Extra-Heavy Crude Oil Degradation by Alternaria sp. Isolated from Deep-Sea Sediments of the Gulf of Mexico
Abstract
:1. Introduction
2. Materials and Methods
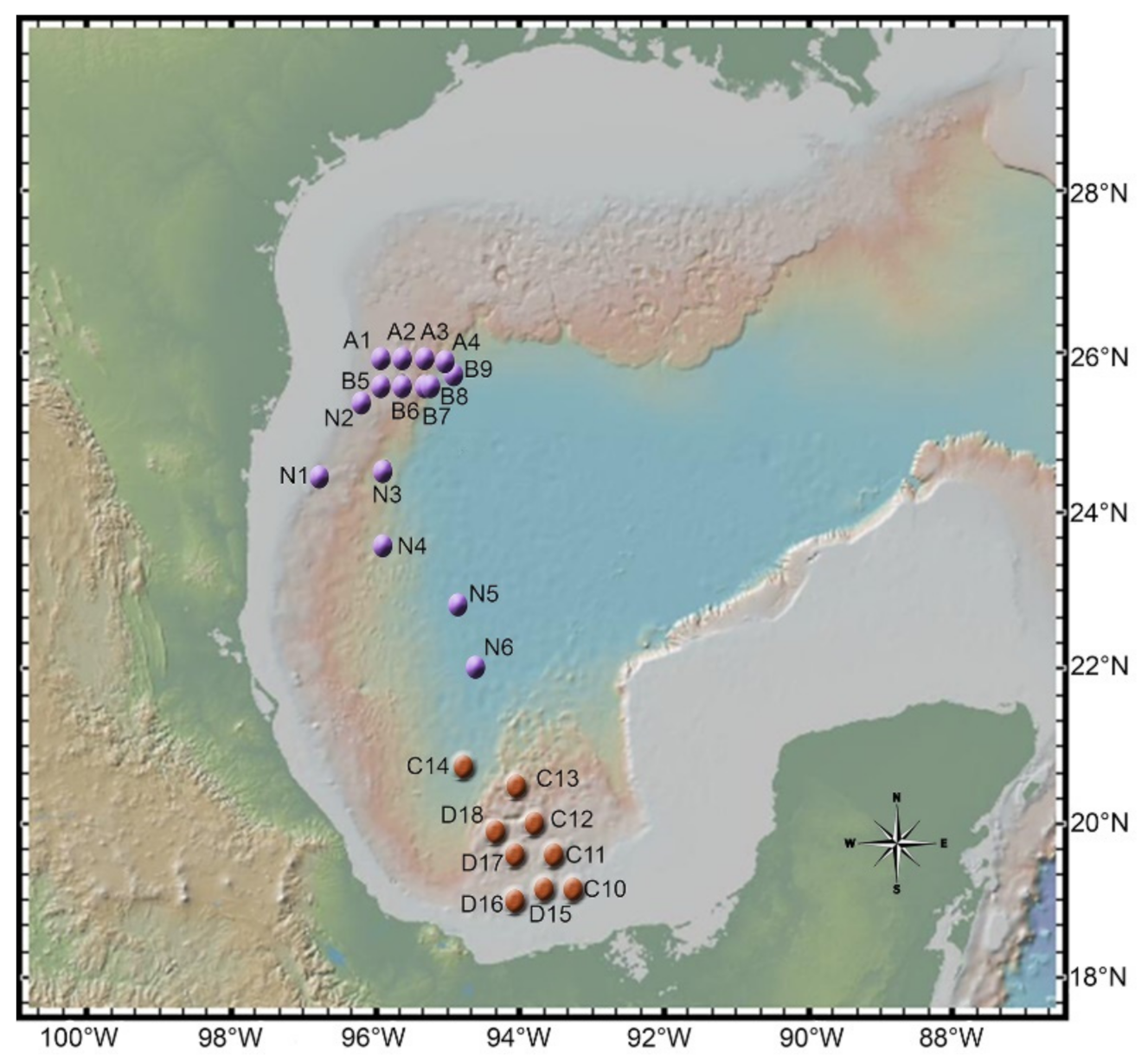
2.1. Sampling
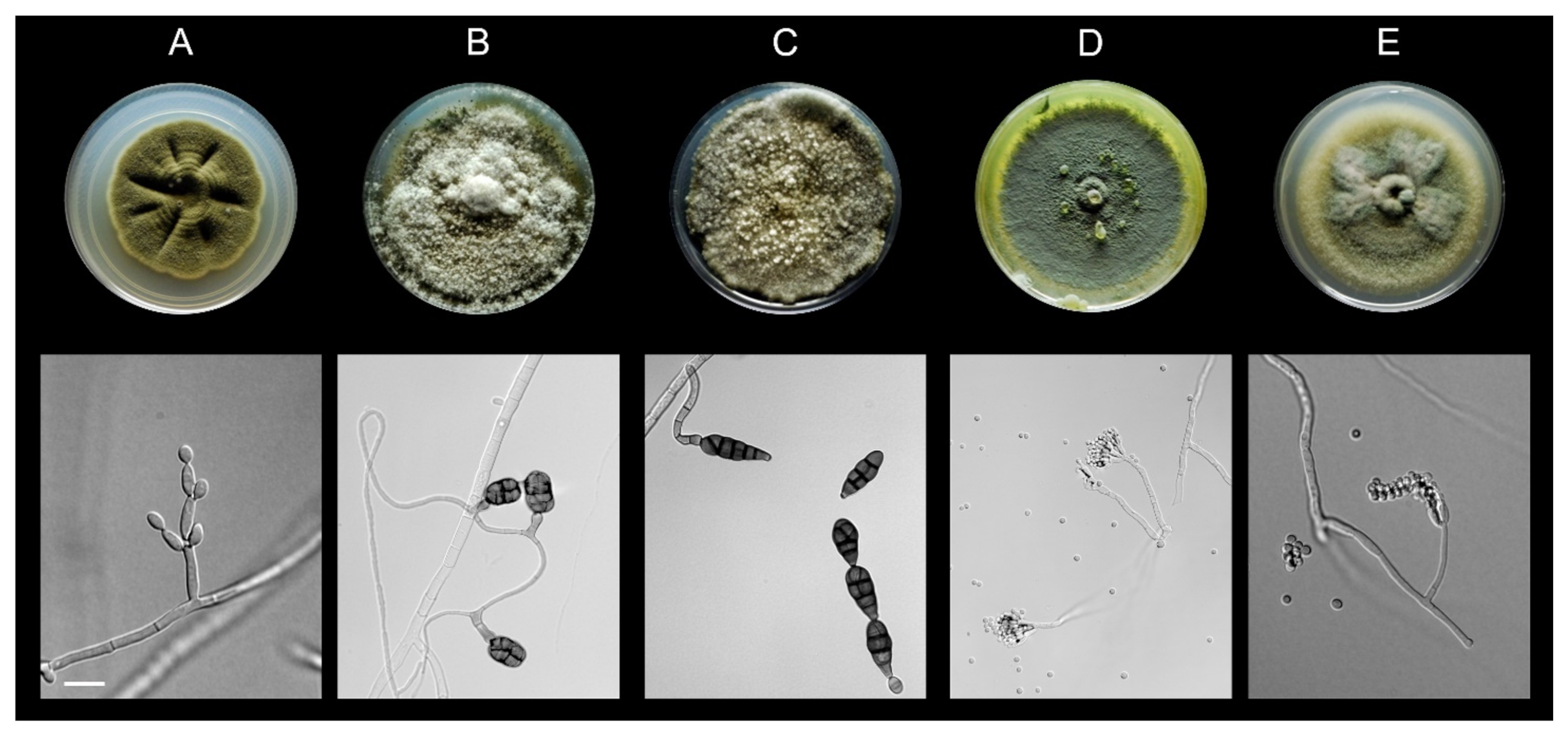
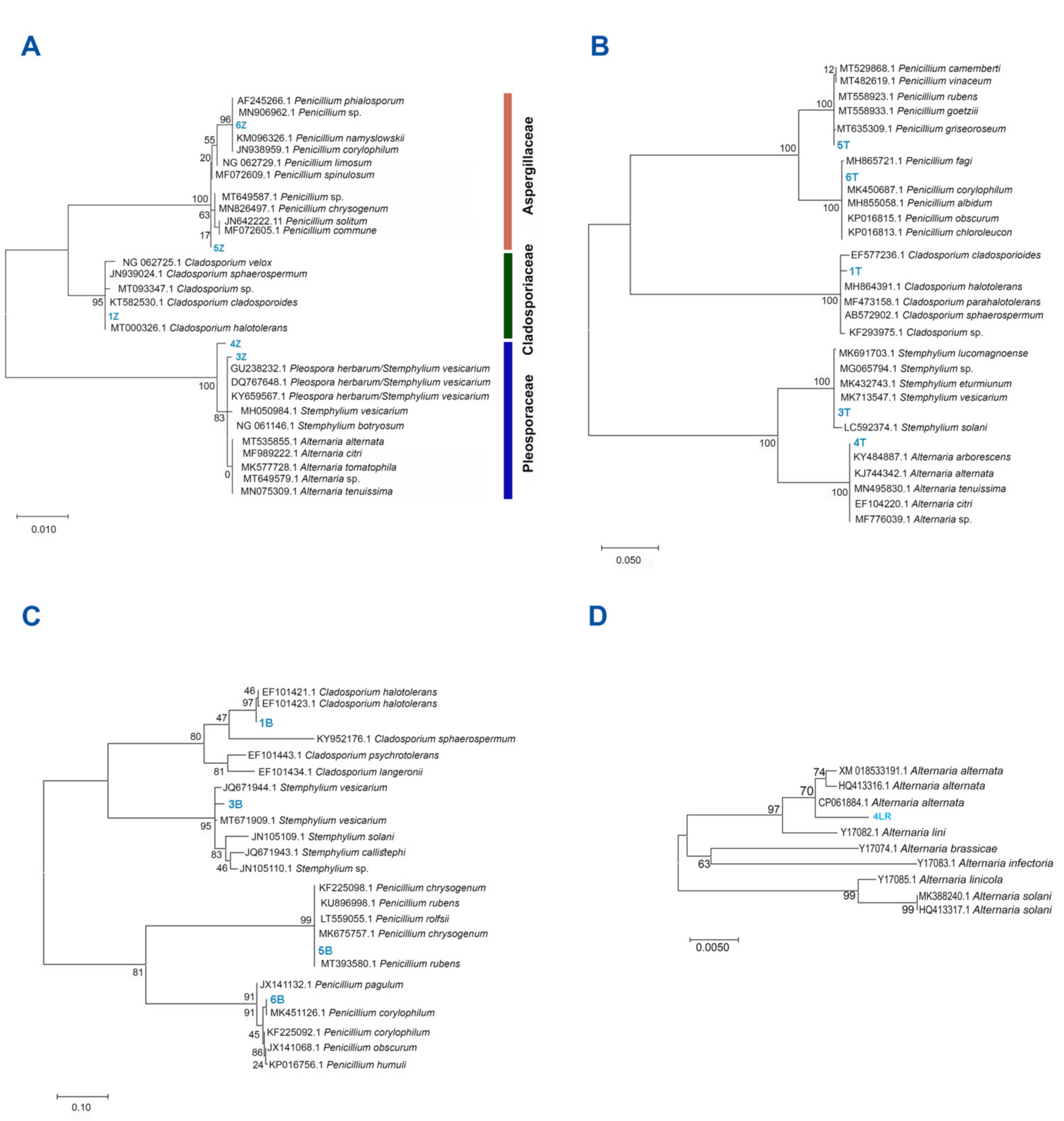
2.2. Fungal Isolation and Identification
2.3. Growth with Crude Oil as a Sole Source of Carbon
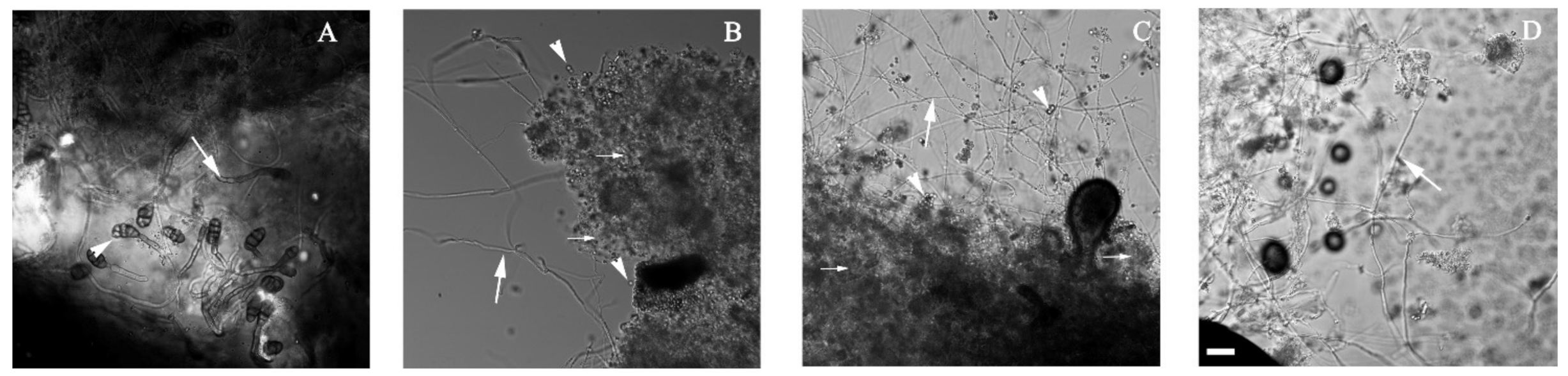
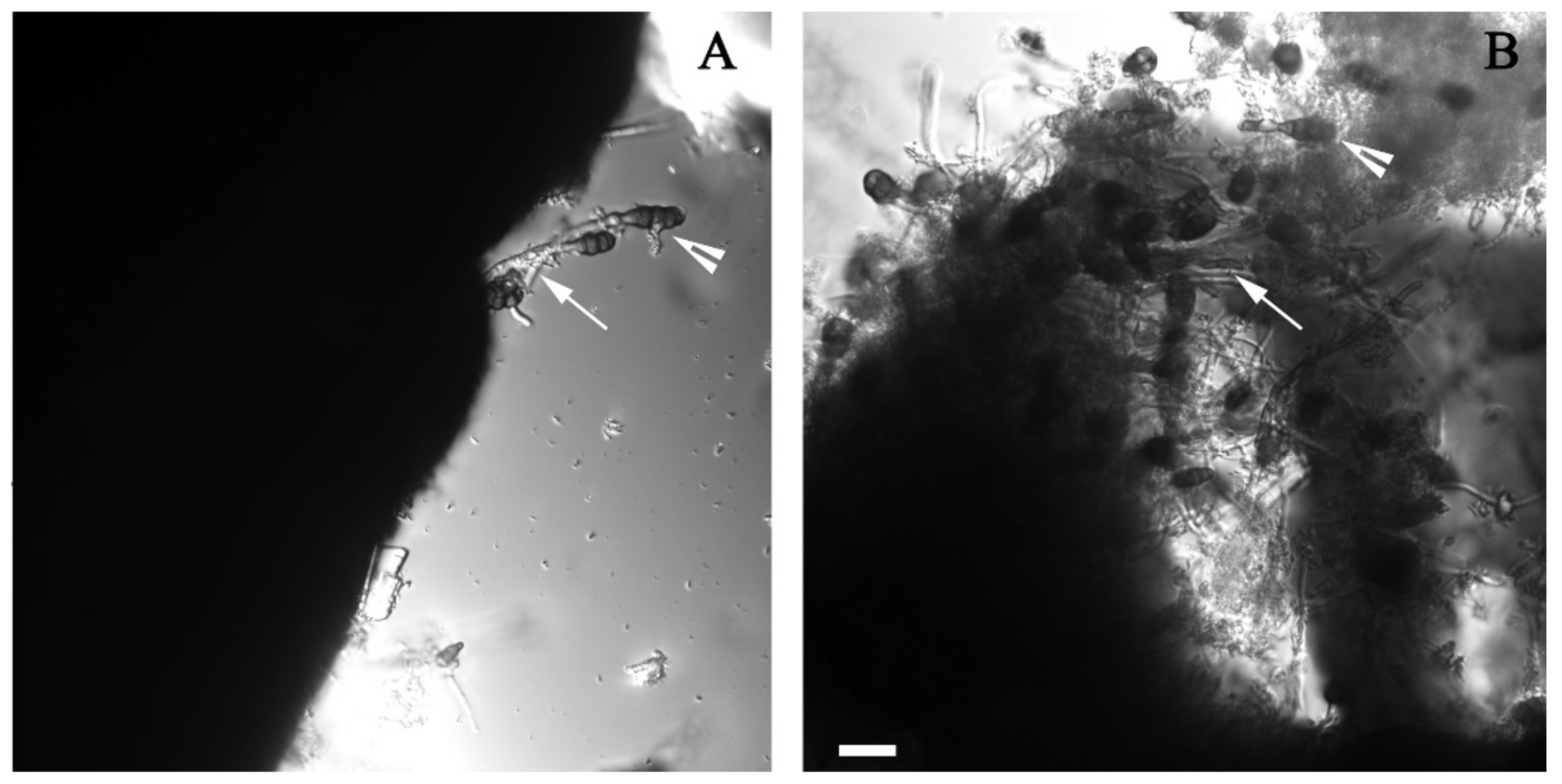
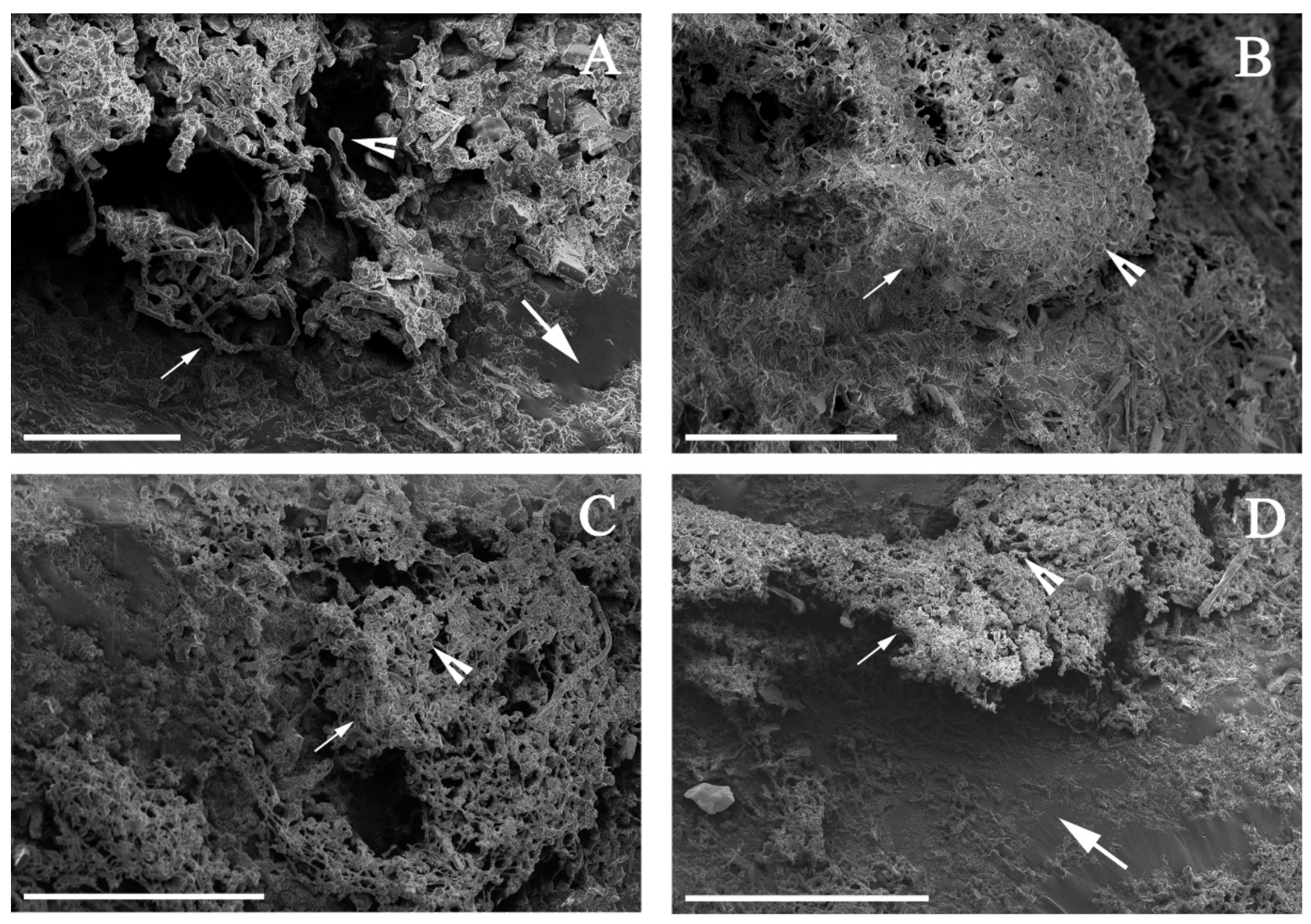
2.4. Microscopic Analysis
2.5. Fungal Growth Quantification
2.6. Liquid–Liquid Extraction
2.7. SAP Analysis
3. Results
3.1. All Fungal Isolates Recovered belonged to the Ascomycota
3.2. Alternaria sp. Was Able to Grow with Extra-Heavy Crude Oil as the Sole Carbon Source
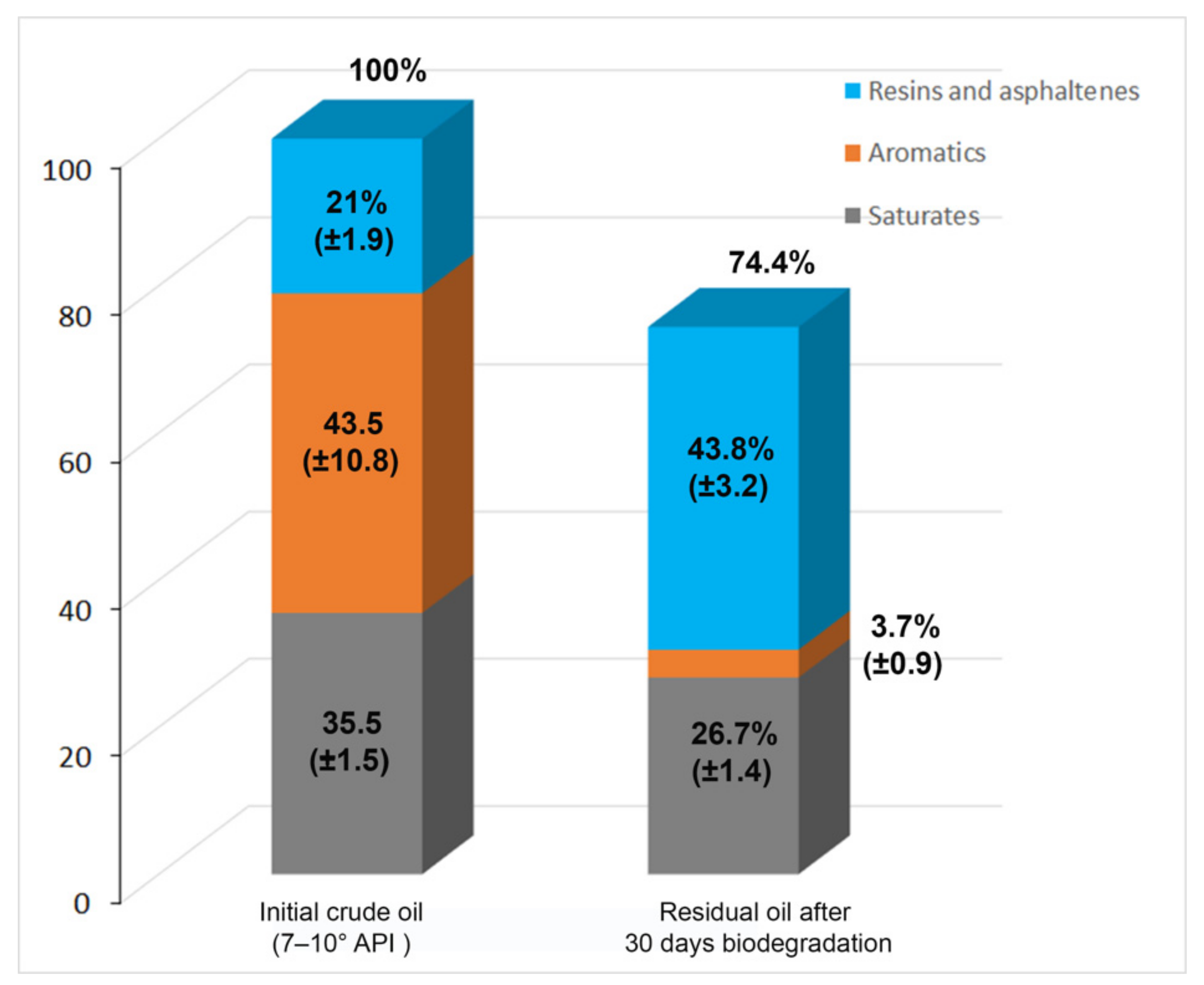
3.3. Alternaria sp. Mainly Metabolizes the Aromatic Components of the EHCO
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Gulf of Mexico | GoM |
| Heavy Crude Oil | HCO |
| Extra-heavy Crude Oil | EHCO |
| Polycyclic Aromatic Hydrocarbons | PAHs |
| Metagenomics Campaign | MET-II |
| Potato Dextrose Agar | PDA |
| Differential Interference Contrast | DIC |
| Internal Transcribed Spacer | ITS |
| β-tubulin gene | benA |
| Maximum Likelihood | ML |
| Maximum Composite Likelihood | MCL |
| American Petroleum Institute | API |
| Total Petroleum Hydrocarbons | TPH |
| Flame Ionization Detector | FID |
| Scanning Electron Microscopy | SEM |
| Saturate, Aromatic and Polar | SAP |
| Economic Exclusive Zone | EZZ |
| Gulf of Mexico Research Consortium | CIGoM |
References
- Matsuo, Y.; Yanagisawa, A.; Yamashita, Y. A global energy outlook to 2035 with strategic considerations for Asia and Middle East energy supply and demand interdependencies. Energy Strat. Rev. 2013, 2, 79–91. [Google Scholar] [CrossRef]
- British Petroleum Company. BP Energy Outlook 2035; British Petroleum Company: London, UK, 2015. [Google Scholar]
- Kuppusamy, S.; Maddela, N.R.; Megharaj, M.; Venkateswarlu, K. Total Petroleum Hydrocarbons. Environ. Fate Toxic. Remediat. 2020. [Google Scholar] [CrossRef] [Green Version]
