Abstract
Fetal alcohol spectrum disorder (FASD) is an umbrella term for children’s conditions due to their mother having consumed alcohol during pregnancy. These conditions can be mild to severe, affecting the subject’s quality of life. An earlier diagnosis of FASD is crucial for an improved quality of life of children by allowing a better inclusion in the educational system. New trends in computer-based diagnosis to detect FASD include using Machine Learning (ML) tools to detect this syndrome. However, most of these studies rely on children’s images that can be invasive and costly. Therefore, this paper presents a study that focuses on evaluating an ANN to classify children with FASD using non-invasive and more accessible data. This data used comes from a battery of tests obtained from children, including psychometric, saccade eye movement, and diffusion tensor imaging (DTI). We study the different configurations of ANN with dense layers being the psychometric data that correctly perform the best with 75% of the outcome. The other models include a feature layer, and we used it to predict FASD using every test individually. Model obtained obtained an accuracy of 88.46% (psychometric, 74.07% (Antisaccadic), 72.24% (Prosaccadic), 88% (Memory guide saccade), and 75% (DTI). These results suggest that the ANN approach is a competitive and efficient methodology to detect FASD. These results are an improvement on Zhang’s 2019 model, which used the same data with less accuracy level.
1. Introduction
The consumption of alcohol during pregnancy is an indisputable generator of mental health problems in children [1,2]. Conditions that emerge from this consumption are grouped under the concept of Fetal Alcohol Spectrum Disorder (FASD). This concept was coined in Canada, and included effects may include physical, mental, behavioral, and learning disabilities with possible lifelong implications diagnosis. The most common conditions are Fetal Alcohol Syndrome (FAS), partial Fetal Alcohol Syndrome (pFAS), and the Alcohol-Related Neuro-Developmental Disorder (ARND) [3,4]. Being FAS is the most severe consequence of mother alcohol consumption since the infants would have potential mental retardation. Most of the common outcomes include significant hyperactivity, language delay, and school learning problems. There is an observed higher rate of mental health problems in adolescence, as well as problematic use of alcohol and other drugs.
An earlier diagnosis of these conditions could reduce the long-term health and psychosocial outcome, especially cognitive difficulties, thus improving the quality of life of young children and adolescents. In addition, people who are diagnosed with FASD report higher unemployment, law-breaking, and other social behavior problems. These problems result in a higher cost to the public welfare and society at large. In spite of the importance of the diagnosis of FASD, there are currently no direct clinical characteristics that can be used to diagnose a child with FASD. The only distinct difference compared to other childhood or adolescent conditions is the mother’s alcohol consumption during pregnancy [5,6,7]. If there is no certainty about the mother’s alcohol consumption, some indirect effects are associated with FAS, such as facial dysmorphia, growth deficiency, or central nervous system abnormalities, can be used as proxy for the diagnosis. However, when none of these effects are present, the diagnosis becomes challenging [8].
In the state of the art, we can find an increased effort to develop tools on specific behavior and profiles to allow an early diagnosis of FASD, where these tools are failing to make a specific diagnosis of FASD compared to other neurodevelopmental conditions [6,9]. Among these tools, we find tools in Machine Learning (ML) [10,11] and Computer-Aided Detection (CAD) [12,13]. However, there are tools to improve FASD detection [14]; therefore, this paper addresses the use of ML techniques for an improved FASD diagnosis. Specifically, this work focuses on developing computational algorithms based on ANN to classify children with FASD. The research question is whether an ANN model could be used for medical diagnosis. One important reason for testing ANN as a diagnostic tool is the easy implementation and simplicity in applying existing data. This model will be evaluated based on its performance, in terms of precision, completeness, and performance benchmark to other machine learning techniques. This model will be evaluate the goodness of fitness of using psychometric, saccadic eye movement, and diffusion tensor imaging (DTI) tests). It is believed that techniques based on machines learning are a good tool for grouping and comparing the clinical information obtained from a set of psychometric data, thus expanding the possibility of detecting differences in FASD patients. To our knowledge, the use of machine learning as FASD diagnostic tools has not been tested. However, there are current studies that have been used them, such as Zhang et al. (2019) [15]. Nevertheless, these authors have used support vector machine regression (SVMR) algorithms to analyze psychometric data and DTI and a linear regression for the rest of the data.
The rest of this paper is organized as follows. The next section presents the required background to understand this study. Section 3 describes materials and methods are used to carry out this study and additionally describes the experiment execution. Section 4 presents the results obtained from the experiment. Section 5 discusses these results, and Section 6 provides the conclusions of this researchand other methods for the remaining tests.
Availability: The ANN algorithms and data used to carry out this study are available at https://github.com/vjduarte/ANN_FASD (accessed on 1 May 2021).
2. Related Work
This section discusses two different approaches, Machine Learning (ML) and Computer-Aided Detection (CAD), that could be used to detect fetal alcohol spectrum disorders.
2.1. Machine Learning
Machine Learning (ML) can be used to improve FASD detection [14], which are computational algorithms designed to emulate human intelligence at learning from the surrounding environment [10]. The areas of application in medicine are varied, such as optical character recognition, face recognition, scientific image analysis, biological signal analysis, and others. In the case of diagnosis, machine learning techniques that have been used for medical diagnosis are: Pattern classification, [16], Principal Component Analysis (PCA) [17], Support Vector Machines (SVMs) [18], Deep Learning [3,19,20], and, especially, Artificial Neural Networks (ANN) [21,22,23]. The latter has become popular for its model estimation transparency and has been used to diagnose diseases, such as cancer and even FASD, from brain volume images [24]. In addition, tools has also been used to diagnose diseases, such as cancer and even FASD, from brain volume image [24]. Unlike ANNs, we can use diverse tools to find and detect patterns (e.g., clustering); however, most of tools require that data satisfy conditions to carry out the operation successfully.
