An Insight into Probiotics Bio-Route: Translocation from the Mother’s Gut to the Mammary Gland
Abstract
1. Introduction
2. General Features of Human Milk Microbiota
3. Microbial Partners of Human Milk ‘Factory’
3.1. Predominance of Skin Microflora
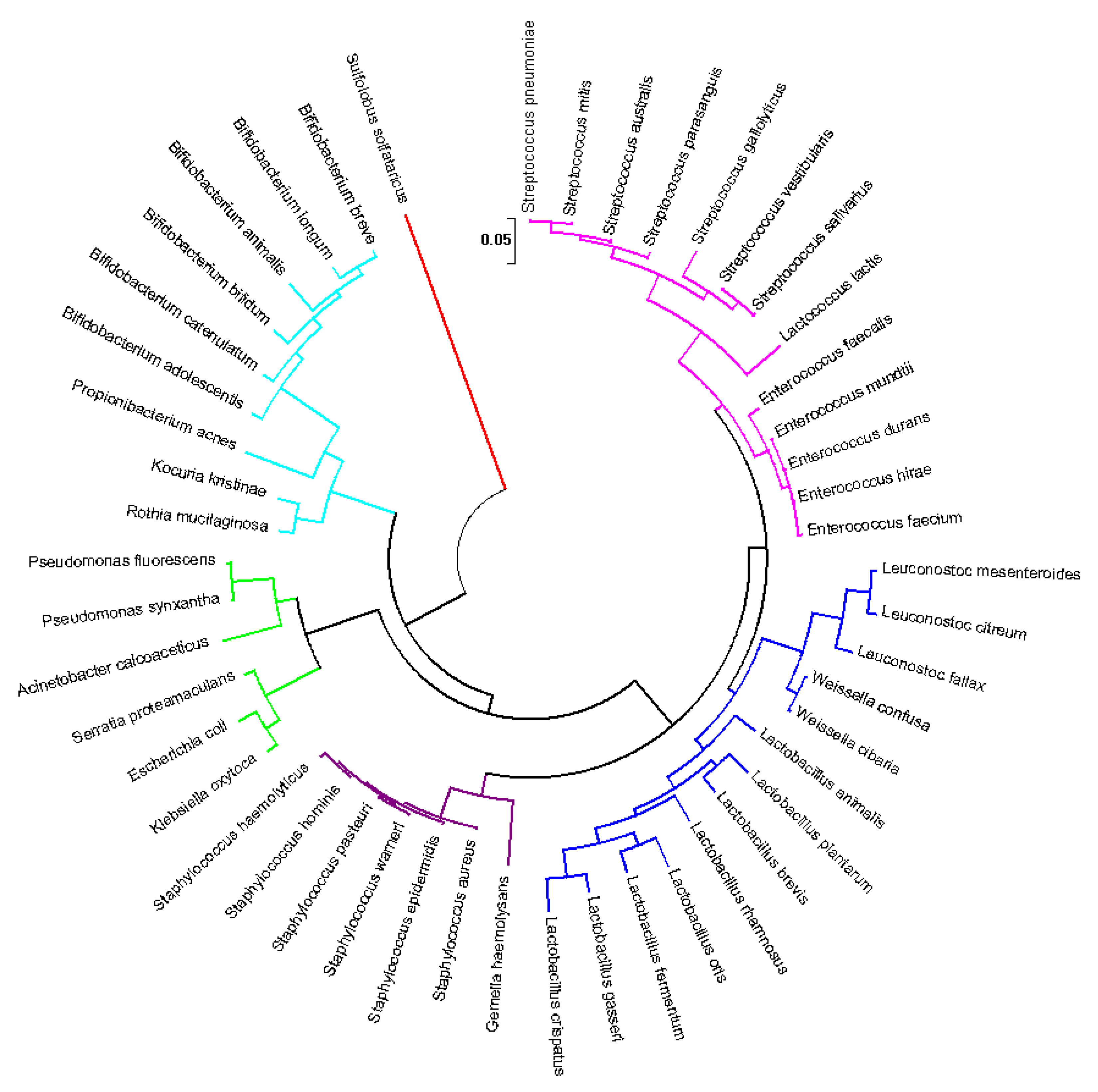
3.2. Human Milk Is a Probiotic Consortium
3.3. Presence of Other Microorganisms
3.4. Predicting the Core of Human Milk Microbiota
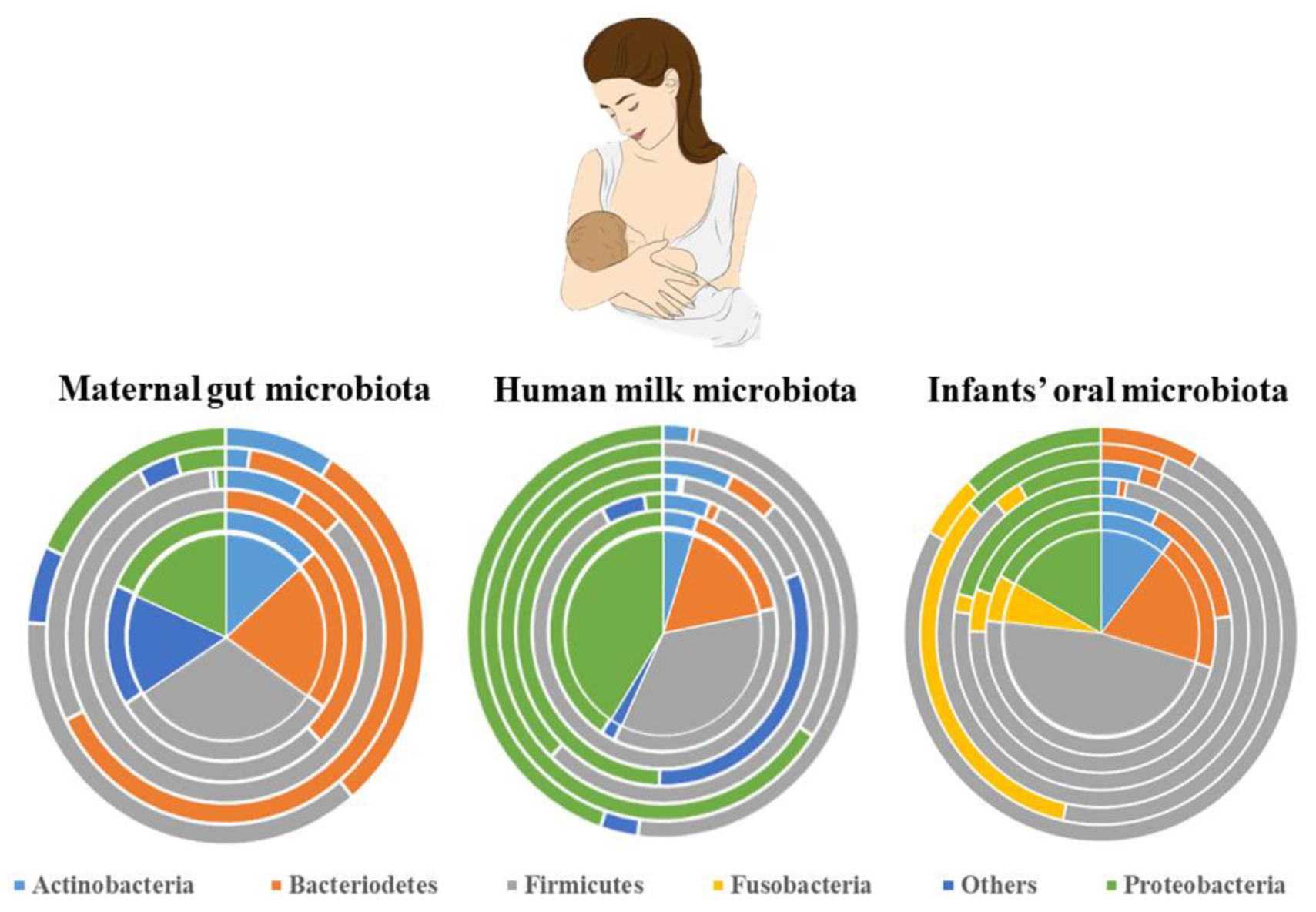
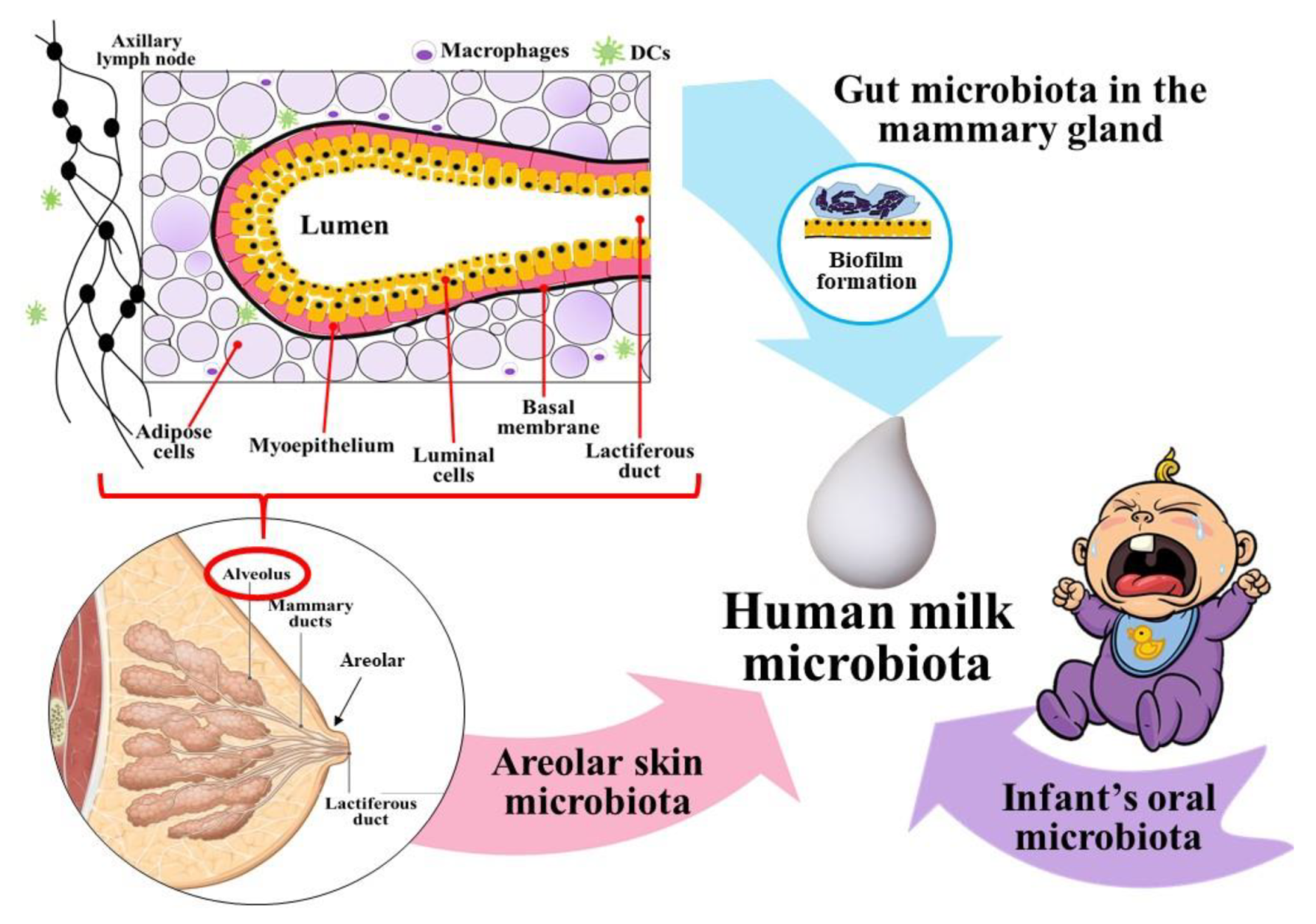
4. Microbial Transmission: Solving the Labyrinth Path
4.1. From the Areolar Skin
4.2. Cross-Contamination from Infants’ Oral Cavity
4.3. Entero-Mammary Pathway: The ‘Silk Route’ to Trade Microbiota from Maternal Gut to Infant
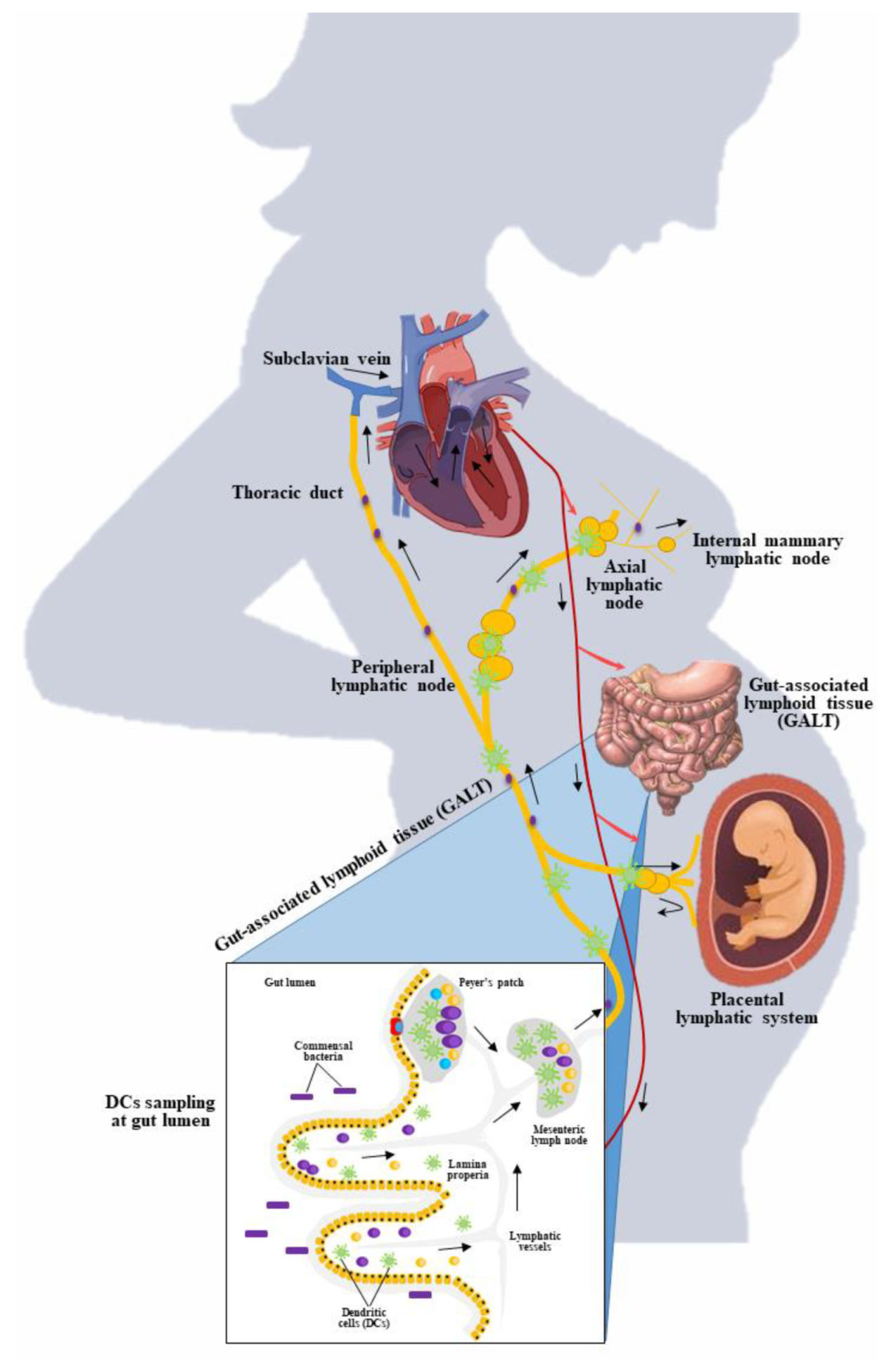
4.3.1. Link between Maternal Gut and Breast Milk
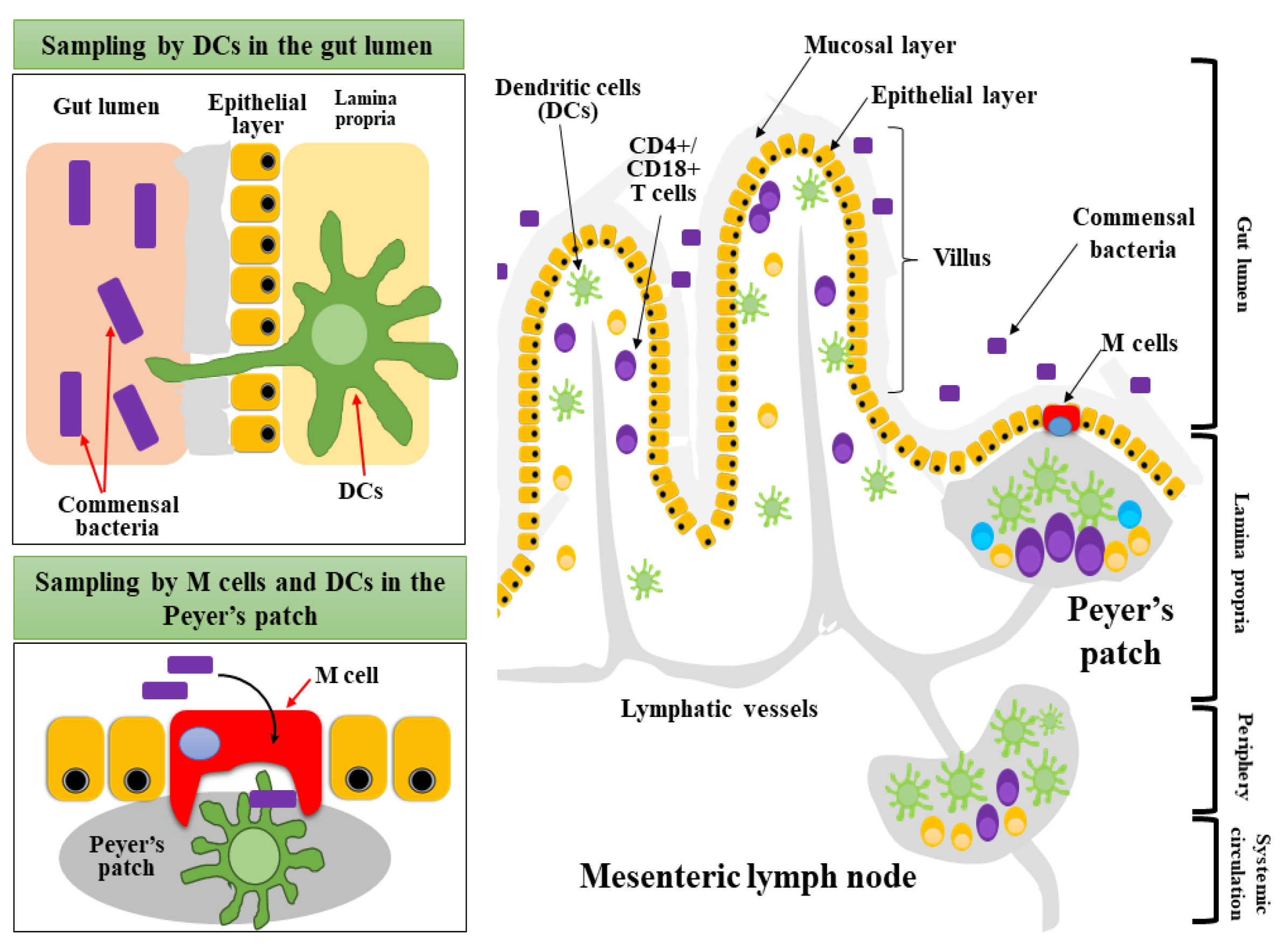
4.3.2. Mucosal Sampling and Migration of Dendritic Cells
4.3.3. Evidence on Gut Transmission to Breast Milk
4.4. Final Microbial Consortia of Human Milk
5. Factors Affecting Microbial Load in the Breast Milk
5.1. Mode of Delivery
5.2. Lactation Period
5.3. Maternal Nutrition
5.4. Maternal Health Status
5.5. Breast Feeding Practices
6. Functionality of Human Milk Microbiota
6.1. Anti-Infections
6.2. Anti-Inflammation
6.3. Metabolic Functions
6.4. Allergic Prevention
6.5. Enterocolitis Prevention
6.6. Growth and Development of Immune System
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatric Clin. 2013, 60, 49–74. [Google Scholar]
- Bardanzellu, F.; Fanos, V.; Strigini, F.A.L.; Artini, P.G.; Peroni, D.G. Human breast milk: Exploring the linking ring among emerging components. Front. Pediatr. 2018, 6, 215. [Google Scholar] [CrossRef] [PubMed]
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 2016, 7, 492. [Google Scholar] [CrossRef] [PubMed]
- Civardi, E.; Garofoli, F.; Tzialla, C.; Paolillo, P.; Bollani, L.; Stronati, M. Microorganisms in human milk: Lights and shadows. J. Matern. Neonatal Med. 2013, 26, 30–34. [Google Scholar] [CrossRef]
- Damaceno, Q.S.; Souza, J.P.; Nicoli, J.R.; Paula, R.L.; Assis, G.B.; Figueiredo, H.; Azevedo, V.; Martins, F.S. Evaluation of potential probiotics isolated from human milk and colostrum. Probiotics Antimicrob. Proteins 2017, 9, 371–379. [Google Scholar] [CrossRef]
- De Andrés, J.; Jiménez, E.; Chico-Calero, I.; Fresno, M.; Fernández, L.; Rodríguez, J.M. Physiological translocation of lactic acid bacteria during pregnancy contributes to the composition of the milk microbiota in mice. Nutrients 2017, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Ricks, N.M.; Panzer, A.; McCoy, A.N.; Azcarate-Peril, M.A.; Keku, T.O.; Boggess, K.A.; Smid, M.C. Maternal gut microbiome biodiversity in pregnancy. Am. J. Perinatol. 2018, 35, 024–030. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Palacio, S.D.; Montes, S.A.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial changes during pregnancy, birth, and infancy. Front. Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M. The origin of human milk bacteria: Is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gallego, C.; Kumar, H.; García-Mantrana, I.; du Toit, E.; Suomela, J.-P.; Linderborg, K.M.; Zhang, Y.; Isolauri, E.; Yang, B.; Salminen, S.; et al. Breast milk polyamines and microbiota interactions: Impact of mode of delivery and geographical location. Ann. Nutr. Metab. 2017, 70, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.; Francis, K.P.; et al. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [PubMed]
- McGuire, M.K.; McGuire, M.A. Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr. Opin. Biotechnol. 2017, 44, 63–68. [Google Scholar] [CrossRef]
- Fitzstevens, J.L.; Smith, K.C.; Hagadorn, J.I.; Caimano, M.J.; Matson, A.P.; Brownell, E.A. Systematic review of the human milk microbiota. Nutr. Clin. Pract. 2016, 32, 354–364. [Google Scholar] [CrossRef]