- Guo, K.; Li, H.; Yu, Z. In situ heavy and extra-heavy oil recovery: A review. Fuel 2016, 185, 886–902. [Google Scholar] [CrossRef]
- Demirbas, A.; Bafail, A.; Nizami, A.-S. Heavy oil upgrading: Unlocking the future fuel supply. Pet. Sci. Technol. 2016, 34, 303–308. [Google Scholar] [CrossRef]
- Covert, T.; Greenstone, M.; Knittel, C.R. Will We Ever Stop Using Fossil Fuels? J. Econ. Perspect. 2016, 30, 117–138. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Lin, F.; Li, X.; Sui, H.; Xu, Z. Interfacial sciences in unconventional petroleum production: From fundamentals to ap-plications. Chem. Soc. Rev. 2015, 44, 5446–5494. [Google Scholar] [CrossRef]
- Farrington, J.W. Need to update human health risk assessment protocols for polycyclic aromatic hydrocarbons in seafood after oil spills. Mar. Pollut. Bull. 2020, 150, 110744. [Google Scholar] [CrossRef] [PubMed]
- Warnock, A.M.; Hagen, S.C.; Passeri, D.L. Marine Tar Residues: A Review. Water Air Soil Pollut. 2015, 226, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Matchinski, E.J.; Chen, B.; Ye, X.; Jing, L.; Lee, K. Marine Oil Spills—Oil Pollution, Sources and Effects. In World Seas: An Environmental Evaluation; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 391–406. [Google Scholar]
- Harwell, M.A.; Gentile, J.H. Ecological significance of residual exposures and effects from the Exxon Valdez oil spill. Integr. Environ. Assess. Manag. 2006, 2, 204. [Google Scholar] [CrossRef]
- Mei, H.; Yin, Y. Studies on marine oil spills and their ecological damage. J. Ocean. Univ. China 2009, 8, 312–316. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Frias-Lopez, J.; Shi, Y.; Tyson, G.; Coleman, M.; Schuster, S.C.; Chisholm, S.W.; DeLong, E.F. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. USA 2008, 105, 3805–3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Hawash, A.B.; Dragh, M.A.; Li, S.; Alhujaily, A.; Abbood, H.A.; Zhang, X.; Ma, F. Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt. J. Aquat. Res. 2018, 44, 71–76. [Google Scholar] [CrossRef]