2.2. Computer-Aided Detection
Computer-aided detection (CADe), and its related term computer-aided diagnosis (CADx), focuses on the detection or classification of different pathologies (e.g., cancer) based on the digital processing of images [13,25]. In the body of literature, a significant number of studies discuss the precision to detect or classify different kinds of pathologies or lesions [26,27,28,29,30,31,32,33]. For example, the authors in Reference [31] review several studies that use CAD to detect breast cancer. Recently, some of these studies have used ML in CAD to process digital images [25,30]; however, to the best of our knowledge, there are few studies in the use of CAD to detect FASD. We next discuss a study about the application of CAD in FASD.
The authors in Reference [34] use the Face2Gene software, which is based on the the facial dysmorphology novel analysis (FDNA) technology, to detect FASD through the analysis of digital images of children’s faces (5–9 years old). The authors conclude that Face2Gene achieves 88% of precision compared to 86% by the manual method (i.e., processed by a medical doctor). The approach followed by the study differs from our study because we do not use images; we process data obtained from several tasks applied to children (e.g., Psychometric task).
3. Materials and Methods
In this section, we analyze the data available and evaluate the performance of the ANN method and the configuration of the neural network with reasonable accuracy.
3.1. Data
The data used in this research is based on an open-access dataset collected by Zhang [15]. We use psychometric, saccadic eye movement, and DTI data from an open dataset collected and analyzed by Zhang et al. (2019) [15] to address the research question. This dataset contains children’s information, aged 3 to 18 years old, including subjects that are clinically diagnosed with or without FASD. The subjects are from five different communities in Canada. The data was obtained from a foundation (Kids Brain Health Network, formerly called NeuroDevNet). We compared results with those models using Support Vector Machine Regression (SVMR) and other methods. The data were divided into two groups: a control group and another that includes children who have been diagnosed with FASD. The FASD patient data consists of different neurological condition diagnoses within the FASD group, such as FAS, pFAS, and ARND.
Table 1 shows summary data for each of the test divided by FASD and Control. The data used describes the performance of children in solving particular task. Psychometric tests (NEPSY-II) [8] are tests designed regarding the psychosocial, intellectual behavior, memory, and performance of children. Saccadic eye movements is a simple method to infer structural or functional brain deficiencies, present in neurological disorders [35]. Ocular behavior, such as fixation and saccades, are crucial for efficient visual perception. Diffusion tensor image (DTI) is the only non-invasive method for characterizing the microstructural organization of brain tissue [36]. It measures the white matter connectivity in the corpus callosum in great detail with advanced diffusion magnetic resonance (MR) imaging schemes [37].

Table 1.
Data summary from all tests.
Prenatal alcohol exposure causes brain damage with serious potential neuropsychological consequences. These potential issues in cognitive functions include difficulties in planning, organization, and attention, consequential learning failures, and memory deficiencies. Some subjects have experienced speech and/or language difficulties, visuospatial functions, and spatial memory, that are increased by exposure to prenatal alcohol [38,39,40]. Those characteristics could be seen in the task and the evaluation. It is a vital diagnosis based on the abnormalities founded and not only in the traditional facial characteristic [41]. The novelty of the Zhang’s dataset [15] is that it includes numeric and image data. We used the psychometric tests, and it is noted that some of the characteristics present in the test correctly differentiate individuals with FASD and Control (see Figure 1a). These tests focuses on reading and math ability (Figure 1b). Moreover, studies have shown that children with FASD possess memory and functional problems [42]. For that reason, this data was chosen to make a preliminary algorithm for FASD detection in children.
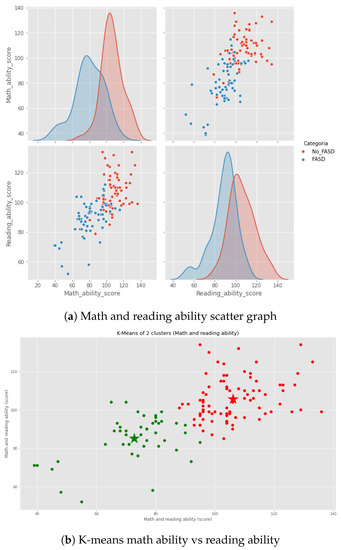
Figure 1.
Psychometric data analysis (characteristic math and reading ability).
This numerical data observes a deep learning operation, providing a novel approach compared to the existing image-based FASD diagnosis. One advantage of this approach is the data collection process is not invasive compared to the image-based data. In addition, it does not require us to know whether the mother consumed alcohol during pregnancy. The psychometric test measures the performance of the subject in skills as memory and mathematics, among others. The saccadic eye movement measures the response to a visual stimulus. All these tests would be made at any age and are not invasive nor expensive. Successful diagnosis could help the teacher and other education providers ensure environmental modification in schools and teach children skills to monitor and modify their behavior [38,39].
3.2. Data Analysis Using Machine Learning
Machine learning (ML) is a sub-discipline of artificial intelligence (AI). ML techniques focus on developing algorithms that are capable of learning or adapting to their structure based on observed data. This learning occurs when the optimization function adjusting the weights by calculating the gradient of the loss function. This is also known as the cost function, which tells us how good the model is [43]. Deep Learning within the ML methods framework have a theoretical foundation centered on classical neural networks. Unlike traditional neural networks, these have a group of hidden neurons and layers () (Figure 2, green square) [20,44]. The input data () passes through the layers in a non-linear combination of their outputs. Besides, it can approximate any arbitrary function through a learning process to a set of parameters in output projected from input space () [45,46]. The output of each neuron is described as a mathematical formula [47] (see Equation (1)), where are the weights without apathetic which the weight inputs, is the neuron activation function, and n is the number of neurons connected to the input (Figure 2, blue square).
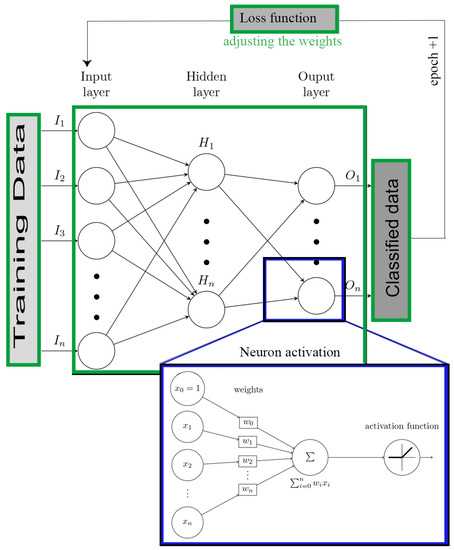
Figure 2.