- Jiménez, E.; de Andrés, J.; Manrique, M.; Pareja-Tobes, P.; Tobes, R.; Martínez-Blanch, J.F.; Codoñer, F.M.; Ramón, D.; Fernández, L.; Rodríguez, J.M. Metagenomic analysis of milk of healthy and mastitis-suffering women. J. Hum. Lact. 2015, 31, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; McMillan, A.; Angelini, M.; Gloor, G.B.; Sumarah, M.; Burton, J.P.; Reid, G. Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome 2014, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Mangifesta, M.; Mancabelli, L.; Lugli, G.A.; James, K.; Duranti, S.; Turroni, F.; Ferrario, C.; Ossiprandi, M.C.; van Sinderen, D.; et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017, 11, 2834–2847. [Google Scholar] [CrossRef]
- Heikkila, M.P.; Saris, P.E.J. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 2003, 95, 471–478. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef]
- Murphy, K.; Curley, D.; O’Callaghan, T.; O’Shea, C.-A.; Dempsey, E.M.; O’Toole, P.; Ross, R.; Ryan, C.A.; Stanton, C. The composition of human milk and infant faecal microbiota over the first three months of life: A pilot study. Sci. Rep. 2017, 7, 40597. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Vaidya, Y.H.; Patel, R.; Pandit, R.J.; Joshi, C.G.; Kunjadiya, A.P. Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Sakwinska, O.; Moine, D.; Delley, M.; Combremont, S.; Rezzonico, E.; Descombes, P.; Vinyes-Pares, G.; Zhang, Y.; Wang, P.; Thakkar, S.K. Microbiota in breast milk of chinese lactating mothers. PLoS ONE 2016, 11, e0160856. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association between breast milk bacterial communities and establishment and development of the infant gut micro-biome. JAMA Pediatrics 2017, 171, 647–654. [Google Scholar] [CrossRef]
- Pevzner, M.; Dahan, A. Mastitis while breastfeeding: Prevention, the importance of proper treatment, and potential complications. J. Clin. Med. 2020, 9, 2328. [Google Scholar] [CrossRef] [PubMed]
- Hunt, K.M.; Preuss, J.; Nissan, C.; Davlin, C.A.; Williams, J.E.; Shafii, B.; Richardson, A.; McGuire, M.K.; Bode, L. Human milk oligosaccharides promote the growth of Staphylococci. Appl. Environ. Microbiol. 2012, 78, 4763–4770. [Google Scholar] [CrossRef]
- Marín, M.; Arroyo, R.; Espinosa-Martos, I.; Fernández, L.; Rodríguez, J. Identification of emerging human mastitis pathogens by MALDI-TOF and assessment of their antibiotic resistance patterns. Front. Microbiol. 2017, 8, 1258. [Google Scholar] [CrossRef]
- Jiménez, E.; Arroyo, R.; Cárdenas, N.; Marín, M.; Serrano, P.; Fernández, L.; Rodríguez, J.M. Mammary candidiasis: A medical condition without scientific evidence? PLoS ONE 2017, 12, e0181071. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2018, 95, 241. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B. Bioactive proteins in human milk: Health, nutrition, and implications for infant formulas. J. Pediatr. 2016, 173, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Hunt, K.M.; Foster, J.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Sonnenburg, J. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 2012, 18, 12–15. [Google Scholar] [CrossRef]
- Thomson, P.; Medina, D.; Garrido, D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018, 75, 37–46. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Chassard, C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE 2012, 7, e44595. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.; Kariyawasam, V.C.; Mogan, S.B.; Patel, K.V.; Pantelidou, M.; Sobczyńska-Malefora, A.; Porté, F.; Griffin, N.; Anderson, S.H.C.; Harrington, D.J.; et al. Prevalence and risk factors for functional vitamin B12 deficiency in patients with Crohn’s disease. Inflamm. Bowel Dis. 2015, 21, 2839–2847. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. BioMed Res. Int. 2017, 2017, 1–7. [Google Scholar] [CrossRef]