- Naranjo-Briceño, L.; Pernía, B.; Perdomo, T.; González, M.; Inojosa, Y.; De Sisto, Á.; Urbina, H.; León, V. Potential role of extremophilic hydrocarbonoclastic fungi for extra-heavy crude oil bioconversion and the sustainable development of the pe-troleum industry. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 559–586. [Google Scholar] [CrossRef]
- Uribe-Alvarez, C.; Ayala, M.; Perezgasga, L.; Naranjo, L.; Urbina, H.; Vazquez-Duhalt, R. First evidence of mineralization of petroleum asphaltenes by a strain of Neosartorya fischeri. Microb. Biotechnol. 2011, 4, 663–672. [Google Scholar] [CrossRef] [Green Version]
- López, E.L.H.; Perezgasga, L.; Huerta-Saquero, A.; Mouriño-Pérez, R.R.; Vazquez-Duhalt, R. Biotransformation of petroleum asphaltenes and high molecular weight polycyclic aromatic hydrocarbons by Neosartorya fischeri. Environ. Sci. Pollut. Res. 2016, 23, 10773–10784. [Google Scholar] [CrossRef]
- Naranjo-Briceño, L.; Pernia, B.; Guerra, M.; Demey, J.R.; De Sisto, Á.; Inojosa, Y.; Gonzalez, M.; Fusella, E.; Freites, M.; Yegres, F. Potential role of oxidative exoenzymes of the extremophilic fungus Pestalotiopsis palmarum BM-04 in biotransformation of extra-heavy crude oil. Microb. Biotechnol. 2013, 6, 720–730. [Google Scholar] [CrossRef]
- Chaillan, F.; Le Flèche, A.; Bury, E.; Phantavong, Y.-H.; Grimont, P.; Saliot, A.; Oudot, J. Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms. Res. Microbiol. 2004, 155, 587–595. [Google Scholar] [CrossRef]
- Ayala, M.; López, E.L.H.; Perezgasga, L.; Vazquez-Duhalt, R. Reduced coke formation and aromaticity due to chloroperoxidase-catalyzed transformation of asphaltenes from Maya crude oil. Fuel 2012, 92, 245–249. [Google Scholar] [CrossRef]
