Abstract
Nineteen tosylated acyl hydrazone derivatives were synthesized, and their inhibitory activities against monoamine oxidases (MAOs), acetylcholinesterase (AChE), butyrylcholinesterase (BChE), and β-secretase (BACE-1) were evaluated. Compound 3o was the most potent inhibitor of MAO-A, with an IC50 value of 1.54 µM, followed by 3a (IC50 = 3.35 µM). A structural comparison with 3a indicated that the 3-F group in 3o increased its inhibitory activity against MAO-A. Compound 3s was the most potent inhibitor of MAO-B, with an IC50 value of 3.64 µM, followed by 3t (IC50 = 5.69 µM). The MAO-B inhibitory activity increased in the order of 3- > 4- > 2-NO2 groups in 3s, 3t, and 3r, respectively. All the compounds weakly inhibited AChE and BChE, which retained >50% residual activity at 10 µM, except for 3a, which inhibited BChE with an IC50 value of 16.1 µM. Interestingly, 3e, 3f, and 3n inhibited BACE-1 with IC50 values of 8.63, 9.92, and 8.47 µM, respectively, which were lower than the IC50 of the quercetin reference. Compounds 3o and 3s were found to be reversible competitive inhibitors of MAO-A and MAO-B, respectively, with Ki values of 0.35 ± 0.074 and 1.97 ± 0.65 µM, respectively. Moreover, compounds 3e, 3f, and 3n were effective BACE-1 inhibitors. The lead molecules were further investigated by molecular docking studies to elucidate the binding interactions with the target enzymes.
1. Introduction
Dementia is a serious disease that predominantly affects aging patients, especially the elderly population. Alzheimer’s disease (AD) is one of the most common causes of dementia and is related to the loss of cognitive functions. Although the mechanism of AD pathogenesis is not fully understood, AD is principally caused by low acetylcholine (ACh) levels, oxidative stress, and β-amyloid plaque deposits [1]. Numerous studies have reported that cognitive functions are related to the cholinesterase (ChE) system, by breaking down the neurotransmitter ACh, and, therefore, ChE inhibition is effective for the treatment of AD [2]. ChE inhibitors suppress the degradation of this chemical messenger in the brain, which is important for memory and learning [3]. The cholinergic hypothesis suggests that the role played by ChE inhibitors in AD is related to neurotransmitter transfer [4].
AD is associated with decreases in neurotransmitter, particularly ACh, and significant increases in acetylcholinesterase (AChE) and butyrylcholinesterase (BChE) levels [5]. In addition, monoamine oxidase (MAO) is responsible for the oxidative deamination of endogenous amines such as serotonin, epinephrine, and other neurotransmitters [6]. The amount of MAO increases with aging and neuronal damage, suggesting that MAO inhibition could be effective in treating AD. Importantly, oxidative stress, associated with free radicals and tissue damage, creates an obstacle in the repair of cognitive functions [7]. On the other hand, β-secretase (β-site amyloid precursor protein cleaving enzyme 1, BACE-1) is a protease that catalyzes the production and deposition of amyloid-β peptide, and it has been considered a promising target for the treatment of AD [8].
Drugs approved by the FDA for the treatment of AD are AChE inhibitors (donepezil, rivastigmine, and galantamine) and N-methyl-D-aspartate receptor antagonists (memantine). However, these drugs are not able to stop the progression of AD and only alleviate the symptoms; therefore, further studies are needed in the field of medicinal chemistry to develop agents, which are more effective and show less side effects than the drugs currently used [1].
We examined the structures and possible interactions with AChE and planned to provide the amine group required for enzyme interactions with hydrazide–hydrazone moiety [1]. Therefore, we designed the compounds by taking advantage of the possible activities of parabens, hydrazide hydrazone moieties.
Parabens are widely used as preservatives in the food, cosmetic, and pharmaceutical industries. They have low toxicity and effective antimicrobial activity [9], and the most frequently used are methyl-/ethyl-/propyl-paraben [10]. Hydrazides are remarkably effective in the field of pharmaceutical chemistry because they engage in many types of reactions to form different derivatives possessing diverse pharmacological profiles, including antiviral [11], antibacterial [12], antimalarial [13], and antitumor [14] activities. These activities highlight the importance of these compounds and derivatives in drug design.
Hydrazones carrying an azomethine structure constitute an important class of compounds and are target molecules for various biological activities [15]. The hydrazide–hydrazone structure is known to have antimycobacterial, antibacterial, antifungal, and carbonic anhydrase inhibition activities [16,17,18,19,20]. With their wide range of activities, these compounds have been recognized as important building blocks in medicinal chemistry studies.
In this study, hydrazide derivatives of ethyl paraben were synthesized in reactions with various benzaldehydes, resulting in 19 tosylated acyl hydrazide derivatives (3a–t), and their potential biological activities were evaluated by analyzing their inhibitory activities against MAOs, AChE, BChE, and BACE-1 enzymes, as well as their antioxidant activities. In addition, the structure–activity relationship between tosylated acyl hydrazide derivatives and enzymes was characterized, and inhibition kinetic experiments and docking simulations were performed.