- Li, S.-W.; Watanabe, K.; Hsu, C.-C.; Chao, S.-H.; Yang, Z.-H.; Lin, Y.-J.; Chen, C.-C.; Cao, Y.-M.; Huang, H.-C.; Chang, C.-H.; et al. Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and mainland China. Front. Microbiol. 2017, 8, 965. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Heilig, H.G.; Zoetendal, E.G.; Jiménez, E.; Fernández, L.; Smidt, H.; Rodríguez, J. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res. Microbiol. 2007, 158, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Sinkiewicz, G.; Ljunggren, L. Occurrence of Lactobacillus reuteri in human breast milk. Microb. Ecol. Health Dis. 2008, 20, 122–126. [Google Scholar] [CrossRef]
- Langa, S.; Barragán, A.M.; Delgado, S.; Martin, R.; Martin, V.; Jiménez, E.; Ruíz-Barba, J.L.; Mayo, B.; Connor, R.I.; Suarez-Fernandez, J.E.; et al. Characterization of Lactobacillus salivarius CECT 5713, a strain isolated from human milk: From genotype to phenotype. Appl. Microbiol. Biotechnol. 2012, 94, 1279–1287. [Google Scholar] [CrossRef]
- Martín, R.; Langa, S.; Reviriego, C.; Jiménez, E.; Marín, M.L.; Olivares, M.; Boza, J.; Jiménez, J.; Fernández, L.; Xaus, J.; et al. The commensal microflora of human milk: New perspectives for food bacteriotherapy and probiotics. Trends Food Sci. Technol. 2004, 15, 121–127. [Google Scholar] [CrossRef]
- Ozgun, D.; Vural, H.C. Identification of Lactobacillus strains isolated from faecal specimens of babies and human milk colostrum by API 50 CHL system. J. Med. Genet. Genom. 2011, 3, 46–49. [Google Scholar]
- Soto, A.; Martin, V.; Jiménez, E.; Mader, I.; Rodríguez, J.; Fernández, L. Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. J. Pediatric Gastroenterol. Nutr. 2014, 59, 78. [Google Scholar] [CrossRef]
- Biagi, E.; Quercia, S.; Aceti, A.; Beghetti, I.; Rampelli, S.; Turroni, S.; Faldella, G.; Candela, M.; Brigidi, P.; Corvaglia, L. The bacterial ecosystem of mother’s milk and infant’s mouth and gut. Front. Microbiol. 2017, 8, 1214. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.M.; Malcata, F. Bifidobacterium spp. and Lactobacillus acidophilus: Biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci. Technol. 1999, 10, 139–157. [Google Scholar] [CrossRef]
- Moossavi, S.; Miliku, K.; Sepehri, S.; Khafipour, E.; Azad, M.B. The prebiotic and probiotic properties of human milk: Implications for infant immune development and pediatric asthma. Front. Pediatr. 2018, 6, 197. [Google Scholar] [CrossRef] [PubMed]
- Rajoka, M.S.R.; Zhao, H.; Lu, Y.; Lian, Z.; Li, N.; Hussain, N.; Shao, D.; Jin, M.-L.; Li, Q.; Shi, J.L. Anticancer potential against cervix cancer (HeLa) cell line of probiotic Lactobacillus casei and Lactobacillus paracasei strains isolated from human breast milk. Food Funct. 2018, 9, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Bond, D.M.; Morris, J.M.; Nassar, N. Study protocol: Evaluation of the probiotic Lactobacillus fermentum CECT5716 for the pre-vention of mastitis in breastfeeding women: A randomised controlled trial. BMC Pregnancy Childbirth 2017, 17, 148. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, J.A.; Maldonado-Lobón, J.A.; Díaz-Ropero, M.P.; Flores-Rojas, K.; Uberos, J.; Leante, J.L.; Affumicato, L.; Couce, M.L.; Garrido, J.M.; Olivares, M.; et al. Oral administration to nursing women of Lactobacillus fermentum CECT5716 prevents lactational mastitis development: A randomized controlled trial. Breastfeed. Med. 2017, 12, 202–209. [Google Scholar] [CrossRef]
- Jeurink, P.; van Bergenhenegouwen, J.; Jiménez, E.; Knippels, L.; Fernández, L.; Garssen, J.; Knol, J.; Rodríguez, J.; Martín, R. Human milk: A source of more life than we imagine. Benef. Microbes 2013, 4, 17–30. [Google Scholar] [CrossRef]
- Mueller, N.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef]