- Pourfakhraei, E.; Badraghi, J.; Mamashli, F.; Nazari, M.; Saboury, A.A. Biodegradation of asphaltene and petroleum compounds by a highly potent Daedaleopsi ssp. J. Basic Microbiol. 2018, 58, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Murawski, S.A.; Hollander, D.J.; Gilbert, S.; Gracia, A. Deepwater Oil and Gas Production in the Gulf of Mexico and Related Global Trends. In Scenarios and Responses to Future Deep Oil Spills; Springer: Berlin/Heidelberg, Germany, 2020; pp. 16–32. [Google Scholar]
- Velez, P.; Gasca-Pineda, J.; Riquelme, M. Cultivable fungi from deep-sea oil reserves in the Gulf of Mexico: Genetic signatures in response to hydrocarbons. Mar. Environ. Res. 2020, 153, 104816. [Google Scholar] [CrossRef] [PubMed]
- Hickey, P.C.; Swift, S.R.; Roca, M.G.; Read, N.D. Live-cell imaging of filamentous fungi using vital fluorescent dyes and con-focal microscopy. Methods Microbiol. 2004, 34, 63–87. [Google Scholar] [CrossRef]
- Kohlmeyer, J.; Kohlmeyer, E. Marine Mycology: The Higher Fungi; Academic Press: Cambridge, MA, USA, 1979. [Google Scholar]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and direct sequencing of fungal ribo-somal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Gargas, A.; Taylor, J.W. Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia 1992, 84, 589–592. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. Base-Calling of Automated Sequencer Traces UsingPhred. I. Accuracy Assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D.; Desmarais, C.; Green, P. Automated Finishing with Autofinish. Genome Res. 2001, 11, 614–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M. The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Abalos, A.; Viñas, M.; Sabate, J.; Manresa, A.; Solanas, A. Enhanced Biodegradation of Casablanca Crude Oil by A Microbial Consortium in Presence of a Rhamnolipid Produced by Pseudomonas Aeruginosa AT10. Biodegradation 2004, 15, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Simister, R.; Poutasse, C.; Thurston, A.; Reeve, J.; Baker, M.; White, H. Degradation of oil by fungi isolated from Gulf of Mexico beaches. Mar. Pollut. Bull. 2015, 100, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydrocarbon-Degrading Bacteria and the Bacterial Community Response in Gulf of Mexico Beach Sands Impacted by the Deepwater Horizon Oil Spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef] [Green Version]