ANN block diagram.
3.2.1. Neural Network Configuration Model
The first model used numerical data from the psychometric test. We have 20 numerical variables and two categorical variables (FASD and Control). Then, the Keras library of TensorFlow was used in Python to implement the algorithm [48]. In addition, we designed various models with dense layer connections, changing the number of neurons in each input layer, hidden layer, and output layer. Besides, the network was configured the optimization function with “Adam” and loss function “sparse categorical cross-entropy” to compile the model. Finally, we added two neurons in the output layer for classifying children with FASD or Control. We employed the dropout regularization, i.e., disabled neurons in each iteration to prevent neuron dependency to each other, thus avoiding overfitting. We added a dropout layer before each hidden layer with 20% of the neurons disable, to improve the data correlation independence of the neural network. For more information on the code, please refer to Appendix A.
Equation (2) shows the “Leaky ReLU” neurons activation function. This function improve the performance of the Rectified Lineal Unit (ReLU), widely used in ANN estimation. In this case, the psychometric data are not in the same range; some are between 1–10, and others of 70–100, and have 58 registers of children with FASD.
Our approach introduces the use of an ANN as improved technique for FASD diagnosis without the need of images. This ANN would require that the prediction model should have a improved performance compared to previous techniques without overfitting. To do this, we made variations in the number of hidden layers and neurons per layer to improve the model’s prediction and performance. We employed K-fold cross-validation [49] as criteria to choose each model and estimate their potential performance. The data used was split randomly into K-folds (K = 10) for each configuration of the net. We used nine folds for training and one for testing, repeating the process K times with different folds per time (group of data). The latter was used to validate the results of the K-fold cross-validation to estimate the best configuration of the network.
Table 2 shows layered network neuron configurations. The results are the average accuracy of ten evaluations, calculating the standard deviation, and average loss. We made a variation in each model in terms of the number of hidden layers and the number of neurons. Later, for each of the combinations, the accuracy was measured and compared by the accuracy metric and loss function. The loss function is the sum of errors made for each example in testing.

Table 2.
Network configuration for performance evaluation with psychometric data.
Our initial models showed a performance between 55% to 75.55%. Therefore, the cross-validation results was analyzed to shed light of each configuration’s performance. Combining 25 neurons in the input layer (IL) and only one hidden layer (HL) with 15 neurons (0.92) or 20 neurons (0.98) presents the lower values average of loss, meaning how well a model behaves after each iteration of optimization compares with others. Then, we evaluated the accuracy metric to measure the model’s performance. The combination of 25 (IL), 20 (HL) performed 77.37% average with the best performance confidence level of 66.28% to 88.46%. Due to its performance, this mode was selected to be used in the data analysis. We validated this model using same data to obtain the best performance, and it was possible to obtain a 75.55% accuracy compared to the real data.
3.2.2. Enhanced Neural Network Model
We changed the parameters to enhance the model, since the initial configuration only included psychometric. We reduced the number of training periods (number of epoch), loss function type, and batch size. We changed the model by using BinaryCrossentropy loss function, activation function ReLU, and Sigmoid and dropout of 10% between layers. The latter is a Dense feature layer with numerical columns to extract information from the input data. This feature layer allows the network to learn directly from those characteristics and propagate the features extracted from all the other layers [50]. The neural network architecture involves one input layer with the number of exploratory variables to be entered into the model, at least one hidden layer with 64 or 256 neurons per layer, and an output layer with only one neuron for binary classification (0 Control and 1 FASD).
Our initial estimation used a simple approach of splitting the data into three chunks: train, validation, and test sets. However, this technique does not allow us to evaluate the performance thoroughly, especially when the amount of data is not enough (roughly less than 100). We used a K-fold cross-validation technique with K = 10 to corroborate the model’s performance on data not used during training (Table 3). First, we improved the psychometric-based model by including a training period of 50 epochs with four hidden layers with 64 and 128 neurons per layer and a batch size of 32 (see Appendix A). Following this enhancement, we created a new model based on eye movement tests, including data regarding sex and age, to analyze the influence of these characteristics on the likelihood to predict FASD.

Table 3.
K-fold cross-validation of models.
We used the same model configuration and replaced the data with the antisaccades and memory-guided saccadic tests. In the prosaccade test data, the model was trained using 100 epochs with only two hidden layers with 64 and 128 neurons. All the models mentioned before used the activation function rectified linear unit (ReLU) and Sigmoid [51]. DTI was a particular case because we do not have enough data. Therefore, we used two hidden layers with 128 neurons, activation function Leaky ReLu, and training for 200 epochs, in this particular case, and a batch size of 16.
The model’s prediction power depends on the data using for training and testing. Therefore, we shuffle the dataset once splitting the dataset into K folds to avoid any issues with the learning process. Exploratory analysis shows that the data is imbalanced in prosaccades, antisaccades, and memory-guided saccades. This issue may lead to unexpected mistakes and severe consequences in the data analysis, primarily to predict children with FASD. The skewed distribution of FASD and control may force the models described above to obtain biased outcomes, especially in this case of the control. This issue would delay the best treatment selection for children with FASD. Whether or not the subject constitutes the minority class, the algorithm classifies the children as healthy. To overcome this issue, we balanced the data by dismissing some control data observations, resulting in a more balanced dataset.
Table 3 shows model’s loss, accuracy, and Mean Absolute Error (MAE) for benchmarking comparison. The average MAE in all the models is below 0.4, which indicates the generalization capacity and that the model avoided the overfitting. The models perform better with less epochs than the first model; a possible justification is using the batch size and feature dense layer. Almost all accuracy levels are over 50%, and the value of the losses are below one on average. After the analysis, we used the model’s weight to show better performance in that fold for further analysis.