- Cacho, N.T.; Lawrence, R.M. Innate immunity and breast milk. Front. Immunol. 2017, 8, 584. [Google Scholar] [CrossRef]
- Fernández, L.; Langa, S.; Martin, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef]
- Bergmann, H.; Rodríguez, J.M.; Salminen, S.; Szajewska, H. Probiotics in human milk and probiotic supplementation in infant nutrition: A workshop report. Br. J. Nutr. 2014, 112, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Björkstén, B.; Sepp, E.; Julge, K.; Voor, T.; Mikelsaar, M. Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 2001, 108, 516–520. [Google Scholar] [CrossRef]
- Johansson, M.A.; Sjögren, Y.M.; Persson, J.-O.; Nilsson, C.; Sverremark-Ekström, E. Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PLoS ONE 2011, 6, e23031. [Google Scholar] [CrossRef] [PubMed]
- Munblit, D.; Verhasselt, V. Allergy prevention by breastfeeding: Possible mechanisms and evidence from human cohorts. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 427–433. [Google Scholar] [CrossRef]
- Van den Elsen, L.W.; Garssen, J.; Burcelin, R.; Verhasselt, V. Shaping the gut microbiota by breastfeeding: The gateway to allergy prevention? Front. Pediatrics 2019, 7, 47. [Google Scholar] [CrossRef]
- Sung, V.; D’Amico, F.; Cabana, M.D.; Chau, K.; Koren, G.; Savino, F.; Szajewska, H.; Deshpande, G.; Dupont, C.; Indrio, F.; et al. Lactobacillus reuteri to treat infant colic: A meta-analysis. Pediatrics 2017, 141, e20171811. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Koyfman, A.; Gottlieb, M. Lactobacillus reuteri for treatment of infant colic. Acad. Emerg. Med. 2020, 27, 1059–1060. [Google Scholar] [CrossRef] [PubMed]
- Toscano, M.; de Grandi, R.; Grossi, E.; Drago, L. Role of the human breast milk-associated microbiota on the newborns’ immune system: A mini review. Front. Microbiol. 2017, 8, 2100. [Google Scholar] [CrossRef]
- Olszak, T.; An, D.; Zeissig, S.; Pinilla-Vera, M.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef]
- Seddik, H.A.; Ceugniez, A.; Bendali, F.; Cudennec, B.; Drider, D. Yeasts isolated from Algerian infants’s feces revealed a burden of Candida albicans species, non-albicans Candida species and Saccharomyces cerevisiae. Arch. Microbiol. 2016, 198, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Mutschlechner, W.; Karall, D.; Hartmann, C.; Streiter, B.; Baumgartner-Sigl, S.; Orth-Höller, D.; Lass-Flörl, C. Mammary candidiasis: Molecular-based detection of Candida species in human milk samples. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1309–1313. [Google Scholar] [CrossRef]
- Boix-Amorós, A.; Martinez-Costa, C.; Querol, A.; Collado, M.C.; Mira, A. Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Moossavi, S.; Azad, M.B. Origins of human milk microbiota: New evidence and arising questions. Gut Microbes 2020, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.E.; Carrothers, J.M.; Lackey, K.A.; Beatty, N.F.; York, M.A.; Brooker, S.L.; Shafii, B.; Price, W.J.; Settles, M.L.; McGuire, M.A.; et al. Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J. Nutr. 2017, 147, 1739–1748. [Google Scholar] [CrossRef]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling interactions in the microbiome: A network perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef]
- Drago, L.; Toscano, M.; de Grandi, R.; Grossi, E.; Padovani, E.M.; Peroni, D.G. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 2017, 11, 875–884. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Schommer, N.N.; Gallo, R.L. Structure and function of the human skin microbiome. Trends Microbiol. 2013, 21, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, J.; Wu, L.; Luo, J.; Liang, X.; Xiao, B.; Zhu, Y. The impacts of delivery mode on infant’s oral microflora. Sci. Rep. 2018, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zeng, D.-N.; Chi, L.; Tan, Y.; Galzote, C.; Cardona, C.; Lax, S.; Gilbert, J.; Quan, Z.-X. The Influence of age and gender on skin-associated microbial communities in urban and rural human populations. PLoS ONE 2015, 10, e0141842. [Google Scholar] [CrossRef]
