.S.; McKirdy, J.; Hall, J.; Sussmann, J.E.; Bastin, M.E.; Clayden, J.D.; Johnstone, E.C.; Lawrie, S.M. White matter tractography in bipolar disorder and schizophrenia. Biol. Psychiatry 2008, 64, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Mamah, D.; Conturo, T.E.; Harms, M.P.; Akbudak, E.; Wang, L.; McMichael, A.R.; Gado, M.H.; Barch, D.M.; Csernansky, J.G. Anterior thalamic radiation integrity in schizophrenia: A diffusion-tensor imaging study. Psychiatry Res. Neuroimaging 2010, 183, 144–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ćurčić-Blake, B.; Nanetti, L.; van der Meer, L.; Cerliani, L.; Renken, R.; Pijnenborg, G.H.; Aleman, A. Not on speaking terms: Hallucinations and structural network disconnectivity in schizophrenia. Brain Struct. Funct. 2015, 220, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Koshiyama, D.; Fukunaga, M.; Okada, N.; Morita, K.; Nemoto, K.; Yamashita, F.; Yamamori, H.; Yasuda, Y.; Fujimoto, M.; Kelly, S.; et al. Role of frontal white matter and corpus callosum on social function in schizophrenia. Schizophr. Res. 2018, 202, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Epstein, K.A.; Cullen, K.R.; Mueller, B.A.; Robinson, P.; Lee, S.; Kumra, S. White matter abnormalities and cognitive impairment in early-onset schizophrenia-spectrum disorders. J. Am. Acad. Child Adolesc. Psychiatry 2014, 53, 362–372. [Google Scholar] [CrossRef] [Green Version]
- Karlsgodt, K.H.; van ERP, T.G.; Poldrack, R.A.; Bearden, C.E.; Nuechterlein, K.H.; Cannon, T.D. Diffusion tensor imaging of the superior longitudinal fasciculus and working memory in recent-onset schizophrenia. Biol. Psychiatry 2008, 63, 512–518. [Google Scholar] [CrossRef]
- Glahn, D.C.; Laird, A.R.; Ellison-Wright, I.; Thelen, S.M.; Robinson, J.L.; Lancaster, J.L.; Bullmore, E.; Fox, P.T. Meta-analysis of gray matter anomalies in schizophrenia: Application of anatomic likelihood estimation and network analysis. Biol. Psychiatry 2008, 64, 774–781. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.Y.; Suzuki, M.; Hagino, H.; Takahashi, T.; Kawasaki, Y.; Matsui, M.; Seto, H.; Kurachi, M. Volumetric analysis of sulci/gyri-defined in vivo frontal lobe regions in schizophrenia: Precentral gyrus, cingulate gyrus, and prefrontal region. Psychiatry Res. Neuroimaging 2005, 139, 127–139. [Google Scholar] [CrossRef]
- Barta, P.E.; Pearlson, G.D.; Powers, R.E.; Richards, S.S.; Tune, L.E. Auditory hallucinations and smaller superior temporal gyral volume in schizophrenia. Am. J. Psychiatry 1990, 147, 1457–1462. [Google Scholar]
