Xylose Metabolism in Bacteria—Opportunities and Challenges towards Efficient Lignocellulosic Biomass-Based Biorefineries
Abstract
1. Introduction
2. Biorefinery Concept
2.1. Lignocellulosic Biomass as Raw Material
2.2. Lignocellulosic Biomass Treatments
3. Major Routes of Xylose Transport and Metabolism in Bacteria
3.1. Mechanisms of Xylose Transport
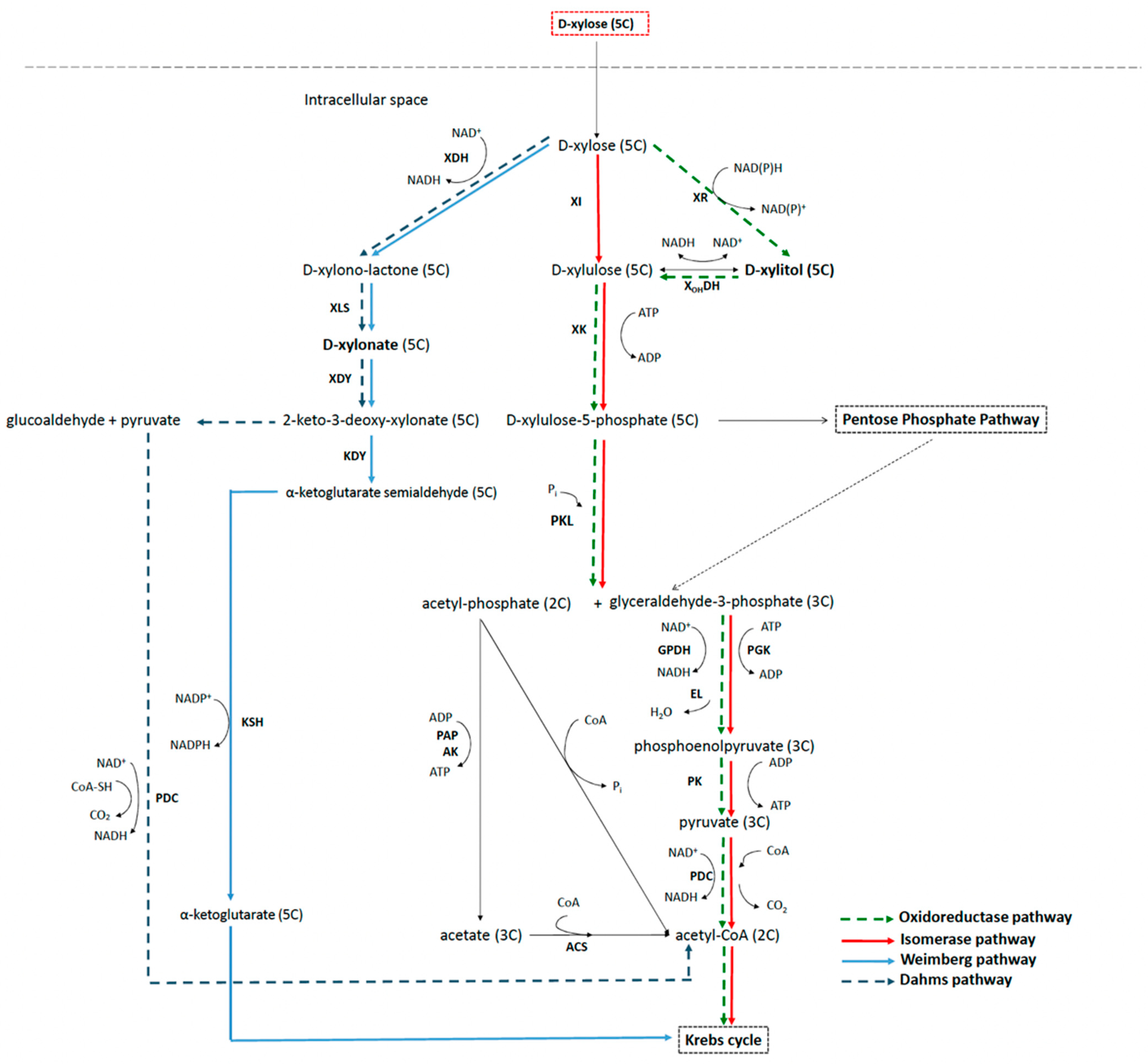
3.2. Xylose Metabolic Network in Bacteria
3.3. Metabolic Pathways to Xylitol and Xylonic Acid
3.3.1. Xylitol
3.3.2. Xylonic Acid
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uniyal, S.; Paliwal, R.; Kaphaliya, B.; Sharma, R.K. Human overpopulation: Impact on environment. In Environmental Issues Surrounding Human Overpopulation; Singh, R.P., Singh, A., Srivastava, V., Eds.; IGI Global: Hershey PA, USA, 2017; pp. 1–11. ISBN 9781522516835. [Google Scholar]
- Song, Q.; Li, J.; Zeng, X. Minimizing the increasing solid waste through zero waste strategy. J. Clean. Prod. 2015, 104, 199–210. [Google Scholar] [CrossRef]
- Minghua, Z.; Xiumin, F.; Rovetta, A.; Qichang, H.; Vicentini, F.; Bingkai, L.; Giusti, A.; Yi, L. Municipal solid waste management in Pudong New Area, China. Waste Manag. 2009, 29, 1227–1233. [Google Scholar] [CrossRef]
- Zaman, A.U. A comprehensive review of the development of zero waste management: Lessons learned and guidelines. J. Clean. Prod. 2015, 91, 12–25. [Google Scholar] [CrossRef]
- Zero Waste Definition. Available online: https://zwia.org/zero-waste-definition/ (accessed on 22 April 2021).