2. Materials and Methods
2.1. Chemistry
All reactions were monitored by analytical thin layer chromatography using Merck pre-coated silica gel plates with F254 indicator. Melting points were determined using a Thermoscientific 9200 capillary melting point apparatus (Philadelphia, PA, USA). 1H and 13C NMR spectra were recorded on a Varian Mercury 400 Digital FT-NMR (400 MHz) instrument with tetramethylsilane as an internal standard in DMSO-d6 solutions. Chemical shifts (δ) are reported in parts per million (ppm) and coupling constants (J) in Hertz with multiplicities, described as s = singlet, d = doublet, t = triplet, q = quartet, and m = multiplet. Mass spectrometry was conducted using a Micromass ZQ LC-MS spectrometer (ESI-mode) connected with Waters Alliance HPLC.
The purities of the compounds were evaluated by Prominence series of a high-pressure liquid chromatography (HPLC) system with photodiode array detector (DAD). The chromatographic conditions were as follows: column, Raptor ARC-18 (100 × 4.6 mm, 2.7 µm); mobile phase, 0.1% formic acid/methanol mixture (35:65, v/v); flow rate, 0.8 mL/min; oven temperature, 25 °C, injection volume, 10 µL. Column eluate was monitored at 254 nm. Working solutions of target compounds were prepared at different concentrations.
Compound 3a was registered in ScienceFinder with CAS number 889795-07-7 and was synthesized here with this method. The chemical structures of the compounds were established by using 1H- and 13C-NMR, and mass analysis spectroscopic methods, and their purities were determined with HPLC (Supplementary Materials Tables S1–S76).
NOE spectra were recorded to identify the isomers obtained, and the compounds were E isomers (Figure 1).
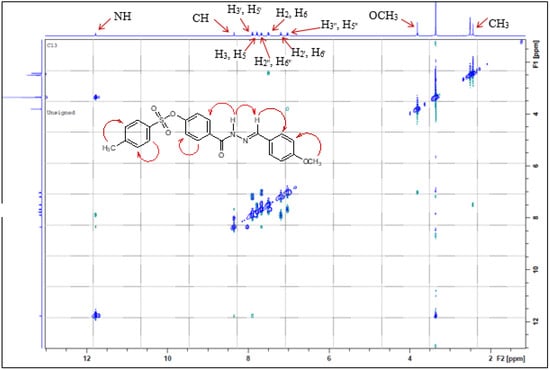
Figure 1.
NOE spectra for compound 3g.
Synthesis of Ethyl Paraben Hydrazide (1). Hydrazine hydrate (5.8 mL) was added portion-wise to a solution of p-hydroxybenzoic acid ethyl ester (0.02 mol) in ethanol (25 mL), and the mixture was heated in a water bath under reflux until the reaction was complete, as described previously [21]. The product was purified by recrystallization from methanol.
Synthesis of N′-(4-/3-/2-/non-substituted benzylidene)-4-(hydroxy)benzohydrazides (2a–t). p-Hydroxybenzoic acid hydrazide (compound 1) (3 mmol) was dissolved in ethanol (15 mL) and heated in a water bath under reflux with 4-/3-/2-/non-substituted benzaldehyde (3 mmol) until the reaction was complete, as described previously [22]. It was stirred for approximately 20 h at room temperature, and the solid obtained was recrystallized from methanol.
Synthesis of N′-(4-/3-/2-/non-substituted benzylidene)-4-[(4-methylphenyl)sulfonyloxy] benzohydrazides (3a–t). p-Hydroxybenzoic acid 4-[(4-/3-/2-/non-substituted phenyl)methylene]hydrazide–hydrazone (compound 2a–t) (7 mmol) was mixed in dichloromethane solvent (10 mL) with K2CO3 (10 mmol) and p-toluenesulfonyl chloride (7 mmol) in an ice bath, as described previously [23]. The progress of the final product formation was monitored by thin-layer chromatography (TLC; chloroform–methanol = 90:10). The solid obtained was purified by recrystallization from methanol–water.
N′-(Phenylmethylidene)-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3a). Yield 65.0%. mp 199.8–200.5 °C. HPLC tR: 5.844 min, purity: 98.44%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.40 (s, 3H, C1-CH3), 7.19–7.21 (d, 2H, J = 9.2 Hz, H2′, H6′), 7.44–7.47 (t, 3H, J = 6.0 Hz, H3″, H4″, H5″), 7.47–7.49 (d, 2H, J = 8.4 Hz, H2, H6), 7.72–7.74 (d, 2H, J = 8.0 Hz, H2″, H6″), 7.76–7.79 (d, 2H, J = 8.4 Hz, H3, H5), 7.90–7.93 (d, 2H, J = 8.4 Hz, H3′, H5′), 8.42 (s, 1H, N=CH), 11.90 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.18 (C1-CH3), 122.14 (C2′, C6′), 127.14 (C2″, C6″), 128.27 (C4′), 128.84 (C3″, C5″), 129.67 (C3, C5), 130.18 (C4″), 130.33 (C2, C6), 131.19 (C4), 132.46 (C3′, C5′), 134.19 (C1″), 146.03 (C1), 148.11 (N=CH), 151.21 (C1′), 162.01 (C=O). MS (ESI) m/z calcd. for C21H18N2O4S [M]+ 394.09, found 393.92.