4. Results
This research evaluated the performance of a ANN-based classification algorithm to detect FASD in children using the result of psychometric, DTI, and saccade tests and comparing the accuracy of the model with the SVMR developed in Reference [15]. The cross-validation process showed that the psychometric data model outperformed the rest. After this process, we saved the best model weight for each of the different data, and then we estimated the accuracy with all the data available. Table 4 shows the the accuracy level for each of the data type and its comparison to results obtained by Zhang’s model.

Table 4.
Accuracy model performance comparison.
In the next subsection, the results of each (Psycometric, Antisaccad, Prosaccade, and DTI) of the tests are shown and discussed.
4.1. Psychometric Data Classification
The data used for testing included psychometric tests (20 input features), associated with analyzing social behavior, memory activities, language delay, and all altered behavioral factors in FASD children. In addition, the dataset included other developmental diseases, making the classification process more difficult. This paper used two models of dense networks using this type of data. The first model archived an accuracy of 75.55%. However, the model did not show significant improvements, despite the variation in the layers and neurons in the layers. In the second model, a feature layer was added, achieving an accuracy of 88.46%.
Once the network configuration was chosen, training and testing or validation behavior are shown in the model precision and loss functions. The loss function with a high result indicates that the neural network has a poor performance and a low result, and that it is doing a good job. Figure 3a shows the accuracy for each of the dataset. In terms of the number of that were aspects found and related to the number of individuals evaluated. We attempted to find a model with no significant difference between the labeled data and the prediction. Nonetheless, with the implemented configuration, the training data increased accuracy. In addition, the validation data also increased in accuracy.
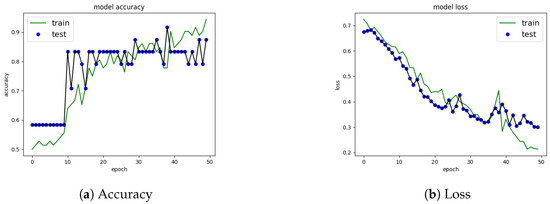
Figure 3.
Precision and Loss using feature dense layer on psychometric data.
Figure 3a shows the loss function on the psychometric data. In this case, the loss function has a decreasing curve in both training and testing. This result suggests that the network may have issues classifying some data not used during training but not significantly.
The confusion matrix (Figure 4) shows the performance of the model with test data. The model has a good performance in classifying children with FASD since the correctness is 38.46% over 42.31% in positive hits (FASD having FASD), representing 90.9%, and it has 7.69% over 42.31% false positives (a control having FASD). It should be noted that the total percentage of children diagnosed with FASD is 42.31%. The model allows classifying FASD in most cases. In classifying control patients, the model classifies 51.0% over 57.69% of control patients who are really in control (86.67% of the cases control). And it fails in 3.85% to classify patients with FASD as controls (13.32% mistake of cases control).
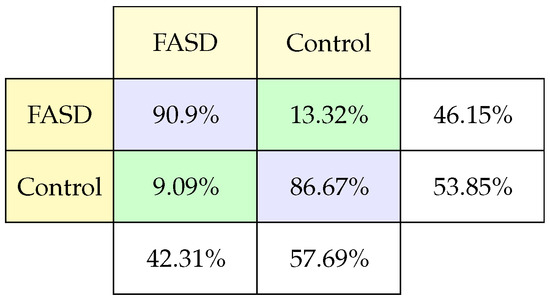
Figure 4.
Classification confusion matrix using psychometric test.
4.2. Antisaccade Data Classification
The results from the Antisaccade tasks correspond to measurements of successes between the number of tests achieved by each individual, where the failures in the automatic saccade inhibition with a peripheral objective are measured. In this case, initially, there were 173 records, but we evaluated the model with 136 records. The characteristics of the Antisaccadic test are: 15 include correct and wrong trails, number of express saccades, the interval between the target appearance, and saccade onset, velocity, amplitude, angle, and other measurements derived from those.
The confusion matrix (Figure 5) results show a high rate of prediction of classifying FASD compared to the amount of data evaluated (100%). However, some of the classification characteristics tend to confuse control children with the possibility of having FASD. These results make it necessary to evaluate the degree of FASD these children have and the reason for this classification (33.33% subject control were classified FASD).
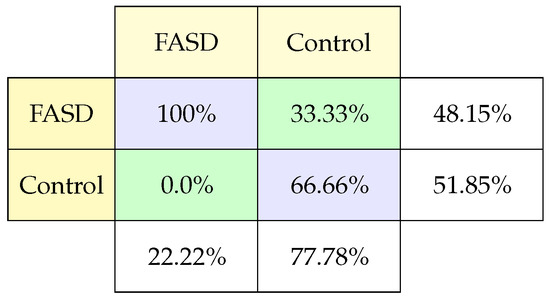
Figure 5.
Classification confusion matrix using Antisaccade task.
4.3. Prosaccade Task Data Classification
Prosaccade tests measure reaction time, performance accuracy, viability, and parameters which corresponding to the main sequence. In this particular case, 18 characteristics were measured in the study, and the data collected is similar to the antisaccades test but include duration, deceleration, acceleration, and skew. For the classification of FASD patients with these data from the 186 records, only 38% correspond to patients were diagnosed with FASD. This study achieved an accuracy of 72.41%.
The confusion matrix (Figure 6) for this case shows that the model predicts FASD patients well, but only in 50% of the cases control patients being controls. This confusion matrix allows us to observe that we just need to make some adjustments to improve the exactness to predict control patients or verify the data. We should better understand the present differentiating characteristics from control patients to adequately identified during training reflected in the test data.
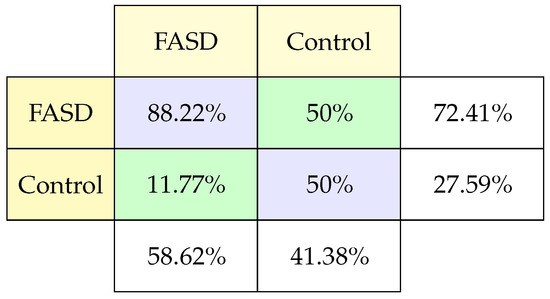
Figure 6.
Classification confusion matrix using Prosaccade task.