- Kamm, B.; Kamm, M. Principles of biorefineries. Appl. Microbiol. Biotechnol. 2004, 64, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Vernès, L.; Li, Y.; Chemat, F.; Abert-Vian, M. Biorefinery Concept as a Key for Sustainable Future to Green Chemistry—The Case of Microalgae. In Plant Based “Green Chemistry 2.0”; Li, Y., Kemat, F., Eds.; Springer: Singapore, 2019; pp. 15–50. ISBN 978-981-13-3810-6. [Google Scholar]
- Ferreira, A.F. Biorefinery concept. In Lecture Notes in Energy—Biorefineries: Targeting Energy, High Value Products and Waste Valorisation; Rabaçal, M., Ferreira, A.F., Silva, C.A.M., Costa, M., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 1–20. ISBN 978-3-319-48286-6. [Google Scholar]
- 2050 Long-Term Strategy. Available online: https://ec.europa.eu/clima/policies/strategies/2050_en (accessed on 1 July 2021).
- United Nations Framework Convention on Climate Change—Paris Agreement. Available online: https://unfccc.int/process-and-meetings/the-paris-agreement/the-paris-agreement (accessed on 1 July 2021).
- Cherubini, F. The biorefinery concept: Using biomass instead of oil for producing energy and chemicals. Energy Convers. Manag. 2010, 51, 1412–1421. [Google Scholar] [CrossRef]
- Moncada, J.; Aristizábal, V. Design strategies for sustainable biorefineries. Biochem. Eng. J. 2016, 116, 122–134. [Google Scholar] [CrossRef]
- Irmak, S. Biomass as Raw Material for Production of High-Value Products. In Biomass Volume Estimation and Valorization for Energy; Tumuluru, J.S., Ed.; IntechOpen: Rijeka, Croatia, 2017; pp. 201–225. ISBN 978-953-51-2938-7. [Google Scholar]
- Isikgor, F.H.; Becer, C.R. Lignocellulosic biomass: A sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 2015, 6, 4497–4559. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, J. Hydrolysis of lignocellulosic materials for ethanol production: A review. Bioresour. Technol. 2002, 83, 1–11. [Google Scholar] [CrossRef]
- Zhou, C.-H.; Xia, X.; Lin, C.-X.; Tong, D.-S.; Beltramini, J. Catalytic conversion of lignocellulosic biomass to fine chemicals and fuels. Chem. Soc. Rev. 2011, 40, 5588–5617. [Google Scholar] [CrossRef] [PubMed]
- Cesário, M.T.F.; de Almeida, M.C.M.D. Lignocellulosic Hydrolysates for the Production of Polyhydroxyalkanoates. In Microorganisms in Biorefineries; Kamm, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 79–104. [Google Scholar]
- Curia, S.; Biundo, A.; Fischer, I.; Braunschmid, V.; Gübitz, G.M.; Stanzione, J.F., 3rd. Towards Sustainable High-Performance Thermoplastics: Synthesis, Characterization, and Enzymatic Hydrolysis of Bisguaiacol-Based Polyesters. ChemSusChem 2018, 11, 2529–2539. [Google Scholar] [CrossRef]
- Stamm, A.; Biundo, A.; Schmidt, B.; Brücher, J.; Lundmark, S.; Olsén, P.; Fogelström, L.; Malmström, E.; Bornscheuer, U.T.; Syrén, P.-O. A Retro-biosynthesis-Based Route to Generate Pinene-Derived Polyesters. ChemBioChem 2019, 20, 1664–1671. [Google Scholar] [CrossRef]
- Romaní, A.; Rocha, C.M.R.; Michelin, M.; Domingues, L.; Teixeira, J.A. Chapter 20—Valorization of lignocellulosic-based wastes. In Current Developments in Biotechnology and Bioengineering—Resource Recovery from Wastes; Varjani, S., Pandey, A., Gnansounou, E., Khanal, S.K., Raveendran, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 383–410. ISBN 978-0-444-64321-6. [Google Scholar]
- Meng, Q.; Yan, J.; Wu, R.; Liu, H.; Sun, Y.; Wu, N.; Xiang, J.; Zheng, L.; Zhang, J.; Han, B. Sustainable production of benzene from lignin. Nat. Commun. 2021, 12, 4534. [Google Scholar] [CrossRef]
- Ciliberti, C.; Biundo, A.; Albergo, R.; Agrimi, G.; Braccio, G.; de Bari, I.; Pisano, I. Syngas Derived from Lignocellulosic Biomass Gasification as an Alternative Resource for Innovative Bioprocesses. Processes 2020, 8, 1567. [Google Scholar] [CrossRef]
- Devarapalli, M.; Atiyeh, H.K. A review of conversion processes for bioethanol production with a focus on syngas fermentation. Biofuel Res. J. 2015, 2, 268–280. [Google Scholar] [CrossRef]
- Olsson, L.; Hahn-Hägerdal, B. Fermentation of lignocellulosic hydrolysates for ethanol production. Enzyme Microb. Technol. 1996, 18, 312–331. [Google Scholar] [CrossRef]
- Hassan, S.S.; Williams, G.A.; Jaiswal, A.K. Lignocellulosic Biorefineries in Europe: Current State and Prospects. Trends Biotechnol. 2019, 37, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, S.; Carus, M.; Carrez, D. European Bioeconomy in Figures. Ind. Biotechnol. 2016, 12, 78–82. [Google Scholar] [CrossRef]
- Jørgensen, H.; Kristensen, J.B.; Felby, C. Enzymatic conversion of lignocellulose into fermentable sugars: Challenges and opportunities. Biofuels Bioprod. Biorefining 2007, 1, 119–134. [Google Scholar] [CrossRef]
- Mussatto, S.; Teixeira, J. Lignocellulose as raw material in fermentation processes. In Current Research, Technology and Education Topics in Applied Microbiology an Microbial Biotechnology; Méndez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2010; Volume 2, pp. 897–907. ISBN 978-84-614-6195-0. [Google Scholar]
- Mosier, N.; Wyman, C.; Dale, B.; Elander, R.; Lee, Y.Y.; Holtzapple, M.; Ladisch, M. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour. Technol. 2005, 96, 673–686. [Google Scholar] [CrossRef]