N′-[(2-Methylphenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3b). Yield 94.0%. mp 202.8–203.2 °C. HPLC tR: 8.656 min, purity: 98.85%. 1H NMR (DMSOd6, 400 MHz) δ: 2.40 (s, 3H, C2″-CH3), 2.41 (s, 3H, C1-CH3), 7.17–7.19 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.22–7.24 (d, 1H, J = 7.6 Hz, H3″), 7.26–7.30 (q, 2H, J1 = 7.2 Hz, J2 = 6.4 Hz, H4″, H5″), 7.45–7.47 (d, 2H, J′ = 8.0 Hz, H2, H6), 7.75–7.77 (d, 2H, J = 8.0 Hz, H3, H5), 7.81–7.83 (d, 1H, J = 8.4 Hz, H6″), 7.89–7.91 (d, 2H, J = 8.8 Hz, H3′, H5′), 8.70 (s, 1H, N=CH), 11.83 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 19.43 (C2″-CH3), 21.63 (C1-CH3), 122.58 (C2′, C6′), 126.29 (C5″), 126.63 (C3″), 128.70 (C4′), 130.05 (C3, C5), 130.29 (C2, C6), 130.75 (C4″), 131.31 (C4), 131.65 (C6″), 132.60 (C3′, C5′), 132.92 (C1″), 137.35 (C2″), 146.46 (C1), 147.18 (N=CH), 151.64 (C1′), 162.29 (C=O). MS (ESI) m/z calcd. for C22H20N2O4S [M+H]+ 409.12, found 409.62.
N′-[(3-Methylphenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3c). Yield 98.1%. mp 162.8–163.2 °C. HPLC tR: 9.047 min, purity: 96.07%. 1H NMR (DMSOd6, 400 MHz) δ: 2.35 (s, 3H, C3″-CH3), 2.42 (s, 3H, C1-CH3), 7.18–7.20 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.24–7.25 (d, 1H, J = 7.2 Hz, H4″), 7.32–7.35 (t, 1H, J1 = 8.0 Hz, J2 = 7.2 Hz), 7.48- 7.50 (d, 3H, J = Hz, H2, H6, H2″), 7.55 (s, 1H, H6″) 7.77–7.79 (d, 2H, J = 8.4 Hz, H3, H5), 7.90–7.92 (d, 2H, J = 8.4 Hz, H3′, H5′), 8.38 (s, 1H, N=CH), 11.82 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 20.84 (C3″-CH3), 21.16 (C1-CH3), 122.10 (C2′, C6′), 124.54 (C5″), 127.36 (C6″), 128.23 (C4′), 128.71 (C4″), 129.64 (C3, C5), 130.30 (C2, C6), 130.88 (C2″), 131.20 (C4), 132.46 (C3′, C5′), 134.12 (C1″), 138.07 (C3″), 146.01 (C1), 148.14 (N=CH), 151.18 (C1′), 161.97 (C=O). MS (ESI) m/z calcd. for C22H20N2O4S [M]+ 408.11, found 408.04.
N′-[(4-Methylphenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3d). Yield 88.5%. mp 209.2–210.0 °C. HPLC tR: 8.614 min, purity: 98.15%. 1H NMR (DMSOd6,400 MHz) δ: 2.33 (s, 3H, C4″-CH3), 2.42 (s, 3H, C1-CH3), 7.18–7.20 (d, 2H, J = 7.6 Hz, H2′, H6′), 7.25–7.27 (d, 2H, J = 8.4 Hz, H3″, H5″), 7.47–7.49 (d, 2H, J = 8.0 Hz, H2, H6), 7.60–7.62 (d, 2H, J = 8.4 Hz, H2″, H6″), 7.77–7.79 (d, 2H, J = 8.0 Hz, H3, H5), 7.89–7.92 (d, 2H, J = 8.8 Hz, H3′, H5′), 8.39 (s, 1H, N=CH), 11.78 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.03 (C4″-CH3), 21.19 (C1-CH3), 122.13 (C2′, C6′), 127.14 (C2″, C6″), 128.28 (C4′), 129.46 (C3″, C5″), 129.65 (C3, C5), 130.34 (C2, C6), 131.20 (C4), 131.48 (C1″), 132.54 (C3′, C5′), 140.04 (C4″), 146.04 (C1), 148.15 (N=CH), 151.17 (C1′), 161.93 (C=O). MS (ESI) m/z calcd. for C22H20N2O4S [M]+ 408.11, found 408.10.
N′-[(2-Methoxyphenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3e). Yield 97.2%. mp 199.3–199.8 °C. HPLC tR: 7.147 min, purity: 97.79%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.41 (s, 3H, C1-CH3), 3.85 (s, 3H, OCH3), 6.99–7.03 (t, 1H, J1 = J2 = 7.6 Hz, H5″), 7.09–7.11 (d, 1H, J = 8.4 Hz, H3″), 7.17–7.19 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.39–7.43 (t, 1H, J1 = J2 = 7.2 Hz, H4″), 7.47–7.49 (d, 2H, J = 8.4 Hz, H2, H6), 7.76–7.78 (d, 2H, J = 8.0 Hz, H3, H5), 7.85–7.87 (d, 1H, J = 7.6 Hz, H6″), 7.92–7.94 (d, 2H, J = 8.8 Hz, H3′, H5′), 8.78 (s, 1H, N=CH). 11.89 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.18 (C1-CH3), 55.67 (C2″-OCH3), 111.83 (C3″), 120.74 (C5″), 122.09 (C2′, C6′), 122.16 (C1″), 125.50 (C6″), 128.57 (C4′), 129.64 (C3, C5), 130.32 (C2, C6), 131.19 (C4), 131.67 (C4″), 132.42 (C3′, C5′), 143.55 (N=CH), 146.03 (C1), 151.17 (C1′), 157.78 (C2″), 161.76 (C=O). MS (ESI) m/z calcd. for C22H20N2O5S [M + H]+ 424.11, found 423.96.