- Prescott, S.L.; Larcombe, D.-L.; Logan, A.C.; West, C.; Burks, W.; Caraballo, L.; Levin, M.; van Etten, E.; Horwitz, P.; Kozyrskyj, A.; et al. The skin microbiome: Impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 2017, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Szabó, K.; Erdei, L.; Bolla, B.S.; Tax, G.; Bíró, T.; Kemény, L. Factors shaping the composition of the cutaneous microbiota. Br. J. Dermatol. 2017, 176, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.A.; Müller, K.M.; Weese, J.S.; Neufeld, J.D. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc. Natl. Acad. Sci. USA 2018, 115, E5786–E5795. [Google Scholar] [CrossRef]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef]
- Wilczyńska, P.; Skarżyńska, E.; Lisowska-Myjak, B. Meconium microbiome as a new source of information about long-term health and disease: Questions and answers. J. Matern. Fetal Neonatal Med. 2019, 32, 681–686. [Google Scholar] [CrossRef]
- Korpela, K.; Costea, P.I.; Coelho, L.P.; Kandels-Lewis, S.; Willemsen, G.; Boomsma, D.I.; Segata, N.; Bork, P. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018, 28, 561–568. [Google Scholar] [CrossRef]
- Schmidt, T.; Raes, J.; Bork, P. The human gut microbiome: From association to modulation. Cell 2018, 172, 1198–1215. [Google Scholar] [CrossRef]
- Gohir, W.; Whelan, F.J.; Surette, M.G.; Moore, C.; Schertzer, J.D.; Sloboda, D.M. Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother’s periconceptional diet. Gut Microbes 2015, 6, 310–320. [Google Scholar] [CrossRef]
- Barko, P.; McMichael, M.; Swanson, K.; Williams, D. The gastrointestinal microbiome: A Review. J. Vet. Intern. Med. 2017, 32, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Bäckhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef]
- Martín, R.; Langella, P. Emerging health concepts in the probiotics field: Streamlining the definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed]
- Albesharat, R.; Ehrmann, M.A.; Korakli, M.; Yazaji, S.; Vogel, R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011, 34, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferretti, P.; Gorfer, V.; et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015, 81, 7078–7087. [Google Scholar] [CrossRef]
- Perez, P.F.; Doré, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J.; Donnet-Hughes, A. bacterial imprinting of the neonatal immune system: Lessons from maternal cells? Pediatrics 2007, 119, e724–e732. [Google Scholar] [CrossRef]
- Palmeira, P.; Quinello, C.; Silveira-Lessa, A.L.; Zago, C.A.; Carneiro-Sampaio, M. IgG placental transfer in healthy and pathological pregnancies. Clin. Dev. Immunol. 2011, 2012, 1–13. [Google Scholar] [CrossRef]
- Nagendran, V.; Emmanuel, N.; Bansal, A.S. Does the maternal serum IgG level during pregnancy in primary antibody deficiency influence the IgG level in the newborn? Case Rep. Immunol. 2015, 2015, 1–4. [Google Scholar] [CrossRef][Green Version]
- Brandtzaeg, P. Mucosal immunity: Integration between mother and the breast-fed infant. Vaccine 2003, 21, 3382–3388. [Google Scholar] [CrossRef]
- Brandtzaeg, P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010, 156, S8–S15. [Google Scholar] [CrossRef]
- Rodríguez, J.; Fernández, L.; Verhasselt, V. The gut‒breast axis: Programming health for life. Nutrients 2021, 13, 606. [Google Scholar] [CrossRef] [PubMed]
- Wilson, E.; Butcher, E.C. CCL28 controls immunoglobulin (Ig)A plasma cell accumulation in the lactating mammary gland and IgA antibody transfer to the neonate. J. Exp. Med. 2004, 200, 805–809. [Google Scholar] [CrossRef]