- Agbor, V.B.; Cicek, N.; Sparling, R.; Berlin, A.; Levin, D.B. Biomass pretreatment: Fundamentals toward application. Biotechnol. Adv. 2011, 29, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Nandal, P.; Singh, J.; Verma, M.L. Nanobiotechnological advancements in lignocellulosic biomass pretreatment. Mater. Sci. Energy Technol. 2020, 3, 308–318. [Google Scholar] [CrossRef]
- Kuenz, A.; Jäger, M.; Niemi, H.; Kallioinen, M.; Mänttäri, M.; Prüße, U. Conversion of Xylose from Birch Hemicellulose Hydrolysate to 2,3-Butanediol with Bacillus vallismortis. Fermentation 2020, 6, 86. [Google Scholar] [CrossRef]
- Nitzsche, R.; Budzinski, M.; Gröngröft, A. Techno-economic assessment of a wood-based biorefinery concept for the production of polymer-grade ethylene, organosolv lignin and fuel. Bioresour. Technol. 2016, 200, 928–939. [Google Scholar] [CrossRef]
- FitzPatrick, M.; Champagne, P.; Cunningham, M.F.; Whitney, R.A. A biorefinery processing perspective: Treatment of lignocellulosic materials for the production of value-added products. Bioresour. Technol. 2010, 101, 8915–8922. [Google Scholar] [CrossRef]
- Stark, A. Ionic liquids in the biorefinery: A critical assessment of their potential. Energy Environ. Sci. 2011, 4, 19–32. [Google Scholar] [CrossRef]
- Kucharska, K.; Rybarczyk, P.; Hołowacz, I.; Łukajtis, R.; Glinka, M.; Kamiński, M. Pretreatment of lignocellulosic materials as substrates for fermentation processes. Molecules 2018, 23, 2937. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Peng, P.; Xu, F.; Sun, R.C. Fractional purification and bioconversion of hemicelluloses. Biotechnol. Adv. 2012, 30, 879–903. [Google Scholar] [CrossRef]
- Gírio, F.M.; Fonseca, C.; Carvalheiro, F.; Duarte, L.C.; Marques, S.; Bogel-Łukasik, R. Hemicelluloses for fuel ethanol: A review. Bioresour. Technol. 2010, 101, 4775–4800. [Google Scholar] [CrossRef]
- Dietrich, K.; Dumont, M.-J.; Del Rio, L.F.; Orsat, V. Producing PHAs in the bioeconomy—Towards a sustainable bioplastic. Sustain. Prod. Consum. 2017, 9, 58–70. [Google Scholar] [CrossRef]
- Izaguirre, J.K.; Barañano, L.; Castañón, S.; Santos, J.A.L.; Cesário, M.T.; da Fonseca, M.M.R.; Alkorta, I.; Garbisu, C. Economic and environmental assessment of bacterial poly(3-hydroxybutyrate) production from the organic fraction of municipal solid waste. Bioresour. Bioprocess. 2021, 8, 39. [Google Scholar] [CrossRef]
- McClintock, M.K.; Zhang, K. Xylose Metabolism and Its Metabolic Engineering Applications. In Engineering Microbial Metabolism for Chemical Synthesis; World Scientific (Europe): Hackensack, NJ, USA, 2017; pp. 209–235. ISBN 978-1-78634-429-8. [Google Scholar]
- Cesário, M.T.; Raposo, R.S.; de Almeida, M.C.M.D.; van Keulen, F.; Ferreira, B.S.; da Fonseca, M.M.R. Enhanced bioproduction of poly-3-hydroxybutyrate from wheat straw lignocellulosic hydrolysates. N. Biotechnol. 2013, 31, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Werpy, T.; Petersen, G. Top Value Added Chemicals from Biomass: Volume I—Results of Screening for Potential Candidates from Sugars and Synthesis Gas; National Renewable Energy Lab.: Golden, CO, USA, 2004. [Google Scholar]
- Gu, Y.; Ding, Y.; Ren, C.; Sun, Z.; Rodionov, D.A.; Zhang, W.; Yang, S.; Yang, C.; Jiang, W. Reconstruction of xylose utilization pathway and regulons in Firmicutes. BMC Genom. 2010, 11, 255. [Google Scholar] [CrossRef]
- Davis, E.O.; Henderson, P.J. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J. Biol. Chem. 1987, 262, 13928–13932. [Google Scholar] [CrossRef]
- Khankal, R.; Chin, J.W.; Cirino, P.C. Role of xylose transporters in xylitol production from engineered Escherichia coli. J. Biotechnol. 2008, 134, 246–252. [Google Scholar] [CrossRef]
- Shamanna, D.K.; Sanderson, K.E. Uptake and catabolism of D-xylose in Salmonella typhimurium LT2. J. Bacteriol. 1979, 139, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Park, C. Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 1997, 179, 7025–7032. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, S.; Bor, Y.C.; Batt, C.A.; Postma, P.W.; Pouwels, P.H. Molecular cloning and functional expression in lactobacillus plantarum 80 of xylT, encoding the D-xylose-H+ symporter of Lactobacillus brevis. Appl. Environ. Microbiol. 1998, 64, 4720–4728. [Google Scholar] [CrossRef]
- Chaillou, S.; Pouwels, P.H.; Postma, P.W. Transport of D-Xylose in Lactobacillus pentosus, Lactobacillus casei, and Lactobacillus plantarum: Evidence for a Mechanism of Facilitated Diffusion via the Phosphoenolpyruvate:Mannose Phos. J. Bacteriol. 1999, 181, 4768–4773. [Google Scholar] [CrossRef]
- Schmiedel, D.; Hillen, W. A Bacillus subtilis 168 mutant with increased xylose uptake can utilize xylose as sole carbon source. FEMS Microbiol. Lett. 1996, 135, 175–178. [Google Scholar] [CrossRef]
- Schmiedel, D.; Kintrup, M.; Küster, E.; Hillen, W. Regulation of expression, genetic organization and substrate specificity of xylose uptake in Bacillus megaterium. Mol. Microbiol. 1997, 23, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Sievert, C.; Nieves, L.M.; Panyon, L.A.; Loeffler, T.; Morris, C.; Cartwright, R.A.; Wang, X. Experimental evolution reveals an effective avenue to release catabolite repression via mutations in XylR. Proc. Natl. Acad. Sci. USA 2017, 114, 7349–7354. [Google Scholar] [CrossRef]