- Otero-Blanca, A.; Folch-Mallol, J.L.; Lira-Ruan, V.; del Rayo Sánchez-Carbente, M.; Batista-García, R.A. Phytoremediation and Fungi: An Underexplored Binomial. In Approaches in Bioremediation; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018; pp. 79–95. [Google Scholar]
- Deshmukh, R.; Khardenavis, A.A.; Purohit, H.J. Diverse Metabolic Capacities of Fungi for Bioremediation. Indian J. Microbiol. 2016, 56, 247–264. [Google Scholar] [CrossRef] [Green Version]
- Chandra, R.; Kumar, V.; Yadav, S. Extremophilic Ligninolytic Enzymes. In Extremophilic Enzymatic Processing of Lignocellulosic Feedstocks to Bioenergy; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2017; pp. 115–154. [Google Scholar]
- Vidali, M. Bioremediation. An overview. Pure Appl. Chem. 2001, 73, 1163–1172. [Google Scholar] [CrossRef]
- Kumar, R.; Kaur, A. Oil spill removal by mycoremediation. In Microbial Action on Hydrocarbons; Kumar, V., Kumar, M., Pra-sad, R., Eds.; Springer: Singapore, 2018; pp. 505–526. [Google Scholar] [CrossRef]
- Kvenvolden, K.A.; Cooper, C.K. Natural seepage of crude oil into the marine environment. Geo-Marine Lett. 2003, 23, 140–146. [Google Scholar] [CrossRef]
- Elshafie, A.; Alkindi, A.Y.; Al-Busaidi, S.; Bakheit, C.; Albahry, S. Biodegradation of crude oil and n-alkanes by fungi isolated from Oman. Mar. Pollut. Bull. 2007, 54, 1692–1696. [Google Scholar] [CrossRef]
- Singh, H. Mycoremediation: Fungal Bioremediation; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Garon, D.; Sage, L.; Seigle-Murandi, F. Effects of fungal bioaugmentation and cyclodextrin amendment on fluorene degrada-tion in soil slurry. Biodegradation 2004, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Saraswathy, A.; Hallberg, R. Mycelial pellet formation by Penicillium ochrochloron species due to exposure to pyrene. Microbiol. Res. 2005, 160, 375–383. [Google Scholar] [CrossRef]