- Rahman, M.; Mohammed, S. Breast cancer metastasis and the lymphatic system. Oncol. Lett. 2015, 10, 1233–1239. [Google Scholar] [CrossRef]
- Suami, H.; Pan, W.-R.; Taylor, G.I. Historical review of breast lymphatic studies. Clin. Anat. 2009, 22, 531–536. [Google Scholar] [CrossRef]
- Adlerberth, I.; Wold, A.E. Establishment of the gut microbiota in Western infants. Acta Paediatr. Int. J. Paediatr. 2009, 98, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Gutzeit, C.; Magri, G.; Cerutti, A. Intestinal IgA production and its role in host-microbe interaction. Immunol. Rev. 2014, 260, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Worbs, T.; Hammerschmidt, S.I.; Förster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2016, 17, 30–48. [Google Scholar] [CrossRef]
- Aliberti, J. Immunity and tolerance induced by intestinal mucosal dendritic cells. Mediat. Inflamm. 2016, 2016, 1–8. [Google Scholar] [CrossRef][Green Version]
- Martín-Fontecha, A.; Lanzavecchia, A.; Sallusto, F. Dendritic cell migration to peripheral lymph nodes. Organotypic Models Drug Dev. 2008, 188, 31–49. [Google Scholar] [CrossRef]
- Farache, J.; Koren, I.; Milo, I.; Gurevich, I.; Kim, K.-W.; Zigmond, E.; Furtado, G.C.; Lira, S.A.; Shakhar, G. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 2013, 38, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Griffin, A.J.; McSorley, S.J. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 2011, 4, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Knoop, K.; Newberry, R.D. Goblet cells: Multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. 2018, 11, 1551–1557. [Google Scholar] [CrossRef]
- Bekiaris, V.; Persson, E.K.; Agace, W.W. Intestinal dendritic cells in the regulation of mucosal immunity. Immunol. Rev. 2014, 260, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Kordy, K.; Gaufin, T.; Mwangi, M.; Li, F.; Cerini, C.; Lee, D.J.; Adisetiyo, H.; Woodward, C.; Pannaraj, P.S.; Tobin, N.H.; et al. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLoS ONE 2020, 15, e0219633. [Google Scholar] [CrossRef]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.-P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Bloom, S.; Bijanki, V.; Nava, G.; Sun, L.; Malvin, N.P.; Donermeyer, D.L.; Dunne, W.M.; Allen, P.M.; Stappenbeck, T.S. Commensal bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 2011, 9, 390–403. [Google Scholar] [CrossRef]
- Franchi, L.; Kamada, N.; Nakamura, Y.; Burberry, A.; Kuffa, P.; Suzuki, S.; Shaw, M.H.; Kim, Y.-G.; Núñez, G. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 2012, 13, 449–456. [Google Scholar] [CrossRef]
- Wendland, M.; Willenzon, S.; Kocks, J.; Davalos-Misslitz, A.C.; Hammerschmidt, S.I.; Schumann, K.; Kremmer, E.; Sixt, M.; Hoffmeyer, A.; Pabst, O.; et al. Lymph node T cell homeostasis relies on steady state homing of dendritic cells. Immunity 2011, 35, 945–957. [Google Scholar] [CrossRef]
- Elias, J.; Bozzo, P.; Einarson, A. Are probiotics safe for use during pregnancy and lactation? Canadian family physician. Can. Fam. Physician 2011, 57, 299–301. [Google Scholar]
- Abrahamsson, T.R.; Sinkiewicz, G.; Jakobsson, T.; Fredrikson, M.; Björkstén, B. Probiotic Lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J. Pediatr. Gastroenterol. Nutr. 2009, 49, 349–354. [Google Scholar] [CrossRef]
- Arroyo, R.; Martin, V.; Maldonado, A.; Jiménez, E.; Fernández, L.; Rodríguez, J. Treatment of Infectious mastitis during lactation: Antibiotics versus oral administration of Lactobacilli isolated from breast milk. Clin. Infect. Dis. 2010, 50, 1551–1558. [Google Scholar] [CrossRef]
- Nasiraii, L.R. Investigation of lactobacilli from mother’s breast milk who were placed on probiotic diet. Afr. J. Microbiol. Res. 2011, 5, 1581–1585. [Google Scholar] [CrossRef]