- Strobel, H.J. Evidence for catabolite inhibition in regulation of pentose utilization and transport in the ruminal bacterium Selenomonas ruminantium. Appl. Environ. Microbiol. 1993, 59, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Takase, K.; Yamato, I.; Abe, K. Sequencing and characterization of the xyl operon of a gram-positive bacterium, Tetragenococcus halophila. Appl. Environ. Microbiol. 1998, 64, 2513–2519. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, C.; Yang, G.; Sun, Z.; Guo, H.; Shao, K.; Gu, Y.; Jiang, W.; Zhang, P. Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria. Proc. Natl. Acad. Sci. USA 2017, 114, 8235–8240. [Google Scholar] [CrossRef] [PubMed]
- Erbeznik, M.; Hudson, S.E.; Herrman, A.B.; Strobel, H.J. Molecular analysis of the xylFGH operon, coding for xylose ABC transport, in Thermoanaerobacter ethanolicus. Curr. Microbiol. 2004, 48, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Shulami, S.; Zaide, G.; Zolotnitsky, G.; Langut, Y.; Feld, G.; Sonenshein, A.L.; Shoham, Y. A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. Appl. Environ. Microbiol. 2007, 73, 874–884. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, Y.; Yang, C.; Yang, S.; Gu, Y.; Jiang, W. A novel three-component system-based regulatory model for D-xylose sensing and transport in Clostridium beijerinckii. Mol. Microbiol. 2015, 95, 576–589. [Google Scholar] [CrossRef]
- Raposo, R.S.; de Almeida, M.C.; de Oliveira, M.D.C.; da Fonseca, M.M.; Cesário, M.T. A Burkholderia sacchari cell factory: Production of poly-3-hydroxybutyrate, xylitol and xylonic acid from xylose-rich sugar mixtures. N. Biotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Goh, E.-B.; Beller, H.R. Engineering, E. coli for simultaneous glucose–xylose utilization during methyl ketone production. Microb. Cell Fact. 2018, 17, 12. [Google Scholar] [CrossRef]
- Desai, T.A.; Rao, C. V Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Microbiol. 2010, 76, 1524–1532. [Google Scholar] [CrossRef]
- Rojo, F. Carbon catabolite repression in Pseudomonas: Optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 2010, 34, 658–684. [Google Scholar] [CrossRef] [PubMed]
- Raposo, R.S.; de Almeida, M.C.M.D.; da Fonseca, M.M.R.; Cesário, M.T. Feeding strategies for tuning poly (3-hydroxybutyrate-co-4-hydroxybutyrate) monomeric composition and productivity using Burkholderia sacchari. Int. J. Biol. Macromol. 2017, 105, 825–833. [Google Scholar] [CrossRef]
- Ren, C.; Gu, Y.; Hu, S.; Wu, Y.; Wang, P.; Yang, Y.; Yang, C.; Yang, S.; Jiang, W. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab. Eng. 2010, 12, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Bruder, M.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Elimination of carbon catabolite repression in Clostridium acetobutylicum--a journey toward simultaneous use of xylose and glucose. Appl. Microbiol. Biotechnol. 2015, 99, 7579–7588. [Google Scholar] [CrossRef] [PubMed]
- Aiba, H.; Nakamura, T.; Mitani, H.; Mori, H. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 1985, 4, 3329–3332. [Google Scholar] [CrossRef] [PubMed]
- Gosset, G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 2005, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Wu, M.; Zhang, Z.; Lin, J.; Yang, L. Efficient production of xylitol from hemicellulosic hydrolysate using engineered Escherichia coli. Metab. Eng. 2015, 31, 112–122. [Google Scholar] [CrossRef]
- Zhao, Z.; Xian, M.; Liu, M.; Zhao, G. Biochemical routes for uptake and conversion of xylose by microorganisms. Biotechnol. Biofuels 2020, 13, 21. [Google Scholar] [CrossRef]
- Jeffries, T.W. Utilization of xylose by bacteria, yeasts, and fungi. In Pentoses and Lignin; Springer: Berlin/Heidelberg, Germany, 1983; pp. 1–32. [Google Scholar]
- Kawaguchi, H.; Vertès, A.A.; Okino, S.; Inui, M.; Yukawa, H. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2006, 72, 3418–3428. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, W.-L.; Liu, X.-Y.; Shen, Y.; Bao, X.-M.; Bai, F.-W.; Qu, Y.-B. Establishment of a xylose metabolic pathway in an industrial strain of Saccharomyces cerevisiae. Biotechnol. Lett. 2004, 26, 885–890. [Google Scholar] [CrossRef]
- Zhang, M.; Eddy, C.; Deanda, K.; Finkelstein, M.; Picataggio, S. Metabolic Engineering of a Pentose Metabolism Pathway in Ethanologenic Zymomonas mobilis. Science 1995, 267, 240–243. [Google Scholar] [CrossRef]
- Lawlis, V.B.; Dennis, M.S.; Chen, E.Y.; Smith, D.H.; Henner, D.J. Cloning and sequencing of the xylose isomerase and xylulose kinase genes of Escherichia coli. Appl. Environ. Microbiol. 1984, 47, 15–21. [Google Scholar] [CrossRef]
- Lokman, B.C.; van Santen, P.; Verdoes, J.C.; Krüse, J.; Leer, R.J.; Posno, M.; Pouwels, P.H. Organization and characterization of three genes involved in D-xylose catabolism in Lactobacillus pentosus. Mol. Gen. Genet. 1991, 230, 161–169. [Google Scholar] [CrossRef]