4.4. Memory-Guided Saccade Task Classification Results
These tasks are related to following the objective order. When there are errors, the subject cannot follow the order that initiates the saccade by the second objective more than on the first. In these tasks, 26 characteristics are measured: correct trial, times between movement and reactions, errors, distance, speed, and others [15]. We took 61 records by classification, achieving 88% in test data, one of the highest in the study.
The confusion matrix (Figure 7) for this model shows a balanced measurement of FASD and control patients greater than 80% per case. And, in the case of FASD, very few mistakes. It is important to note that the studies that achieved the best results have an element of the individual’s memory. This result makes us think that we must pay more attention to this type of test, including memory issues.
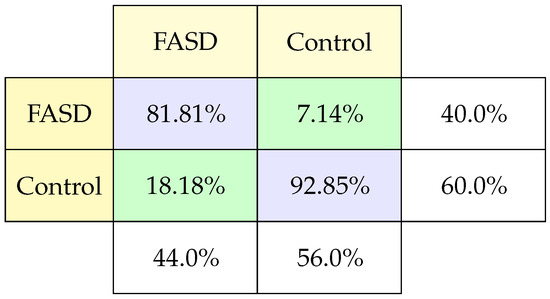
Figure 7.
Classification confusion matrix using Memory-guide saccadic task.
4.5. Diffusion Tensor Imaging (DTI) Data Classification Results
This study measures the connectivity of the white matter in the corpus callosum. It comes from a structural MRI, so it does not involve tests. This study has 76.54% records classified as FASD and 46% control. Having little data means that this study did not achieve a good accuracy (less than 50%). The Leaky-ReLU function was added in the intermediate layers for improvement, managing to rise to 75% in a combination of 4 hidden layers of 64 and 128 neurons, a feature layer, and the training time was 100 epochs compared to the other models with 50 epochs training.
The confusion matrix (Figure 8) shows that the percentage of failures to classify FASD and control is the same. And, concerning the percentage of samples of each type, we observed a good performance.
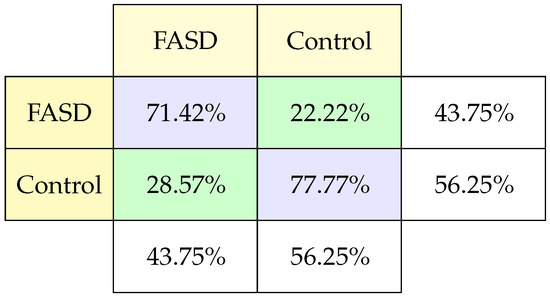
Figure 8.
Classification confusion matrix using DTI.
5. Discussion
This paper evaluated the usage of computational algorithms based on ANN to classify children with FASD for an improved medical diagnosis. This paper estimated the prediction model using numerical psychometric data, DTI, and saccadic eye movement. Unlike most of the current research that implements ML using image-based data to predict FASD, the novelty of this paper is that it uses numerical data to estimate the model. Although Zhang et al. (2019) [15] collected and used the same data, our results show a better performance using ANN instead of SVMR or other methods.
First, for psychometric data, a basic network configuration was obtained with correctness with test data of 75.5% with a Leaky-ReLU activation function, which did not require data normalization. Subsequently, a change was made in the configuration of the networks adding a feature layer. This layer allows identifying the essential characteristics for FASD diagnosis. This feature significantly contributes to the classification, achieving 88% accuracy. It also produced the better results using the K-Fold Cross validation (K = 10) and K-Fold Cross-Validation (K = 5), suggesting that this should be the direction for improving FASD diagnosis using ANN. For the rest of the data, a network configuration with a feature layer was used to evaluate the neural network model performance. Using numerical origin (Saccade, Prosaccade, and DTI) data related to FASD, models achieved an accuracy greater than 70%. Despite this higher rate obtained by all of these configurations, it is not yet considered to be a safe prediction for a medical diagnoses.
The confusion matrices obtained demonstrate that the model predicts at least 71% of patients diagnosed with FASD, from the around 50% that this group represents. However, the prediction of control patients has over 77% of patients who have not been diagnosed with FASD (except Prosaccadic task). These matrices suggest that the model must be recalibrated or overlearned from the data of patients with FASD, or else some control patients may have FASD but have not been diagnosed. These results are not completely demonstrated; thus, the use of ANN as a diagnostic tool must be improved before its use can be suggested.
Results show that the estimated models present a better performance in all cases compared to the results obtained by Zhang’s research. Overall, the use of ANN outperformed by 14.22% (S.D. 10.58) the results obtained by Zhang’s model. Unlike the model obtained by Zhang, all the characteristics obtained from each study are included in the estimated model. This is a distinctive feature since Zhang removed some characteristics due to their view that they did not improve the prediction’s performance. Therefore, our results show that the model indeed improved by including data from sex and age. In addition, we used a feature layer that allows the algorithm to distinguish those characteristics, which contributes to improving pathology’s prediction. In addition, the estimated model shows that, by eliminating sex and age from the studies, the prediction decreased dramatically. Zhang used these variable to normalize the data in Saccade, Antisaccade, and MGsaccade, to increase model’s accuracy from an average of 60% to 70%. The latter is a key feature, since our estimated model uses raw data, keeping the estimation process simple.
Zhang’s model utilized methods, especially SVMR, which require fewer parameters to optimize its performance, reducing the possibility of over-tuning training data, and increasing actual performance. According to the training data results, unlike ANN, the study of different configurations can change its operation and performance in general. The increase in data allows ANN to improve its performance, without changing parameters. Nevertheless, a new training improves the ANN model accuracy, unlike in the case of SVMR. However, SVMR is faster and more stable than other ANNs. Therefore, implementing an ML model depends on the change in data or other characteristics rather than on the accuracy of each one.
One caveat is the occurrence of the false-positive, which is to diagnose a person with FASD that does not have it, since the data between FASD and control patients can be similar. It is essential to mention that some patients are not correctly diagnosed with FASD or have some cognitive problems similar to FASD, which can also lead to many false negatives. This result is because collecting data on alcohol consumption during pregnancy is not accessible. Nevertheless, it requires the mother’s declaration about the consumption. Likewise, over time, the early samples of FASD decrease and are dependent on the time of alcohol exposure. Furthermore, the phase of gestation during which the ingestion occurred and the amount of it. Some patients who present neuropsychological and developmental deficits (between groups diagnosed with FASD and other pathology) do not show significant differences, disabilities, nor behavioral problems. Therefore, the diagnosis of FASD cannot be made lightly and should be considered aside other results, such as magnetic resonance imaging and eye movement, among others.