N′-[(3-Methoxyphenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3f). Yield 96.1%. mp 147.1–147.8 °C. HPLC tR: 7.106 min, purity: 99.42%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.42 (s, 3H, C1-CH3), 3.80 (s, 3H, OCH3), 7.00–7.02 (dd, 1H, J1 = 8.0 Hz, J2 = 1.6 Hz, H4″), 7.19–7.21 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.27–7.28 (d, 2H, J = 4.0 Hz, H2″, H6″), 7.35–7.39 (t, 1H, J1 = 8.4 Hz, J2 = 7.6 Hz, H5″), 7.47–7.49 (d, 2H, J = 8.0 Hz, H2, H6), 7.77–7.79 (d, 2H, J = 8.4 Hz, H3, H5), 7.90–7.92 (d, 2H, J = 8.8 Hz, H3′, H5′), 8.39 (s, 1H, N=CH), 11.90 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.15 (C1-CH3), 55.14 (C3″-OCH3), 111.23 (C2″), 116.30 (C4″), 120.07 (C6″), 122.11 (C2′, C6′), 128.23 (C4′), 129.65 (C3, C5), 129.92 (C5″), 130.30 (C2, C6), 131.21 (C4), 132.42 (C3′, C5′), 135.60 (C1″), 146.00 (C1), 148.00 (N=CH), 151.19 (C1′), 159.53 (C3″), 162.01 (C=O). MS (ESI) m/z calcd. for C22H20N2O5S [M]+ 424.11, found 423.87.
N′-[(4-Methoxyphenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3g). Yield 95.1%. mp 172.1–172.8 °C. HPLC tR: 6.158 min, purity: 99.77%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.41 (s, 3H, C1-CH3), 3.79 (s, 3H, OCH3), 7.00–7.02 (d, 2H, J = 8.4 Hz, H3″, H5″), 7.18–7.20 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.47–7.48 (d, 2H, J = 7.6 Hz, H2, H6), 7.66–7.68 (d, 2H, J = 8.4 Hz, H2″, H6″), 7.76–7.78 (d, 2H, J = 8.4 Hz, H3, H5), 7.90–7.92 (d, 2H, J = 8.8 Hz, H3′, H5′), 8.36 (s, 1H, N=CH), 11.77 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.16 (C1-CH3), 55.26 ((C4″-OCH3), 114.32 (C3″, C5″), 122.08 (C2′, C6′), 126.74 (C1″), 128.24 (C4′), 128.75 (C2″, C6″), 129.58 (C3, C5), 130.30 (C2, C6), 131.23 (C4), 132.62 (C3′, C5′), 146.00 (C1), 148.02 (N=CH), 151.12 (C1′), 160.91 (C4″), 161.83 (C=O). MS (ESI) m/z calcd. for C22H20N2O5S [M]+ 424.11, found 423.94.
N′-[(2-Bromophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3h). Yield 99.1%. mp 192.3–193.0 °C. HPLC tR: 10.518 min, purity: 98.25%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.40 (s, 3H, C1-CH3), 7.18–7.20 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.34–7.37 (t, 1H, J1 = 8.0 Hz, J2 = 7.4 Hz, H4″), 7.42–7.45 (t, 1H, J = 8.0 Hz, H5″), 7.45–7.47 (d, 2H, J = 8.0 Hz, H2, H6), 7.65–7.67 (d, 1H, J = 7.2 Hz, H3″), 7.75–7.77 (d, 2H, J = 8.4 Hz, H3, H5), 7.91–7.93 (d, 2H, J = 9.2 Hz, H3′, H5′), 7.92–7.99 (d, 1H, J = 7.2 Hz, H6″), 8.77 (s, 1H, N=CH), 12.12 (s,1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.63 (C1-CH3), 122.63 (C2′, C6′), 124.07 (C2″), 127.71 (C5″), 128.54 (C4′), 128.70 (C6″), 130.18 (C3, C5), 130.76 (C2, C6), 131.63 (C4), 132.26 (C4″), 132.63 (C3′, C5′), 133.38 (C3″), 133.61 (C1″), 146.47 (C1), 146.79 (N=CH), 151.76 (C1′), 162.51 (C=O). MS (ESI) m/z calcd. for C21H17BrN2O4S [M + 2H]+ 474.02, found 473.91.
N′-[(3-Bromophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3i). Yield 99.2%. mp 187.0–187.5 °C. HPLC tR: 11.956 min, purity: 97.63%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.42 (s, 3H, C1-CH3), 7.19–7.21 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.39–7.43 (t, 1H, J1 = 8.0 Hz, J2 = 7.6 Hz, H5″), 7.47–7.50 (d, 2H, J = 8.8 Hz, H2, H6), 7.61–7.62 (d, 1H, J = 7.6 Hz, H4″), 7.71–7.72 (d, 1H, J = 7.2 Hz, H6″), 7.76–7.79 (d, 2H, J = 8.4 Hz, H3, H5), 7.90–7.92 (d, 3H, J = 7.2 Hz, H3′, H5′, H2″), 8.38 (s, 1H, N=CH), 11.98 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.17 (C1-CH3), 122.15 (C2′, C6′), 122.15 (C3″), 126.24 (C2″), 128.23 (C4′), 129.15 (C6″), 129.71 (C3, C5), 130.30 (C2, C6), 130.98 (C5″), 131.19 (C4), 132.24 (C4″), 132.65 (C3′, C5′), 136.65 (C1″), 146.02 (C1), 146.25 (N=CH), 151.26 (C1′), 162.13 (C=O). MS (ESI) m/z calcd. for C21H17BrN2O4S [M]+ 472.01, found 472.02.