- Rygus, T.; Scheler, A.; Allmansberger, R.; Hillen, W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus megaterium encoded regulon for xylose utilization. Arch. Microbiol. 1991, 155, 535–542. [Google Scholar] [CrossRef]
- Stephens, C.; Christen, B.; Fuchs, T.; Sundaram, V.; Watanabe, K.; Jenal, U. Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 2007, 189, 2181–2185. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Bondar, M.; da Fonseca, M.M.R.; Cesário, M.T. Xylonic acid production from xylose by Paraburkholderia sacchari. Biochem. Eng. J. 2021, 170, 107982. [Google Scholar] [CrossRef]
- Wong, H.C.; Ting, Y.; Lin, H.C.; Reichert, F.; Myambo, K.; Watt, K.W.; Toy, P.L.; Drummond, R.J. Genetic organization and regulation of the xylose degradation genes in Streptomyces rubiginosus. J. Bacteriol. 1991, 173, 6849–6858. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yamanaka, K. Inhibition of d-xylose isomerase by pentitols and d-lyxose. Arch. Biochem. Biophys. 1969, 131, 502–506. [Google Scholar] [CrossRef]
- Kovalevsky, A.; Hanson, B.L.; Mason, S.A.; Forsyth, V.T.; Fisher, Z.; Mustyakimov, M.; Blakeley, M.P.; Keen, D.A.; Langan, P. Inhibition of D-xylose isomerase by polyols: Atomic details by joint X-ray/neutron crystallography. Acta Crystallogr. Sect. D 2012, 68, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Moat, A.G.; Foster, J.W.; Spector, M.P. Microbial Physiology, 4th ed.; Wiley-Liss, Inc.: New York, NY, USA, 2002; ISBN 0-471-39483-1. [Google Scholar]
- Gu, Y.; Jiang, Y.; Yang, S.; Jiang, W. Utilization of economical substrate-derived carbohydrates by solventogenic clostridia: Pathway dissection, regulation and engineering. Curr. Opin. Biotechnol. 2014, 29, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, L.; Tang, W.; Gu, Y.; Hua, Q.; Yang, S.; Jiang, W.; Yang, C. Phosphoketolase pathway for Xylose Catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J. Bacteriol. 2012, 194, 5413–5422. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Komiyama, A.; Sonomoto, K.; Ishizaki, A.; Hall, S.; Stanbury, P. Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Appl. Microbiol. Biotechnol. 2002, 60, 160–167. [Google Scholar] [CrossRef]
- Kwak, S.; Jin, Y.-S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Fact. 2017, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- McClintock, M.K.; Wang, J.; Zhang, K. Application of Nonphosphorylative Metabolism as an Alternative for Utilization of Lignocellulosic Biomass. Front. Microbiol. 2017, 8, 2310. [Google Scholar] [CrossRef] [PubMed]
- Weimberg, R. Pentose oxidation by Pseudomonas fragi. J. Biol. Chem. 1961, 236, 629–635. [Google Scholar] [CrossRef]
- Köhler, K.A.K.; Blank, L.M.; Frick, O.; Schmid, A. D-Xylose assimilation via the Weimberg pathway by solvent-tolerant Pseudomonas taiwanensis VLB120. Environ. Microbiol. 2015, 17, 156–170. [Google Scholar] [CrossRef]
- Franden, M.A.; Jayakody, L.N.; Li, W.-J.; Wagner, N.J.; Cleveland, N.S.; Michener, W.E.; Hauer, B.; Blank, L.M.; Wierckx, N.; Klebensberger, J.; et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018, 48, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Nunn, C.E.M.; Johnsen, U.; Schönheit, P.; Fuhrer, T.; Sauer, U.; Hough, D.W.; Danson, M.J. Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J. Biol. Chem. 2010, 285, 33701–33709. [Google Scholar] [CrossRef]
- Borgström, C.; Wasserstrom, L.; Almqvist, H.; Broberg, K.; Klein, B.; Noack, S.; Lidén, G.; Gorwa-Grauslund, M.F. Identification of modifications procuring growth on xylose in recombinant Saccharomyces cerevisiae strains carrying the Weimberg pathway. Metab. Eng. 2019, 55, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Kohlhaas, M.; Enoki, J.; Meier, R.; Schönenberger, B.; Wohlgemuth, R.; Kourist, R.; Niemeyer, F.; van Niekerk, D.; Bräsen, C.; et al. A combined experimental and modelling approach for the Weimberg pathway optimisation. Nat. Commun. 2020, 11, 1098. [Google Scholar] [CrossRef]
- Almqvist, H.; Jonsdottir Glaser, S.; Tufvegren, C.; Wasserstrom, L.; Lidén, G. Characterization of the Weimberg Pathway in Caulobacter crescentus. Fermentation 2018, 4, 44. [Google Scholar] [CrossRef]
- Bañares, A.B.; Nisola, G.M.; Valdehuesa, K.N.G.; Lee, W.K.; Chung, W.J. Understanding D-xylonic acid accumulation: A cornerstone for better metabolic engineering approaches. Appl. Microbiol. Biotechnol. 2021, 105, 5309–5324. [Google Scholar] [CrossRef] [PubMed]
- Salusjärvi, L.; Havukainen, S.; Koivistoinen, O.; Toivari, M. Biotechnological production of glycolic acid and ethylene glycol: Current state and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, Y.; Wang, Y.; Gu, J.; Lu, X.; Liao, X.; Shi, J.; Kim, C.H.; Lye, G.; Baganz, F.; et al. Ethylene glycol and glycolic acid production from xylonic acid by Enterobacter cloacae. Microb. Cell Fact. 2020, 19, 89. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Woo, H.M. Deciphering bacterial xylose metabolism and metabolic engineering of industrial microorganisms for use as efficient microbial cell factories. Appl. Microbiol. Biotechnol. 2018, 102, 9471–9480. [Google Scholar] [CrossRef] [PubMed]
- Melaja, A.; Hämäläinen, L.; Heikkilä, H. Menetelmä Ksylitolin Suhteen Rikastuneen Polyolin. FI Patent 589,388, 10 August 1981. [Google Scholar]