Future work should be done on using these types of ANN to predict other deceases or conditions that have cognitive development issues, such as Autism or ADHD. For instance, some cases of autism are classified by FASD; therefore, the treatment is not the correct. The introduction of the ANN would allow to a better diagnosis, thus improving their quality of life. The data is currently limited and the testing process is in early stages. To our knowledge, there are no studies that have gone in this direction.
6. Conclusions
There is currently an underestimation of people with FASD. This syndrome affects a significant percentage of the world population. However, its diagnosis requires a certain consumption during embryonic development. It could be undiagnosed in the control population due to the clinical similarities with other neuro-development diseases. This issue is a critical factor for developing a new technique for FASD diagnosis since image-based Artificial Neural Networks (ANNs) and Computer-Aided Detection tools (CADs) studies are invasive and cannot be repeated. For this reason, the use of numerical data from non-invasive tests can be an excellent alternative to diagnose FASD. This paper developed ANN algorithms to determine FASD likelihood in children using numerical psychometric, DTI, and Saccadic eye movement data on children/young people diagnosed with FASD. It is essential to mention that the best results were obtained using psychometric data. It is crucial to carry out a more in-depth study of this test and even incorporate data from children with a neuro-behavioral disorder that helps to classify better and identify some syndromes.
In conclusion, we suggest that ANN algorithms improve their performance with a suitable network configuration by increasing the number of neurons, hidden layers, changing optimization functions, adding a feature layer, and the activation algorithm used to incorporate new data into training. The combination with other data groups even allows us to increase its performance, indicating that ANNs can be a good alternative to detect FASD. This paper evaluated the usage of computational algorithms based on ANN to classify children with FASD for an improved medical diagnosis. As a result, this paper estimated the prediction model using numerical psychometric data, DTI, and saccadic eye movement. Although other studies, like Zhang et al. (2019) [15], collect and use the same data, our results show a better performance using ANN instead of SVMR or other methods. However, a deeper study of the different network configurations must be undertaken to improve performance.
Author Contributions
All authors contributed to this study. Conceptualization, V.D., P.L., S.C.; Algorithm, V.D., P.L., S.C. and H.F.; validation, V.D., P.L., and S.C.; writing—original draft preparation, V.D., P.L., S.C.; writing—V.D., S.C., and P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to express our great appreciation to Gregory Stanton for his valuable and constructive suggestions to this research project.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Listing A1.
ANN Used in This Study.
Listing A1.
ANN Used in This Study.
optimizer=Adam( )
loss_function= sparse_categorical_crossentropy
num_classes=2
no_epoch=1000
verbosity = 0
#ANN model
model = Sequential ( )
model.add (Dense (200, activation=LeakyReLU(alpha =0.2),
input_shape = [20]))
model.add (Dropout (0.2))
model.add (Dense (15, activation=LeakyReLU(alpha =0.2)))
model.add (Dropout (0.2))
model.add (Dense (num_classes, activation= tf.nn.softmax))
#compile the model
model.compile (opt imizer=optimizer ,
loss=loss_function ,
metrics=[’accuracy’])
# Fit data to model
X_train=X.iloc[train ,:]
y_train=y.iloc[train]
history= model.fit(X_train ,y_train ,epochs=no_epoch,
verbose=verbosity)
# generate generalization metrics
X_test = X.iloc[test , :]
y_test = y.iloc[test]
scores =model.evaluate(X_test,y_test,verbose=0)
predict=model.predict(X_test)
|

Listing A2.
Model Configuration with a Feature Layer.
Listing A2.
Model Configuration with a Feature Layer.
for header in [ ’ f1 ’ , ’ f2 ’ , ’ f3 ’ , ’ f4 ’ , ’ f5 ’ , ’ f6 ’ , ’ f7 ’ , ’ f8 ’ , ’ f9 ’ , ’ f10 ’ , ’ f11 ’ ,
’ f12 ’ , ’ f13 ’ , ’ f14 ’ , ’ f15 ’ , ’ f16 ’ , ’ f17 ’ , ’ f18 ’ , ’ f19 ’ , ’ f20 ’ ] :
feature_columns.append(feature_column.numeric_column(header))
feature_layer = tf.keras.layers.DenseFeatures(feature_columns)
batch_size = 18
train_ds = df _ to_dataset(train , batch_size=batch_size)
val_ds = df_to_dataset(val , shuffle=False , batch_size=batch_size)
test_ds = df_to_dataset(test , shuffle=False , batch_ size=batch_size)
train_ds
model = tf.keras.Sequential([
feature_layer ,
layers.Dense(64, activation=’relu’),
layers.Dropout(.1) ,
layers.Dense(128, activation=’sigmoid’),
layers.Dropout(.1),
layers.Dense(64, activation=’sigmoid’),
layers.Dropout(.1),
layers.Dense(128, activation=’relu’),
layers.Dropout(.1),
layers.Dense(1)
])
model.compile(optimizer=’adam’,
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=[’accuracy’])
history=model.fit(train_ds,validation_data=val_ds,epochs=50)
loss, accuracy = model.evaluate(test_ds)
|
References
- O’Leary, C.; Bower, C. Guidelines for pregnancy: What’s an acceptable risk, and how is the evidence (finally) shaping up? Drug Alcohol. Rev. 2012, 31, 170–183. [Google Scholar] [CrossRef] [PubMed]
- McCallum, K.; Holland, K. ‘To drink or not to drink’: Media framing of evidence and debate about alcohol consumption in pregnancy. Crit. Public Health 2018, 28, 412–423. [Google Scholar] [CrossRef]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 843–852. [Google Scholar]
- Cook, J.L.; Green, C.R.; Lilley, C.M.; Anderson, S.M.; Baldwin, M.E.; Chudley, A.E.; Conry, J.L.; LeBlanc, N.; Loock, C.A.; Lutke, J.; et al. Fetal alcohol spectrum disorder: A guideline for diagnosis across the lifespan. CMAJ 2016, 188, 191–197. [Google Scholar] [CrossRef]
- Chudley, A.E. Diagnosis of fetal alcohol spectrum disorder: Current practices and future considerations. Biochem. Cell Biol. 2018, 96, 231–236. [Google Scholar] [CrossRef]
- Popova, S.; Lange, S.; Chudley, A.E.; Reynolds, J.N.; Rehm, J.; May, P.; Riley, E. World Health Organization International Study on the Prevalence of Fetal Alcohol Spectrum Disorder (FASD). Cent. Addit. Ment. Health. Available online: www.camh.ca (accessed on 1 May 2021).