- Melendez-Estrada, J.; Amezcua-Allieri, M.A.; Alvarez, P.; Rodríguez-Vázquez, R. Phenanthrene Removal byPenicillium frequentansGrown on a Solid-State Culture: Effect of Oxygen Concentration. Environ. Technol. 2006, 27, 1073–1080. [Google Scholar] [CrossRef]
- Mohammadian, E.; Arzanlou, M.; Babai-Ahari, A. Diversity of culturable fungi inhabiting petroleum-contaminated soils in Southern Iran. Antonie van Leeuwenhoek 2017, 110, 903–923. [Google Scholar] [CrossRef] [PubMed]
- Bovio, E.; Gnavi, G.; Prigione, V.; Spina, F.; Denaro, R.; Yakimov, M.; Calogero, R.; Crisafi, F.; Varese, G.C. The culturable mycobiota of a Mediterranean marine site after an oil spill: Isolation, identification and potential application in bioremediation. Sci. Total Environ. 2017, 576, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Dhar, K.; Dutta, S.; Anwar, M.N. Biodegradation of petroleum hydrocarbon by indigenous fungi isolated from ship breaking yards of Bangladesh. Int. Res. J. Biol. Sci. 2014, 3, 22–30. [Google Scholar]
- Balaji, V.; Arulazhagan, P.; Ebenezer, P. Enzymatic bioremediation of polyaromatic hydrocarbons by fungal consortia en-riched from petroleum contaminated soil and oil seeds. J. Environ. Biol. 2014, 35, 521–529. [Google Scholar] [PubMed]
- Loretta, O.O.; Samuel, O.; Johnson, G.H.; Emmanuel, S. Comparative Studies on the Biodegradation of Crude Oil-polluted Soil by Pseudomonas aeruginosa and Alternaria Species Isolated from Unpolluted Soil. Microbiol. Res. J. Int. 2017, 19, 1–10. [Google Scholar] [CrossRef]
- Ali, M.I.; Khalil, N.M.; Abd El-Ghany, M.N. Biodegradation of some polycyclic aromatic hydrocarbons by Aspergillus terreus. Afr. J. Microbiol. Res. 2012, 6, 3783–3790. [Google Scholar]
- Vazquez-Duhalt, R.; Westlake, D.W.S.; Fedorak, P.M. Lignin peroxidase oxidation of aromatic compounds in systems con-taining organic solvents. Appl. Environ. Microbiol. 1994, 60, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeiner, C.A.; Purvine, S.; Zink, E.M.; Paša-Tolić, L.; Chaput, D.L.; Haridas, S.; Wu, S.; LaButti, K.; Grigoriev, I.V.; Henrissat, B.; et al. Comparative Analysis of Secretome Profiles of Manganese(II)-Oxidizing Ascomycete Fungi. PLoS ONE 2016, 11, e0157844. [Google Scholar] [CrossRef]