- Delgado Arcaño, Y.; Valmaña García, O.D.; Mandelli, D.; Carvalho, W.A.; Magalhães Pontes, L.A. Xylitol: A review on the progress and challenges of its production by chemical route. Catal. Today 2020, 344, 2–14. [Google Scholar] [CrossRef]
- Härkönen, M.; Nuojua, P. Eri tekijoiden vaikutus ksyloosin katalyyttiseen hydraukseen ksylitoliksi, osa 1 (The effects of different factors on catalytic hydration of xylose into xylitol, part 1). Kemia-Kemi 1979, 6, 445–447. [Google Scholar]
- Dasgupta, D.; Bandhu, S.; Adhikari, D.K.; Ghosh, D. Challenges and prospects of xylitol production with whole cell bio-catalysis: A review. Microbiol. Res. 2017, 197, 9–21. [Google Scholar] [CrossRef]
- de Albuquerque, T.L.; da Silva, I.J.; de Macedo, G.R.; Rocha, M.V.P. Biotechnological production of xylitol from lignocellulosic wastes: A review. Process Biochem. 2014, 49, 1779–1789. [Google Scholar] [CrossRef]
- Izumori, K.; Tuzaki, K. Production of xylitol from D-xylulose by Mycobacterium smegmatis. J. Ferment. Technol. 1988, 66, 33–36. [Google Scholar] [CrossRef]
- Rafiqul, I.S.M.; Sakinah, A.M.M. Processes for the Production of Xylitol—A Review. Food Rev. Int. 2013, 29, 127–156. [Google Scholar] [CrossRef]
- Rangaswamy, S.; Agblevor, F.A. Screening of facultative anaerobic bacteria utilizing D-xylose for xylitol production. Appl. Microbiol. Biotechnol. 2002, 60, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Sugiyama, M.; Mihara, Y.; Hashiguchi, K.; Yokozeki, K. Novel enzymatic method for the production of xylitol from D-arabitol by Gluconobacter oxydans. Biosci. Biotechnol. Biochem. 2002, 66, 2614–2620. [Google Scholar] [CrossRef][Green Version]
- Yoshitake, J.; Ishizaki, H.; Shimamura, M.; Imai, T. Xylitol Production by an Enterobacter Species. Agric. Biol. Chem. 1973, 37, 2261–2267. [Google Scholar] [CrossRef][Green Version]
- Guamán, L.P.; Oliveira-Filho, E.R.; Barba-Ostria, C.; Gomez, J.G.C.; Taciro, M.K.; da Silva, L.F. xylA and xylB overexpression as a successful strategy for improving xylose utilization and poly-3-hydroxybutyrate production in Burkholderia sacchari. J. Ind. Microbiol. Biotechnol. 2018, 45, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Zhang, Z.; Wu, M.; Lin, J.; Yang, L. Construction of plasmid-free Escherichia coli for the production of arabitol-free xylitol from corncob hemicellulosic hydrolysate. Sci. Rep. 2016, 6, 26567. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.-Q.; Xu, W.; Yang, B.; Liu, Z.-Q.; Zheng, Y.-G. Efficient Biosynthesis of Xylitol from Xylose by Coexpression of Xylose Reductase and Glucose Dehydrogenase in Escherichia coli. Appl. Biochem. Biotechnol. 2019, 187, 1143–1157. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, J.; Lin, J.; Yang, L.; Wu, M. Efficient production of xylitol by the integration of multiple copies of xylose reductase gene and the deletion of Embden–Meyerhof–Parnas pathway-associated genes to enhance NADPH regeneration in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2019, 46, 1061–1069. [Google Scholar] [CrossRef]
- Häcker, B.; Habenicht, A.; Kiess, M.; Mattes, R. Xylose utilisation: Cloning and characterisation of the Xylose reductase from Candida tenuis. Biol. Chem. 1999, 380, 1395–1403. [Google Scholar] [CrossRef]
- Pal, S.; Mondal, A.K.; Sahoo, D.K. Molecular strategies for enhancing microbial production of xylitol. Process Biochem. 2016, 51, 809–819. [Google Scholar] [CrossRef]
- Suzuki, T.; Yokoyama, S.; Kinoshita, Y.; Yamada, H.; Hatsu, M.; Takamizawa, K.; Kawai, K. Expression of xyrA gene encoding for D-Xylose reductase of Candida tropicalis and production of xylitol in Escherichia coli. J. Biosci. Bioeng. 1999, 87, 280–284. [Google Scholar] [CrossRef]
- Branco, R.F.; Santos, J.C.; Murakami, L.Y.; Mussatto, S.I.; Dragone, G.; Silva, S.S. Xylitol production in a bubble column bioreactor: Influence of the aeration rate and immobilized system concentration. Process Biochem. 2007, 42, 258–262. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Roberto, I.C. Xylitol production from high xylose concentration: Evaluation of the fermentation in bioreactor under different stirring rates. J. Appl. Microbiol. 2003, 95, 331–337. [Google Scholar] [CrossRef]
- Winkelhausen, E.; Kuzmanova, S. Microbial conversion of d-xylose to xylitol. J. Ferment. Bioeng. 1998, 86, 1–14. [Google Scholar] [CrossRef]
- Preziosi-Belloy, L.; Nolleau, V.; Navarro, J.M. Fermentation of hemicellulosic sugars and sugar mixtures to xylitol by Candida parapsilosis. Enzyme Microb. Technol. 1997, 21, 124–129. [Google Scholar] [CrossRef]
- Dhar, K.S.; Wendisch, V.F.; Nampoothiri, K.M. Engineering of Corynebacterium glutamicum for xylitol production from lignocellulosic pentose sugars. J. Biotechnol. 2016, 230, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Yoshitake, J.; Ohiwa, H.; Shimamura, M.; Imai, T. Production of Polyalcohol by a Corynebacterium sp. Agric. Biol. Chem. 1971, 35, 905–911. [Google Scholar] [CrossRef][Green Version]
- Toivari, M.H.; Nygård, Y.; Penttilä, M.; Ruohonen, L.; Wiebe, M.G. Microbial D-xylonate production. Appl. Microbiol. Biotechnol. 2012, 96, 1–8. [Google Scholar] [CrossRef]