- Macias, H.; Hinck, L. Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 533–557. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Miras, A.D.; Jackson, R.N.; Jackson, S.N.; Goldstone, A.P.; Olbers, T.; et al. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 2016, 4, 1–9. [Google Scholar] [CrossRef]
- Hoashi, M.; Meche, L.; Mahal, L.; Bakacs, E.; Nardella, D.; Naftolin, F.; Bar-Yam, N.; Dominguez-Bello, M.G. Human milk bacterial and glycosylation patterns differ by delivery mode. Reprod. Sci. 2015, 23, 902–907. [Google Scholar] [CrossRef]
- Khodayarpardo, P.; Pascual, L.M.; Collado, M.C.; Martinez-Costa, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 2014, 34, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Nommsen-Rivers, L.A.; Chantry, C.J.; Peerson, J.M.; Cohen, R.J.; Dewey, K.G.; Appleton, K.M.; Rogers, P.J.; Ness, A.R. Delayed onset of lactogenesis among first-time mothers is related to maternal obesity and factors associated with ineffective breastfeeding. Am. J. Clin. Nutr. 2010, 92, 574–584. [Google Scholar] [CrossRef]
- Chu, D.M.; Meyer, K.M.; Prince, A.L.; Aagaard, K.M. Impact of maternal nutrition in pregnancy and lactation on offspring gut microbial composition and function. Gut Microbes 2016, 7, 459–470. [Google Scholar] [CrossRef]
- Prescott, S.L.; Wickens, K.; Westcott, L.; Jung, W.; Currie, H.; Black, P.N.; Stanley, T.V.; Mitchell, E.A.; Fitzharris, P.; Siebers, R.; et al. Supplementation with Lactobacillus rhamnosus or Bifidobacterium lactis probiotics in pregnancy increases cord blood interferon-γ and breast milk transforming growth factor-β and immunoglobin A detection. Clin. Exp. Allergy 2008, 38, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Licciardi, P.V.; Ismail, I.H.; Balloch, A.; Mui, M.; Hoe, E.; Lamb, K.; Tang, M.L.K. Maternal supplementation with LGG reduces vaccine-specific immune responses in infants at high-risk of developing allergic disease. Front. Immunol. 2013, 4, 381. [Google Scholar] [CrossRef] [PubMed]
- Boyle, R.J.; Ismail, I.H.; Kivivuori, S.; Licciardi, P.V.; Robins-Browne, R.M.; Mah, L.-J.; Axelrad, C.; Moore, S.; Donath, S.; Carlin, J.B.; et al. Lactobacillus GG treatment during pregnancy for the prevention of eczema: A randomized controlled trial. Allergy 2010, 66, 509–516. [Google Scholar] [CrossRef]
- Liakopoulou, E.; Blau, C.A.; Li, Q.; Josephson, B.; Wolf, J.A.; Fournarakis, B.; Raisys, V.; Dover, G.; Papayannopoulou, T.; Stamatoyannopoulos, G. Stimulation of fetal hemoglobin production by short chain fatty acids. Blood 1995, 85, 3227–3235. [Google Scholar] [CrossRef]
- Wopereis, H.; Sim, K.; Shaw, A.; Warner, J.O.; Knol, J.; Kroll, J.S. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J. Allergy Clin. Immunol. 2018, 141, 1334–1342.e5. [Google Scholar] [CrossRef]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 2016, 7, 1619. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Páez, A.; Olivares, M.; Szajewska, H.; Pieścik-Lech, M.; Polanco, I.; Castillejo, G.; Nuñez, M.; Ribes-Koninckx, C.; Korponay-Szabó, I.R.; Koletzko, S.; et al. Breast-milk microbiota linked to celiac disease development in children: A pilot study from the prevent cd cohort. Front. Microbiol. 2020, 11, 1335. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. The effect of antibiotics on the composition of the intestinal microbiota—A systematic review. J. Infect. 2019, 79, 471–489. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. Breast milk microbiota: A review of the factors that influence composition. J. Infect. 2020, 81, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.W.; Bernard, J.Y.; Thavamani, G.; Chan, Y.H.; Fok, D.; Soh, S.-E.; Chua, M.C.; Lim, S.B.; Shek, L.P.; Yap, F.; et al. Direct vs. expressed breast milk feeding: Relation to duration of breastfeeding. Nutrients 2017, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Becker, G.E.; Smith, H.A.; Cooney, F. Methods of milk expression for lactating women. Cochrane Database Syst. Rev. 2016, 9, CD006170. [Google Scholar] [CrossRef]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 2019, 25, 324–335.e4. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.B. Infant feeding and the developmental origins of chronic disease in the child cohort: Role of human milk bioactives and gut microbiota. Breastfeed. Med. 2019, 14, S-22–S-24. [Google Scholar] [CrossRef]