- Job, D.E.; Whalley, H.C.; McConnell, S.; Glabus, M.; Johnstone, E.C.; Lawrie, S.M. Structural gray matter differences between first-episode schizophrenics and normal controls using voxel-based morphometry. NeuroImage 2002, 17, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Onitsuka, T.; Shenton, M.E.; Salisbury, D.F.; Dickey, C.C.; Kasai, K.; Toner, S.K.; Frumin, M.; Kikinis, R.; Jolesz, F.A.; McCarley, R.W. Middle and inferior temporal gyrus gray matter volume abnormalities in chronic schizophrenia: An MRI study. Am. J. Psychiatry 2004, 161, 1603–1611. [Google Scholar] [CrossRef]
- Kuroki, N.; Shenton, M.E.; Salisbury, D.F.; Hirayasu, Y.; Onitsuka, T.; Ersner, H.; Yurgelun-Todd, D.; Kikinis, R.; Jolesz, F.A.; McCarley, R.W. Middle and inferior temporal gyrus gray matter volume abnormalities in first-episode schizophrenia: An MRI study. Am. J. Psychiatry 2006, 163, 2103–2110. [Google Scholar] [CrossRef] [Green Version]
- Cascella, N.G.; Fieldstone, S.C.; Rao, V.A.; Pearlson, G.D.; Sawa, A.; Schretlen, D.J. Gray-matter abnormalities in deficit schizophrenia. Schizophr. Res. 2010, 120, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Onitsuka, T.; Shenton, M.E.; Kasai, K.; Nestor, P.G.; Toner, S.K.; Kikinis, R.; Jolesz, F.A.; McCarley, R.W. Fusiform gyrus volume reduction and facial recognition in chronic schizophrenia. Arch. Gen. Psychiatry 2003, 60, 349–355. [Google Scholar] [CrossRef] [PubMed]


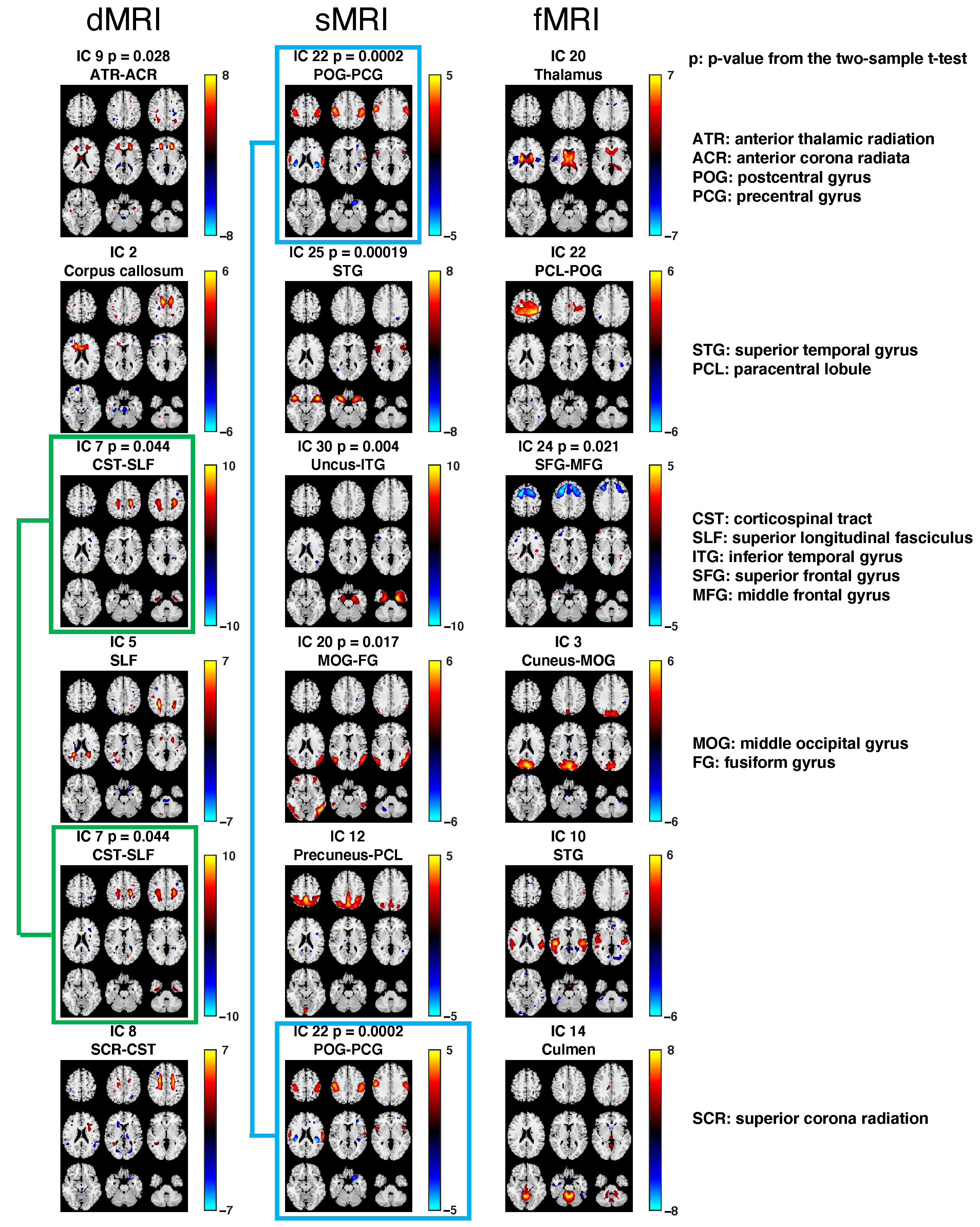
| Triplet # | dMRI–sMRI | dMRI–fMRI | sMRI–fMRI |
|---|---|---|---|
| 1 | |||
| 2 | |||
| 3 | |||
| 4 | |||
| 5 | |||
| 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, C.; Akhonda, M.A.B.S.; Levin-Schwartz, Y.; Long, Q.; Calhoun, V.D.; Adali, T. Consecutive Independence and Correlation Transform for Multimodal Data Fusion: Discovery of One-to-Many Associations in Structural and Functional Imaging Data. Appl. Sci. 2021, 11, 8382. https://doi.org/10.3390/app11188382
Jia C, Akhonda MABS, Levin-Schwartz Y, Long Q, Calhoun VD, Adali T. Consecutive Independence and Correlation Transform for Multimodal Data Fusion: Discovery of One-to-Many Associations in Structural and Functional Imaging Data. Applied Sciences. 2021; 11(18):8382. https://doi.org/10.3390/app11188382
Chicago/Turabian StyleJia, Chunying, Mohammad Abu Baker Siddique Akhonda, Yuri Levin-Schwartz, Qunfang Long, Vince D. Calhoun, and Tülay Adali. 2021. "Consecutive Independence and Correlation Transform for Multimodal Data Fusion: Discovery of One-to-Many Associations in Structural and Functional Imaging Data" Applied Sciences 11, no. 18: 8382. https://doi.org/10.3390/app11188382
APA StyleJia, C., Akhonda, M. A. B. S., Levin-Schwartz, Y., Long, Q., Calhoun, V. D., & Adali, T. (2021). Consecutive Independence and Correlation Transform for Multimodal Data Fusion: Discovery of One-to-Many Associations in Structural and Functional Imaging Data. Applied Sciences, 11(18), 8382. https://doi.org/10.3390/app11188382