- Buchert, J.; Viikari, L.; Linko, M.; Markkanen, P. Production of xylonic acid byPseudomonas fragi. Biotechnol. Lett. 1986, 8, 541–546. [Google Scholar] [CrossRef]
- Buchert, J.; Viikari, L. Oxidative D-xylose metabolism of Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 1988, 29, 375–379. [Google Scholar] [CrossRef]
- Buchert, J.; Puls, J.; Poutanen, K. Comparison of Pseudomonas fragi and Gluconobacter oxydans for production of xylonic acid from hemicellulose hydrolyzates. Appl. Microbiol. Biotechnol. 1988, 28, 367–372. [Google Scholar] [CrossRef]
- Buchert, J.; Viikari, L. The role of xylonolactone in xylonic acid production by Pseudomonas fragi. Appl. Microbiol. Biotechnol. 1988, 27, 333–336. [Google Scholar] [CrossRef]
- Wang, C.; Wei, D.; Zhang, Z.; Wang, D.; Shi, J.; Kim, C.H.; Jiang, B.; Han, Z.; Hao, J. Production of xylonic acid by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2016, 100, 10055–10063. [Google Scholar] [CrossRef]
- Sundar, L.; Susmitha, A.; Soumya, M.; Sasikumar, K.; Nampoothiri, M. Bioconversion of D-xylose to D-xylonic acid by Pseudoduganella danionis. Indian J. Exp. Biol. 2019, 57, 825–838. [Google Scholar]
- Dvořák, P.; Kováč, J.; de Lorenzo, V. Biotransformation of D-xylose to D-xylonic acid coupled to medium chain length polyhydroxyalkanoate production in cellobiose-grown Pseudomonas putida EM42. bioRxiv 2019, 702662. [Google Scholar] [CrossRef]
- Nygård, Y.; Toivari, M.H.; Penttilä, M.; Ruohonen, L.; Wiebe, M.G. Bioconversion of D-xylose to D-xylonate with Kluyveromyces lactis. Metab. Eng. 2011, 13, 383–391. [Google Scholar] [CrossRef]
- Liu, M.; Ding, Y.; Xian, M.; Zhao, G. Metabolic engineering of a xylose pathway for biotechnological production of glycolate in Escherichia coli. Microb. Cell Fact. 2018, 17, 51. [Google Scholar] [CrossRef]
- Liu, H.; Valdehuesa, K.N.G.; Nisola, G.M.; Ramos, K.R.M.; Chung, W.-J. High yield production of d-xylonic acid from d-xylose using engineered Escherichia coli. Bioresour. Technol. 2012, 115, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xian, M.; Zou, H.; Zhang, H. Metabolic engineering of Escherichia coli for the production of xylonate. PLoS ONE 2013, 8, e67305. [Google Scholar] [CrossRef]
- Sundar, M.S.L.; Susmitha, A.; Rajan, D.; Hannibal, S.; Sasikumar, K.; Wendisch, V.F.; Nampoothiri, K.M. Heterologous expression of genes for bioconversion of xylose to xylonic acid in Corynebacterium glutamicum and optimization of the bioprocess. AMB Express 2020, 10, 68. [Google Scholar] [CrossRef]
- Lee, S.S.; Choi, J.-I.; Woo, H.M. Bioconversion of Xylose to Ethylene Glycol and Glycolate in Engineered Corynebacterium glutamicum. ACS Omega 2019, 4, 21279–21287. [Google Scholar] [CrossRef]
- Zhou, X.; Lü, S.; Xu, Y.; Mo, Y.; Yu, S. Improving the performance of cell biocatalysis and the productivity of xylonic acid using a compressed oxygen supply. Biochem. Eng. J. 2015, 93, 196–199. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, G.; Zhang, J.; Bao, J. Fermentative production of high titer gluconic and xylonic acids from corn stover feedstock by Gluconobacter oxydans and techno-economic analysis. Bioresour. Technol. 2016, 219, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhou, X.; Xu, Y. Improvement of fermentation performance of Gluconobacter oxydans by combination of enhanced oxygen mass transfer in compressed-oxygen-supplied sealed system and cell-recycle technique. Bioresour. Technol. 2017, 244, 1137–1141. [Google Scholar] [CrossRef]
- Rodzri, N.A.M.; Zain, W.S.; Hanapiah, R.M.A.; Samah, R.A.; Illias, R. D-Xylonic Acid from Recombinant E. coli BL21 (DE3): Comparison between Shake Flask and Benchtop Bioreactor Fermentation. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2020. [Google Scholar]
- Zhang, H.; Han, X.; Wei, C.; Bao, J. Oxidative production of xylonic acid using xylose in distillation stillage of cellulosic ethanol fermentation broth by Gluconobacter oxydans. Bioresour. Technol. 2017, 224, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhou, X.; Liu, G.; Xu, Y.; Balan, V. Integrated production of gluconic acid and xylonic acid using dilute acid pretreated corn stover by two-stage fermentation. Biochem. Eng. J. 2018, 137, 18–22. [Google Scholar] [CrossRef]
- Zhou, X.; Han, J.; Xu, Y. Electrodialytic bioproduction of xylonic acid in a bioreactor of supplied-oxygen intensification by using immobilized whole-cell Gluconobacter oxydans as biocatalyst. Bioresour. Technol. 2019, 282, 378–383. [Google Scholar] [CrossRef] [PubMed]
| Strains | C-Source | Genetic Modification | Growth Conditions | Xylitol (g L−1) * | Yxylitol/xylose (g g−1) | Productivity (g L−1 h−1) * | Ref. |
|---|---|---|---|---|---|---|---|
| Corynebacterium glutamicum Cg-ax3 | arabinose glucose xylose | Yes | Batch shake flask | 6.7 | n.a. | n.a. | [122] |
| Fed-batch shake flask | 31 | n.a. | 0.28 ggcdw−1 h−1 | ||||
| acid pre-treated liquor of sorghum stover | Fed-batch shake flask | 27 | n.a. | 0.22 g g−1cdw h−1 | |||
| Corynebacterium sp. NRRL B 4247 | xylose | No | Shake flask | 1.7 | 0.57 | 0.071 | [108] |