- Popova, S.; Lange, S.; Probst, C.; Gmel, G.; Rehm, J. Estimation of national, regional, and global prevalence of alcohol use during pregnancy and fetal alcohol syndrome: A systematic review and meta-analysis. Lancet Glob. Health 2017, 5, e290–e299. [Google Scholar] [CrossRef]
- Paolozza, A.; Treit, S.; Beaulieu, C.; Reynolds, J.N. Diffusion tensor imaging of white matter and correlates to eye movement control and psychometric testing in children with prenatal alcohol exposure. Hum. Brain Mapp. 2016, 38, 444–456. [Google Scholar] [CrossRef] [PubMed]
- May, P.A.; Chambers, C.D.; Kalberg, W.O.; Zellner, J.; Feldman, H.; Buckley, D.; Kopald, D.; Hasken, J.M.; Xu, R.; Honerkamp-Smith, G.; et al. Prevalence of Fetal Alcohol Spectrum Disorders in 4 US Communities. JAMA 2018, 319, 474. [Google Scholar] [CrossRef] [PubMed]
- El Naqa, I.; Murphy, M.J. What is machine learning? In Machine Learning in Radiation Oncology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 3–11. [Google Scholar]
- Mohri, M.; Rostamizadeh, A.; Talwalkar, A. Foundations of Machine Learning; MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef]
- Ronald, C. Computer Aided Detection (CAD): An Overview. Cancer Imaging Off. Publ. Int. Cancer Imaging Soc. 2005, 5, 17–19. [Google Scholar]
- Rodriguez, C.I.; Vergara, V.M.; Davies, S.; Calhoun, V.D.; Savage, D.D.; Hamilton, D.A. Detection of prenatal alcohol exposure using machine learning classification of resting-state functional network connectivity data. Alcohol 2021, 93, 25–34. [Google Scholar] [CrossRef]
- Zhang, C.; Paolozza, A.; Tseng, P.H.; Reynolds, J.N.; Munoz, D.P.; Itti, L. Detection of Children/Youth With Fetal Alcohol Spectrum Disorder Through Eye Movement, Psychometric, and Neuroimaging Data. Front. Neurol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Liu, Y.; Huang, J.; Vinci-Booher, S.; Anthony, B.; Zhou, F. Facial Image Classification of Mouse Embryos for the Animal Model Study of Fetal Alcohol Syndrome. Proc. Symp. Appl. Comput. 2009, 2009, 852–856. [Google Scholar] [PubMed]
- Huang, J.; Jain, A.; Fang, S.; Riley, E. Using facial images to diagnose fetal alcohol syndrome (FAS). In Proceedings of the International Conference on Information Technology: Coding and Computing (ITCC’05)—Volume II; IEEE: Las Vegas, NV, USA, 2005; pp. 66–71. [Google Scholar]
- Kim, S.Y.; Moon, S.K.; Jung, D.C.; Hwang, S.I.; Sung, C.K.; Cho, J.Y.; Kim, S.H.; Lee, J.; Lee, H.J. Pre-operative prediction of advanced prostatic cancer using clinical decision support systems: Accuracy comparison between support vector machine and artificial neural network. Korean J. Radiol. 2011, 12, 588–594. [Google Scholar] [CrossRef]
- Li, Y.; Huang, C.; Ding, L.; Li, Z.; Pan, Y.; Gao, X. Deep learning in bioinformatics: Introduction, application, and perspective in the big data era. Methods 2019, 166, 4–21. [Google Scholar] [CrossRef] [PubMed]
- Ravi, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.Z. Deep Learning for Health Informatics. IEEE J. Biomed. Health Inform. 2017, 21, 4–21. [Google Scholar] [CrossRef]
- Al-Shayea, Q.K. Artificial neural networks in medical diagnosis. Int. J. Comput. Sci. Issues 2011, 8, 150–154. [Google Scholar]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Wozniak, J.R.; Muetzel, R.L. What does diffusion tensor imaging reveal about the brain and cognition in fetal alcohol spectrum disorders? Neuropsychol. Rev. 2011, 21, 133–147. [Google Scholar] [CrossRef]
- Little, G.; Beaulieu, C. Multivariate models of brain volume for identification of children and adolescents with fetal alcohol spectrum disorder. Hum. Brain Mapp. 2020, 41, 1181–1194. [Google Scholar] [CrossRef]
- Gao, Y.; Geras, K.J.; Lewin, A.A.; Moy, L. New Frontiers: An Update on Computer-Aided Diagnosis for Breast Imaging in the Age of Artificial Intelligence. AJR Am. J. Roentgenol. 2019, 212, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Massari, L.; Bulletti, A.; Prasanna, S.; Mazzoni, M.; Frosini, F.; Vicari, E.; Pantano, M.; Staderini, F.; Ciuti, G.; Cianchi, F.; et al. A Mechatronic Platform for Computer Aided Detection of Nodules in Anatomopathological Analyses via Stiffness and Ultrasound Measurements. Sensors 2019, 19, 2512. [Google Scholar] [CrossRef]
- Niemeijer, M.; Loog, M.; Abràmoff, M.D.; Viergever, M.A.; Prokop, M.; van Ginneken, B. On Combining Computer-Aided Detection Systems. IEEE Trans. Med. Imaging 2011, 30, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, R.M. Computer-aided Detection and Diagnosis. In Digital Mammography; Bick, U., Diekmann, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 85–106. [Google Scholar]
- Cicerone, M.T.; Camp, C.H. Potential Roles for Spectroscopic Coherent Raman Imaging for Histopathology and Biomedicine. In Neurophotonics and Biomedical Spectroscopy; Alfano, R.R., Shi, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 547–570. [Google Scholar]