N′-[(4-Bromophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3j). Yield 85.7%. mp 202.8–203.5 °C. HPLC tR: 12.025 min, purity: 98.81%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.49 (s, 3H, C1-CH3), 7.19–7.21 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.47–7.49 (d, 2H, J = 8.8 Hz, H2, H6), 7.63–7.65 (d, 2H, J = 8.8 Hz, H3″, H5″), 7.66–7.69 (d, 2H, J = 8.8 Hz, H2″, H6″), 7.76–7.79 (d, 2H, J = 8.4 Hz, H3, H5), 7.90–7.92 (d, 2H, J = 8.4 Hz, H3′, H5′), 8.40 (s, 1H, N=CH), 11.91 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.16 (C1-CH3), 122.12 (C2′, C6′), 123.40 (C4″), 128.23 (C4′), 128.96 (C2″, C6″), 129.67 (C3, C5), 130.30 (C2, C6), 131.20 (C4), 131.82 (C3″, C5″), 132.33 (C3′, C5′), 133.46 (C1″), 146.00 (C1), 146.82 (N=CH), 151.12 (C1′), 162.03 (C=O). MS (ESI) m/z calcd. for C21H17BrN2O4S [M]+ 472.01, found 472.04.
N′-[(2-Chlorophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3k). Yield 98.9%. mp 188.8–189.2 °C. HPLC tR: 11.096 min, purity: 99.47%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.40 (s, 3H, C1-CH3), 7.18–7.20 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.41–7.51 (m, 3H, H3″, H4″, H5″), 7.45–7.47 (d, 2H, J = 8.8 Hz, H3′, H5′), 7.75–7.77 (d, 2H, J = 8.0 Hz, H2, H6), 7.91–7.93 (d, 2H, J = 8.4 Hz, H3, H5), 7.98–8.00 (d, 1H, J = 7.2 Hz, H6″), 8.81 (s, 1H, N=CH), 12.09 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.63 (C1-CH3), 122.61 (C2′, C6′), 127.32 (C3″), 128.06 (C5″), 128.70 (C4′), 130.17 (C3, C5), 130.37 (C6″), 130.76 (C2, C6), 131.64 (C4), 131.88 (C4″), 132.02 (C1″), 132.63 (C3′, C5′), 133.69 (C2″), 144.45 (N=CH), 146.47 (C1), 151.76 (C1′), 162.48 (C=O). MS (ESI) m/z calcd. for C21H17ClN2O4S [M+2H]+ 430.07, found 429.91.
N′-[(3-Chlorophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3l). Yield 98.7%. mp 141.7–142.2 °C. HPLC tR: 10.518 min, purity: 98.96%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.42 (s, 3H, C1-CH3), 7.19–7.21 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.47–7.49 (d, 4H, J = 8.4 Hz, H2, H6, H4″, H6″), 7.68 (s, 1H, H5″), 7.76–7.78 (d, 3H, J = 8.4 Hz, H3, H5, H2″), 7.90–7.92 (d, 2H, J = 7.6 Hz, H3′, H5′), 8.40 (s, 1H, N=CH), 11.98 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.17 (C1-CH3), 122.14 (C2′, C6′), 125.82 (C6″), 126.32 (C2″), 128.24 (C4′), 129.72 (C3, C5), 129.78 (C4″), 130.31 (C5″), 130.72 (C2, C6), 131.19 (C4), 132.24 (C3′, C5′), 133.64 (C1″), 136.42 (C3″), 146.02 (C1), 146.38 (N=CH), 151.27 (C1′), 162.14 (C=O). MS (ESI) m/z calcd. for C21H17ClN2O4S [M]+ 428.06, found 428.05.
N′-[(4-Chlorophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3m). Yield 91.2%. mp 201.8–202.4 °C. HPLC tR: 10.485 min, purity: 99.13%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.48 (s, 3H, C1-CH3), 7.17–7.19 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.21–7.74 (d,2H, J = 8.8 Hz, H2″, H6″), 7.45–7.47 (d, 2H, J = 8.4 Hz, H3″, H5″), 7.48–7.50 (d, 2H, J = 8.8 Hz, H2, H6), 7.75–7.77 (d, 2H, J = 8.4 Hz, H3, H5), 7.88–7.90 (d, 2H, J = 8.4 Hz, H3′, H5′), 8.39 (s, 1H, N=CH), 11.93 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.63 (C1-CH3), 122.58 (C2′, C6′), 128.70 (C4′), 129.19 (C2″, C6″), 129.37 (C3, C5), 130.13 (C3″, C5″), 130.76 (C2, C6), 131.66 (C4), 132.79 (C3′, C5′), 133.58 (C4″), 135.05 (C1″), 147.19 (N=CH), 146.46 (C1), 151.68 (C1′), 162.48 (C=O). MS (ESI) m/z calcd. for C21H17ClN2O4S [M+2H]+ 430.06, found 429.91.