| 6-phosphogluconate (source of NADPH) added to the medium Shake flask | 10 | n.a. | 0.067 | ||||
| Corynebacterium sp. no. 208 | xylose | No | 6-phosphogluconate (source of NADPH) was added to the medium Shake flask | 69 | n.a. | 0.21 | [123] |
| Enterobacter liquefaciens 553 | xylose | No | Shake flask | 33 | n.a. | 0.35 | [110] |
| E. coli BL21(DE3) | xylose | Yes | Shake flask | 202 | 1.0 | 6.37 | [113] |
| Escherichia coli IS5-d | xylose and glucose | Yes | 5 L Batch STR | 110 | n.a. | 3.06 | [112] |
| Escherichia coli IS5-M | corncob hemicellulosic hydrolysate and 24 g L−1 corn steep liquor | Yes | 15 L Fed-batch STR | 144 | n.a. | 1.84 | [112] |
| Escherichia coli HK402 | xylose and glucose | Yes | 15 L Fed-batch STR | 172 | >0.95 | 1.57 | [112] |
| detoxified hemicellulosic hydrolysate and glucose | 150 | >0.95 | 1.40 | ||||
| Escherichia coli WZ51 | detoxified hemicellulosic hydrolysate | Yes | 15 L Fed-batch STR | 132 | 0.95 | 2.09 | [114] |
| Mycobacterium smegmatis | xylose | No | immobilized D-xylose isomerase from Bacillus coagulans and immobilized M. smegmatis Shake flask | 5 g | 0.80 | n.a. | [106] |
| Paraburkholderia sacchari DSM 17165 | xylose | No | 2 L Fed-batch STR | 17 | n.a. | 0.39 | [60] |
| Paraburkholderia sacchari DSM 17165 | xylose | No | 2 L Fed-batch STR | 70 | 0.39 | 0.50 | [80] |
| Strains | C-Source | Genetic Modification | Growth Conditions | Xylonic Acid (g L−1) * | Yxylonic acid/xylose (g g−1) | Productivity (g L−1 h−1) * | Ref. |
|---|---|---|---|---|---|---|---|
| Corynebacterium glutamicum ATCC13032 | xylose | Yes | Shake flask | 50.7 | 0.76 | 0.42 | [136] |
| Corynebacterium glutamicum ATCC31831 | rice straw hydrolysate after dilute sulfuric acid pretreatment | Yes | Shake flask | 42.9 | 1.1 | 0.37 | [136] |
| xylose | 56.3 | 0.84 | 0.47 | ||||
| Escherichia coli BL21 | xylose | Yes | Shake flask | 9.1 | 1.10 | 0.45 | [141] |
| 2 L Batch STR | 6.9 | 0.89 | 0.11 | ||||
| Escherichia coli W3110 | xylose and glucose | Yes | Shake flask | 5.1 | 0.51 | 0.084 | [134] |
| 5 L Fed-batch STR | 39.2 | 0.98 | 1.09 | ||||
| Escherichia coli BL21 | xylose and glycerol | Yes | 5 L Fed-batch STR | 27.3 | n.a. | 1.8 | [135] |
| Gluconobacter oxydans ATCC 621 | xylose | No | 3 L Batch STR | 109 | 0.95 | 2.5 | [127] |
| steamed and enzymatically hydrolyzed birchwood | 12.4 | 0.50 | n.a. | ||||
| Gluconobacter oxydans DSM 2003 | corn stover hydrolysate after dry dilute acid pretreatment | No | 3 L Batch STR | 38.9 | 0.9 | n.a. | [139] |
| Gluconobacter oxydans DSM 2003 | xylose | No | 3 L Batch STR | 66.4 | n.a. | 5.5 | [142] |
| Gluconobacter oxydans NL71 | xylose | No | Compressed oxygen-supplied sealed stirred tank reactor (COS-SSTR); pure oxygen supply | 586.3 | 0.95 | 4.7 | [138] |
| corn stover diluted sulfuric acid hydrolysates without detoxification | 143.9 | 0.97 | 1.0 | ||||
| Gluconobacter oxydans NL71 | xylose in distillation stillage of cellulosic ethanol fermentation broth | No | COS-SSTR; fed-batch addition of xylose with cell-recycling | 1813 g in 6-fold cell recycling; 1 L culture medium | n.a. | 16.8 g h−1 in 108 h | [140] |
| Gluconobacter oxydans NL71 | corn stover hydrolysate after dry diluted acid pretreatment | No | Two-stage fermentation in a 3 L COS-SSTR bioreactor with cell recycling | 167.4 g from 1 kg corn stover | 0.97 | 3.7 | [143] |
| Gluconobacter Oxydans ATCC 621 | xylose | No | Fed-batch bioreactor; Immobilized whole-cells; pressurized pure oxygen supply followed by electrodialysis acid chamber (POA-SSB-OE) | 329.2 g xylonic acid | n.a. | 7.1 g h−1 in 48 h | [144] |
| Klebsiella pneumoniae (modified) | bamboo hydrolysate | Yes | Fed-batch cultivations | 103 g L−1 | 0.98 | n.a. | [129] |
| Paraburkholderia sacchari DSM 17165 | xylose | No | 2 L Fed-batch STR xylose as carbon source; high dissolved oxygen concentration | 150 g L−1 | 0.85 | 1.5 | [80] |
| xylose and glucose | 2 L Fed-batch STR high dissolved oxygen concentration | 390 g L−1 | 1.1 | 6.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domingues, R.; Bondar, M.; Palolo, I.; Queirós, O.; de Almeida, C.D.; Cesário, M.T. Xylose Metabolism in Bacteria—Opportunities and Challenges towards Efficient Lignocellulosic Biomass-Based Biorefineries. Appl. Sci. 2021, 11, 8112. https://doi.org/10.3390/app11178112
Domingues R, Bondar M, Palolo I, Queirós O, de Almeida CD, Cesário MT. Xylose Metabolism in Bacteria—Opportunities and Challenges towards Efficient Lignocellulosic Biomass-Based Biorefineries. Applied Sciences. 2021; 11(17):8112. https://doi.org/10.3390/app11178112
Chicago/Turabian StyleDomingues, Rafael, Maryna Bondar, Inês Palolo, Odília Queirós, Catarina Dias de Almeida, and M. Teresa Cesário. 2021. "Xylose Metabolism in Bacteria—Opportunities and Challenges towards Efficient Lignocellulosic Biomass-Based Biorefineries" Applied Sciences 11, no. 17: 8112. https://doi.org/10.3390/app11178112
APA StyleDomingues, R., Bondar, M., Palolo, I., Queirós, O., de Almeida, C. D., & Cesário, M. T. (2021). Xylose Metabolism in Bacteria—Opportunities and Challenges towards Efficient Lignocellulosic Biomass-Based Biorefineries. Applied Sciences, 11(17), 8112. https://doi.org/10.3390/app11178112