- Sharma, A.; Aggarwal, N.K.; Yadav, A. First Report of Lignin Peroxidase Production from Alternaria alternata ANF238 Isolated from Rotten Wood Sample. Bioeng. Biosci. 2016, 4, 76–87. [Google Scholar] [CrossRef]
- Speight, J.G. The Chemistry and Technology of Petroleum; Marcel Decker: New York, NY, USA, 1999. [Google Scholar]
- Hinkle, A.; Shin, E.-J.; Liberatore, M.W.; Herring, A.M.; Batzle, M. Correlating the chemical and physical properties of a set of heavy oils from around the world. Fuel 2008, 87, 3065–3070. [Google Scholar] [CrossRef]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Van Hamme, J.D.; Singh, A.; Ward, O.P. Recent Advances in Petroleum Microbiology. Microbiol. Mol. Biol. Rev. 2003, 67, 503–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meulenberg, R.; Rijnaarts, H.H.; Doddema, H.J.; A Field, J. Partially oxidized polycyclic aromatic hydrocarbons show an increased bioavailability and biodegradability. FEMS Microbiol. Lett. 2006, 152, 45–49. [Google Scholar] [CrossRef]
- Kulik, N.; Goi, A.; Trapido, M.; Tuhkanen, T. Degradation of polycyclic aromatic hydrocarbons by combined chemical pre-oxidation and bioremediation in creosote contaminated soil. J. Environ. Manag. 2006, 78, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P. Topographic Rossby waves in the Gulf of Mexico. Prog. Oceanogr. 2009, 82, 1–31. [Google Scholar] [CrossRef]
- Portela, E.; Tenreiro, M.; Pallàs-Sanz, E.; Meunier, T.; Ruiz-Angulo, A.; Sosa-Gutiérrez, R.; Cusí, S. Hydrography of the central and western Gulf of Mexico. J. Geophys. Res. Ocean. 2018, 123, 5134–5149. [Google Scholar] [CrossRef]
- Alboudwarej, H.; Felix, J.; Taylor, S.; Badry, R.; Bremner, C.; Brough, B.; Palmer, D.; Pattisson, K.; Beshry, M.; Krawchuk, P.; et al. Highlighting heavy oil. Schlumberger Oilfield Rev. Summer 2006, 18, 34–53. [Google Scholar]