N′-[(2-Fluorophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3n). Yield 93.0%. mp 146.5–147.0 °C. HPLC tR: 7.470 min, purity: 99.77%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.42 (s, 3H, C1-CH3), 7.19–7.21 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.28–7.32 (t, 2H, J1 = 8.4 Hz, J2 = 8.0 Hz, H3″, H5″), 7.48–7.50 (d, 3H, J = 8.0 Hz, H2, H6, H6″), 7.76–7.78 (d, 2H, J = 8.0 Hz, H3, H5), 7.91–7.95 (d, 3H, J = Hz, H3′, H5′, H4″), 8.66 (s, 1H, N=CH), 12.00 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.16 (C1-CH3), 115.89, 116.09 (C3″), 121.66, 121.76 (C1″), 122.15 (C2′, C6′), 124.91 (C5″), 126.30 (C6″), 128.24 (C4′), 129.66 (C3, C5), 130.30 (C2, C6), 131.19 (C4), 132.06, 132.14 (C4″), 132.21 (C3′, C5′), 140.76 (N=CH), 146.02 (C1), 151.27 (C1′), 159.55 (C2″), 161.95 (C=O). MS (ESI) m/z calcd. for C21H17FN2O4S [M]+ 412.09, found 411.99.
N′-[(3-Fluorophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3o). Yield 98.5%. mp 153.8–154.1 °C. HPLC tR: 6.909 min, purity: 98.84%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.42 (s, 3H, C1-CH3), 7.19–7.21 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.24–7.28 (t, 1H, J1 = 8.4 Hz, J2 = 6.8 Hz, H4″), 7.47–7.49 (d, 2H, J = 8.0 Hz, H2, H6), 7.53–7.57 (t, 3H, J1 = 7.2 Hz, J2 = 6.4 Hz, H2″, H5″, H6″), 7.76–7.79 (d, 2H, J = 8.4 Hz, H3, H5), 7.90–7.92 (d, 2H, J = 8.0 Hz, H3′, H5′), 8.43 (s, 1H, N=CH), 11.95 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.16 (C1-CH3), 112.94, 113.16 (C2″), 116.79, 117.01 (C4″), 122.18 (C2′, C6′), 123.47 (C6″), 128.24 (C4′), 129.71 (C3, C5), 130.31 (C2, C6), 130.97, 130.87 (C5″), 131.20 (C4), 132.28 (C3′, C5′), 136.72, 136.79 (C1″), 146.03 (C1), 146.69 (N=CH), 151.27 (C1′), 161.17, 162.12 (C3″), 163.60 (C=O). MS (ESI) m/z calcd. for C21H17FN2O4S [M]+ 412.09, found 412.07.
N′-[(4-Fluorophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3p). Yield 98.8%. mp 174.8–175.5 °C. HPLC tR: 6.625 min, purity: 99.07%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.40 (s, 3H, C1-CH3), 7.18–7.21 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.26–7.30 (d, 2H, J = 9.2, 8.4 Hz, H3″, H5″), 7.46–7.48 (d, 2H, J = 8.4 Hz, H2, H6), 7.76–7.80 (d, 4H, J = 8.4, 7.6 Hz, H3, H5, H2″, H6″), 7.90–7.92 (d, 2H, J = 8.4 Hz, H3′, H5′), 8.41 (s, 1H, N=CH), 11.92 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz): δ 21.19 (C1-CH3), 115.81, 116.03 (C3″, C5″), 122.68 (C2′, C6′), 128.28 (C4′), 129.29, 129.38 (C2″, C6″), 129.69 (C3, C5), 130.33 (C2, C6), 130.80, 130.83 (C1″), 131.22 (C4), 132.44 (C3′, C5′), 146.05 (C1), 146.99 (N=CH), 151.24 (C1′), 161.96, 162.05 (C4″), 164.42 (C=O). MS (ESI) m/z calcd. for C21H17FN2O4S [M]+ 412.09, found 411.99.
N′-[(2-Nitrophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3r). Yield 96.3%. mp 180.0–180.4 °C. HPLC tR: 6.586 min, purity: 99.39%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.41 (s, 3H, C1-CH3), 7.19–7.22 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.47–7.49 (d, 2H, J = 8.0 Hz, H2, H6), 7.65–7.68 (t, 1H, J = 7.6 Hz, H4″), 7.76–7.78 (d, 2H, J = 8.0 Hz, H3, H5), 7.78–7.81 (t, 1H, J = 7.6 Hz, H5″), 7.93–7.95 (d, 2H, J′ = 7.6 Hz, H3′, H5′), 8.05–8.07 (d, 1H, J = 8.0 Hz, H3″), 8.11–8.12 (d, 1H, J = 7.2 Hz, H6″), 8.54 (s, 1H, N=CH), 12.19 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.15 (C1-CH3), 122.14 (C2′, C6′), 124.65 (C3″), 127.93 (C1″), 128.23 (C4′), 128.60 (C6″), 129.79 (C3, C5), 130.29 (C2, C6), 130.71 (C4″), 131.19 (C4), 132.00 (C3′, C5′), 133.69 (C5″), 143.33 (C2″), 146.01 (C1), 148.20 (N=CH), 151.37 (C1′), 162.19 (C=O). MS (ESI) m/z calcd. for C21H17N3O6S [M]+ 439.08, found 439.03.
N′-[(3-Nitrophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3s). Yield 97.7%. mp 182.5–183.1 °C. HPLC tR: 6.820 min, purity: 99.04%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.42 (s, 3H, C1-CH3), 7.20–7.22 (d, 2H, J = 8.8 Hz, H2′, H6′), 7.47–7.49 (d, 2H, J = 8.0 Hz, H2, H6), 7.72–7.74 (t, 1H, J = 7.6 Hz, H5″), 7.76–7.78 (d, 2H, J = 8.4 Hz, H3, H5), 7.91–7.93 (d, 2H, J = 8.4 Hz, H3′, H5′), 8.13–8.15 (d, 1H, J = 7.6 Hz, H6″), 8.24–8.26 (d, 1H, J = 7.6 Hz, H4″), 8.51–8.53 (d, 2H, H2″, N=CH), 12.15 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.16 (C1-CH3), 120.95 (C4″), 122.16 (C2′, C6′), 124.31 (C2″), 128.23 (C4′), 129.76 (C3, C5), 130.30 (C2, C6), 130.43 (C5″), 131.20 (C4), 132.13 (C3′, C5′), 133.38 (C6″), 136.05 (C1″), 145.63 (C3″), 146.03 (C1), 148.20 (N=CH), 151.34 (C1′), 162.26 (C=O). MS (ESI) m/z calcd. for C21H17N3O6S [M]+ 439.08, found 438.97.