- Fan, P.; Li, L.; Rezaei, A.; Eslamfam, S.; Che, D.; Ma, X. Metabolites of dietary protein and peptides by intestinal microbes and their Impacts on gut. Curr. Protein Pept. Sci. 2015, 16, 646–654. [Google Scholar] [CrossRef]
- Grönlund, M.-M.; Gueimonde, M.; Laitinen, K.; Kociubinski, G.; Grönroos, T.; Salminen, S.; Isolauri, E. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin. Exp. Allergy 2007, 37, 1764–1772. [Google Scholar] [CrossRef]
- Chiu, Y.-H.; Tsai, J.-J.; Lin, S.-L.; Chotirosvakin, C.; Lin, M.-Y. Characterisation of bifidobacteria with immunomodulatory properties isolated from human breast milk. J. Funct. Foods 2014, 7, 700–708. [Google Scholar] [CrossRef]
- Nolan, L.S.; Parks, O.B.; Good, M. A review of the immunomodulating components of maternal breast milk and protection against necrotizing enterocolitis. Nutrients 2019, 12, 14. [Google Scholar] [CrossRef]
- Altobelli, E.; Angeletti, P.M.; Verrotti, A.; Petrocelli, R. The impact of human milk on necrotizing enterocolitis: A systematic review and meta-analysis. Nutrients 2020, 12, 1322. [Google Scholar] [CrossRef] [PubMed]
- Bullen, C.L.; Tearle, P.V.; Willis, A.T. Bifidobacteria in the intestinal tract of infants: An in-vivo study. J. Med. Microbiol. 1976, 9, 325–333. [Google Scholar] [CrossRef]
- Laforest-Lapointe, I.; Arrieta, M.-C. Patterns of early-life gut microbial colonization during human immune development: An ecological perspective. Front. Immunol. 2017, 8, 788. [Google Scholar] [CrossRef] [PubMed]
| References | Oral Administration | Method of Detection |
|---|---|---|
| Arroyo et al. [112] | L. fermentum CECT5716 | 16s rRNA sequence |
| Nasiraii et al. [113] | L. rhamnosus strain LC705 | qPCR and 16S rRNA sequencing |
| Fernández et al. [52] | L. salivarius PS2 | MALDI-TOF and PFGE |
| Hurtado et al. [48] | L. fermentum CECT5716 | MALDI-TOF mass spectrometry |
| Jiménez et al. [27] | L. salivarius CECT5713 and L. gasseri CECT5714 | species-specific PCR, 16S rRNA sequencing and PFGE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selvamani, S.; Dailin, D.J.; Gupta, V.K.; Wahid, M.; Keat, H.C.; Natasya, K.H.; Malek, R.A.; Haque, S.; Sayyed, R.Z.; Abomoelak, B.; et al. An Insight into Probiotics Bio-Route: Translocation from the Mother’s Gut to the Mammary Gland. Appl. Sci. 2021, 11, 7247. https://doi.org/10.3390/app11167247
Selvamani S, Dailin DJ, Gupta VK, Wahid M, Keat HC, Natasya KH, Malek RA, Haque S, Sayyed RZ, Abomoelak B, et al. An Insight into Probiotics Bio-Route: Translocation from the Mother’s Gut to the Mammary Gland. Applied Sciences. 2021; 11(16):7247. https://doi.org/10.3390/app11167247
Chicago/Turabian StyleSelvamani, Shanmugaprakasham, Daniel Joe Dailin, Vijai Kumar Gupta, Mohd Wahid, Ho Chin Keat, Khairun Hani Natasya, Roslinda Abd Malek, Shafiul Haque, R. Z. Sayyed, Bassam Abomoelak, and et al. 2021. "An Insight into Probiotics Bio-Route: Translocation from the Mother’s Gut to the Mammary Gland" Applied Sciences 11, no. 16: 7247. https://doi.org/10.3390/app11167247
APA StyleSelvamani, S., Dailin, D. J., Gupta, V. K., Wahid, M., Keat, H. C., Natasya, K. H., Malek, R. A., Haque, S., Sayyed, R. Z., Abomoelak, B., Sukmawati, D., Varzakas, T., & El Enshasy, H. A. (2021). An Insight into Probiotics Bio-Route: Translocation from the Mother’s Gut to the Mammary Gland. Applied Sciences, 11(16), 7247. https://doi.org/10.3390/app11167247