- Ben-Cohen, A.; Greenspan, H. Liver lesion detection in CT using deep learning techniques. In Handbook of Medical Image Computing and Computer Assisted Intervention; Zhou, S.K., Rueckert, D., Fichtinger, G., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 65–90. [Google Scholar]
- Katzen, J.; Dodelzon, K. A review of computer aided detection in mammography. Clin. Imaging 2018, 52, 305–309. [Google Scholar] [CrossRef]
- Suzuki, K. A supervised ’lesion-enhancement’ filter by use of a massive-training artificial neural network (MTANN) in computer-aided diagnosis (CAD). Phys. Med. Biol. 2019, 54, s31–s45. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muhammad Ashfaq Khan, Y.K. Cardiac Arrhythmia Disease Classification Using LSTM Deep Learning Approach. Comput. Mater. Contin. 2021, 67, 427–443. [Google Scholar] [CrossRef]
- Valentine, M.; Bihm, D.C.; Wolf, L.; Hoyme, H.E.; May, P.A.; Buckley, D.; Kalberg, W.; Abdul-Rahman, O.A. Computer-Aided Recognition of Facial Attributes for Fetal Alcohol Spectrum Disorders. Pediatrics 2017, 140, e20162028. [Google Scholar] [CrossRef] [PubMed]
- Green, C.R.; Lebel, C.; Rasmussen, C.; Beaulieu, C.; Reynolds, J.N. Diffusion Tensor Imaging Correlates of Saccadic Reaction Time in Children with Fetal Alcohol Spectrum Disorder. Alcohol. Clin. Exp. Res. 2013, 37, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.K.; Leemans, A. Diffusion Tensor Imaging. In Magnetic Resonance Neuroimaging: Methods and Protocols; Methods in Molecular Biology; Modo, M., Bulte, J.W., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 127–144. [Google Scholar]
- Hagmann, P.; Jonasson, L.; Maeder, P.; Thiran, J.P.; Wedeen, V.J.; Meuli, R. Understanding Diffusion MR Imaging Techniques: From Scalar Diffusion-weighted Imaging to Diffusion Tensor Imaging and Beyond. RadioGraphics 2006, 26, S205–S223. [Google Scholar] [CrossRef]
- Glass, L.; Moore, E.M.; Akshoomoff, N.; Jones, K.L.; Riley, E.P.; Mattson, S.N. Academic Difficulties in Children with Prenatal Alcohol Exposure: Presence, Profile, and Neural Correlates. Alcohol. Clin. Exp. Res. 2017, 41, 1024–1034. [Google Scholar] [CrossRef]
- Green, J.H. Fetal Alcohol Spectrum Disorders: Understanding the Effects of Prenatal Alcohol Exposure and Supporting Students. J. Sch. Health 2007, 77, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, S.; Page, S.J.; Wang, L.; Ishii, S.; Li, P.; Sasaki, T.; Basha, A.; Salzberg, A.; Quezado, Z.; Imamura, F.; et al. Kcnn2 blockade reverses learning deficits in a mouse model of fetal alcohol spectrum disorders. Nat. Neurosci. 2020, 23, 533–543. [Google Scholar] [CrossRef]
- Wozniak, J.R.; Muetzel, R.L.; Mueller, B.A.; McGee, C.L.; Freerks, M.A.; Ward, E.E.; Nelson, M.L.; Chang, P.N.; Lim, K.O. Microstructural Corpus Callosum Anomalies in Children With Prenatal Alcohol Exposure: An Extension of Previous Diffusion Tensor Imaging Findings. Alcohol. Clin. Exp. Res. 2009, 33, 1825–1835. [Google Scholar] [CrossRef]
- Rasmussen, C. Executive Functioning and Working Memory in Fetal Alcohol Spectrum Disorder. Alcohol. Clin. Exp. Res. 2005, 29, 1359–1367. [Google Scholar] [CrossRef]
- Sajda, P. Machine Learning for Detection and Diagnosis of Disease. Annu. Rev. Biomed. Eng. 2006, 8, 537–565. [Google Scholar] [CrossRef]
- Jo, T.; Nho, K.; Saykin, A.J. Deep Learning in Alzheimer’s Disease: Diagnostic Classification and Prognostic Prediction Using Neuroimaging Data. Front. Aging Neurosci. 2019, 11, 220. [Google Scholar] [CrossRef]
- Shukla, P.; Gupta, T.; Saini, A.; Singh, P.; Balasubramanian, R. A Deep Learning Frame-Work for Recognizing Developmental Disorders. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; pp. 705–714. [Google Scholar]
- Deng, L.; Hinton, G.; Kingsbury, B. New types of deep neural network learning for speech recognition and related applications: An overview. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–30 May 2013; pp. 8599–8603. [Google Scholar]
- Villada, F.; Muñoz, N.; García-Quintero, E. Redes Neuronales Artificiales aplicadas a la Predicción del Precio del Oro. Informa. Tecnol. 2016, 27, 143–150. [Google Scholar] [CrossRef]
- Ketkar, N. Deep Learning with Python: A Hands-On Introduction; Apress: New York, NY, USA, 2017. [Google Scholar]
- Stone, M. Cross-Validatory Choice and Assessment of Statistical Predictions. J. R. Stat. Soc. 1974, 36, 111–147. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, C.; Luo, B.; Tang, J.; Wang, X. Dense Feature Aggregation and Pruning for RGBT Tracking. In Proceedings of the 27th ACM International Conference on Multimedia, MM ’19, Nice, France, 21–25 October 2019; Association for Computing Machinery: New York, NY, USA, 2019; pp. 465–472. [Google Scholar]
- Paneiro, G.; Rafael, M. Artificial neural network with a cross-validation approach to blast-induced ground vibration propagation modeling. Undergr. Space 2021, 6, 281–289. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).