N′-[(4-Nitrophenyl)methylidene)]-4-[(4-methylphenyl)sulfonyloxy]benzohydrazide (3t). Yield 98.8%. mp 227.3–228.0 °C. HPLC tR: 6.564 min, purity: 99.20%. 1H NMR (DMSO-d6, 400 MHz) δ: 2.43 (s, 3H, C1-CH3), 7.20–7.22 (d, 2H, J = 8.4 Hz, H2′, H6′), 7.48–7.50 (d, 2H, J = 8.4 Hz, H2, H6), 7.77–7.79 (d, 2H, J = 8.4 Hz, H3, H5), 7.91–7.93 (d, 2H, J = 8.0 Hz, H3′, H5′), 7.97–7.99 (d, 2H, J = 8.0 Hz, H2″, H6″), 8.27–8.29 (d, 2H, J = 8.0 Hz, H3″, H5″), 8.51 (s, 1H, N=CH), 12.13 (s, 1H, CONH). 13C NMR (DMSO-d6, 100 MHz) δ: 21.18 (C1-CH3), 122.19 (C2′, C6′), 124.05 (C3″, C5″), 128.05 (C2″, C6″), 128.25 (C4′), 129.80 (C3, C5), 130.33 (C2, C6), 131.20 (C4), 132.09 (C3′, C5′), 140.47 (C1″), 145.58 (C4″), 146.05 (C1), 147.91 (N=CH), 151.38 (C1′), 162.29 (C=O). MS (ESI) m/z calcd. for C21H17N3O6S [M]+ 439.08, found 439.12.
2.2. Enzyme and Antioxidant Inhibition Assays
MAO activities were assayed with recombinant human MAO-A and MAO-B with 0.06 mM of kynuramine and 0.3 mM of benzylamine substrates in 50 mM sodium phosphate buffer (pH 7.2), respectively, using a spectrophotometer (Optizen Pop, KLAB, Daejeon, Korea), as described previously [24]. The Km values of these substrates were 0.047 and 0.085 mM, respectively; thus, the substrate concentrations were 1.3 × and 3.5 × Km, respectively. AChE and BChE activities were measured using the enzymes of Electrophorus electricus Type VI-S and equine serum, respectively, in the presence of 0.5 mM acetylthiocholine iodide (ATCI) and butyrylthiocholine iodide (BTCI) as substrates, in 100 mM sodium phosphate buffer (pH 7.5), respectively, and 0.5 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) was used as a color developing agent, as described previously [25,26]. Inhibitory activities were measured after preincubating enzymes and inhibitors for 15 min. BACE-1 activity was measured using a BACE-1 activity detection kit, measuring at an excitation wavelength of 320 nm and emission wavelength of 405 nm using a fluorescence spectrometer (FS-2, Scinco, Seoul, Korea) [27]. Antioxidant activity was measured using 0.2 mL of ethanol solution at 517 nm after incubating 50 µM compound and 0.1 mM 2,2-diphenyl-1-picrylhydrazyl (DPPH) for 15 min in the dark, as described previously [27]. Enzymes and chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). The experiments were carried out for duplicate or triplicate.
2.3. Analysis of Enzyme Inhibition and Kinetics
The inhibitory activities of 19 compounds against MAO-A, MAO-B, AChE, BChE, and BACE-1 were first observed at concentrations of 10 µM. The IC50 values of the compounds that resulted in residual enzyme activities of <50% were then determined. Reversibility and kinetic studies were performed on the most potent inhibitors, i.e., 3o for MAO-A and 3s for MAO-B, as previously described [28]. Kinetic experiments were carried out at five substrate concentrations and three inhibitor concentrations.
2.4. Analysis of Inhibitor Reversibility
The reversibility of 3o and 3s was analyzed using a dialysis method after preincubating the compounds with MAO enzymes for 30 min, as previously described [29]. In the reversibility experiments of MAO-A, the concentrations used were 3.0 µM for 3o, 2.0 µM for toloxatone as a reversible MAO-A reference inhibitor, and 0.014 µM for clorgyline as an irreversible MAO-A reference inhibitor. In MAO-B experiments, the concentrations used were 7.0 µM for 3s, 0.20 µM for lazabemide as a reversible MAO-B reference inhibitor, and 0.28 µM for pargyline as an irreversible MAO-B reference inhibitor. The relative activities for undialyzed (AU) and dialyzed (AD) samples were compared to determine the reversibility patterns.