- Kingston, P. Long-term environmental impact of oil spills. Splill Sci. Technol. Bull. 2002, 7, 53–61. [Google Scholar] [CrossRef]
- Atlas, R.M.; Hazen, T.C. Oil Biodegradation and Bioremediation: A Tale of the Two Worst Spills in U.S. History. Environ. Sci. Technol. 2011, 45, 6709–6715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macaulay, B.M.; Rees, D. Bioremediation of oil spills: A review of challenges for research advancement. Ann. Environ. Sci. 2014, 8, 9–37. [Google Scholar]
| Crude Oil Density | |||
|---|---|---|---|
| Identified Isolate | Light 40° API | Heavy 16–20° API | Extra-Heavy 7–10° API |
| Alternaria sp. | + | + | + |
| Cladosporium halotolerans | + | − | − |
| Penicillium sp. 1 | + | + | − |
| Penicillium sp. 2 | + | + | − |
| Stemphylium sp. | + | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-Hernández, L.; Velez, P.; Betanzo-Gutiérrez, I.; Camacho-López, M.D.; Vázquez-Duhalt, R.; Riquelme, M. Extra-Heavy Crude Oil Degradation by Alternaria sp. Isolated from Deep-Sea Sediments of the Gulf of Mexico. Appl. Sci. 2021, 11, 6090. https://doi.org/10.3390/app11136090
Romero-Hernández L, Velez P, Betanzo-Gutiérrez I, Camacho-López MD, Vázquez-Duhalt R, Riquelme M. Extra-Heavy Crude Oil Degradation by Alternaria sp. Isolated from Deep-Sea Sediments of the Gulf of Mexico. Applied Sciences. 2021; 11(13):6090. https://doi.org/10.3390/app11136090
Chicago/Turabian StyleRomero-Hernández, Lucia, Patricia Velez, Itandehui Betanzo-Gutiérrez, María Dolores Camacho-López, Rafael Vázquez-Duhalt, and Meritxell Riquelme. 2021. "Extra-Heavy Crude Oil Degradation by Alternaria sp. Isolated from Deep-Sea Sediments of the Gulf of Mexico" Applied Sciences 11, no. 13: 6090. https://doi.org/10.3390/app11136090
APA StyleRomero-Hernández, L., Velez, P., Betanzo-Gutiérrez, I., Camacho-López, M. D., Vázquez-Duhalt, R., & Riquelme, M. (2021). Extra-Heavy Crude Oil Degradation by Alternaria sp. Isolated from Deep-Sea Sediments of the Gulf of Mexico. Applied Sciences, 11(13), 6090. https://doi.org/10.3390/app11136090