2.5. Molecular Docking
The three-dimensional X-ray structures of MAO-A, MAO-B, and BACE-1 were obtained from the Protein Data Bank (entries 2Z5X, 2V5Z, and 3TPP, respectively) [30,31,32]. Optimization and minimization of the crystal structures were carried out with the Protein Preparation wizard, and ligands were treated with the Ligprep Tool to find all possible tautomers and protonation states at physiological pH, as well as conformation states. GLIDE software was used to perform the molecular docking analysis. Enclosing boxes for each simulation were created by using the center of mass of cognate ligands, and the SP docking protocol was used by setting 50,000 poses per ligand for the initial phase and 4000 poses per ligand for energy minimization with the OPLS3e force field [33,34,35]. To confirm the reliability of docking simulations, redocking analyses were performed on the cognate ligands in their binding sites. Cognate ligands moved back to the original positions with Root Mean Square Deviations (RMSD), based on all the heavy atoms, as small as 0.155 Å, 0.208 Å, and 0.467 Å for BACE-1, MAO-A, and MAO-B cognate ligands, respectively.
3. Results and Discussion
3.1. Synthesis
The general synthesis of 3a–t is depicted in Scheme 1. Compounds were synthesized starting from ethyl paraben hydrazide (compound 1). Then, by the reaction of this compound (1) with various benzaldehydes, hydrazide–hydrazone derivatives (N′-(4-/3-/2-/non-substituted benzylidene)-4-(hydroxy)benzohydrazides; compounds 2a–t) were obtained. By the reaction of these compounds (2a–t) with tosyl chloride, new tosylated acyl hydrazide derivatives (compounds 3a–t) were obtained (Scheme 1).

Scheme 1.
Synthesis of compounds 3a–t.
3.2. Inhibitory Activities against MAOs, AChE, BChE, and BACE-1 and Antioxidant Activity
Among the 19 derivatives, four and three compounds showed effective inhibitory activities against MAO-A and MAO-B, respectively, with residual activities of <50% at 10 µM (Table 1). Compound 3o was the most potent inhibitor of MAO-A, with an IC50 value of 1.54 µM, followed by 3a and 3p (IC50 = 3.35 and 4.77 µM, respectively). A structural comparison with 3a indicated that the 3-F group in 3o increased its inhibitory activity against MAO-A. Compound 3s was the most potent inhibitor of MAO-B, with an IC50 value of 3.64 µM, followed by 3t and 3a (IC50 = 5.69 and 7.69 µM, respectively). It was observed that MAO-B inhibitory activity was increased in the order of 3- > 4- > 2-NO2 groups in 3s, 3t, and 3r, respectively. The selectivity index (SI) values of 3s and 3a for MAO-B over MAO-A were 4.31 and 0.44, respectively, indicating that 3s and 3a are moderately selective for MAO-B and MAO-A, respectively. Benzylidene-4-[(4-methylphenyl)sulfonyloxy] moiety in this study was less effective in potency and MAO-A selectivity than 3-benzylquinoxaline moiety previously reported, in which benzyl moiety strongly bound to MAO-A with aromatic interactions through Val210 and Phe208 [36]. On the other hand, all compounds weakly inhibited AChE and BChE, which retained >50% residual activities at 10 µM, except for 3a, which inhibited BChE with an IC50 value of 16.1 µM.

Table 1.
Inhibition of MAO-A, MAO-B, AChE, BChE, and BACE-1 by N′-(4-/3-/2-/non-substituted benzylidene)-4-[(4-methylphenyl)sulfonyloxy] benzohydrazides (3a–t) a.
Interestingly, 3e, 3f, and 3n inhibited BACE-1 with IC50 values of 8.63, 9.92, and 8.47 µM, respectively, which were lower than the IC50 of the quercetin reference. BACE-1 inhibitors have been studied as pharmaceuticals, and some of them have been submitted to phase I–III clinical trials [37]. The potency of 3n against BACE-1 was slightly weaker than that of compound PC3, a piperazine-substituted chalcone, with an IC50 value of 6.72 µM [38]. These results suggest that 3e, 3f, and 3n can be considered potential agents for the treatment of AD. In an antioxidant assay using DPPH, lead compounds 3a, 3o, 3p, 3s, and 3t showed negligible inhibition (0.18–3.36%).
3.3. Structure–Activity Relationship (SAR)
In addition to evaluating the inhibitory activities of 3a–t against MAOs, AChE, BChE, and BACE-1, we investigated their structures. For the enzyme inhibitory activity against MAO-A (reference compound toloxatone; IC50: 1.08 µM), compound 3o (IC50: 1.54 µM) carrying 3-F and the non-substituted compound 3a (IC50: 3.35 µM) were found to have the highest activity. Compound 3p (IC50: 4.77 µM) carrying 4-F in its structure showed high activity, while compound 3n carrying 2-F (IC50: 13.0 µM) showed low activity. The fluorine atom is inductively electron-attracting and resonantly electron-donating. The fluorine atom increased the activity when it is in the meta position but decreased it when it is in the ortho position. This suggests that the addition of fluorine in meta improves the interactions with the enzyme, perhaps participating in halogen bond or favoring conformations for π-stacking. Otherwise, in the ortho position it probably hinders the interaction with the enzyme.
For the MAO-B enzyme inhibition activity (reference compound pargyline IC50: 0.14 µM), the compounds showing the highest activity were the 3-NO2-carrying compound 3s (IC50: 3.64 µM), the 4-NO2 derivative compound 3t (IC50: 5.69 µM), and the non-substituted compound 3a (IC50: 7.69 µM). The activity of compound 3r, which carries a 2-NO2 substituent (IC50: 27.6 µM), was found to be quite low. The higher activity of the compound carrying the nitro group in the meta position (compound 3s), compared to the non-substituted derivative (compound 3a), suggested that the inductive effect of nitro played an important role. In addition, the activity of the compound having the nitro substituent in the para position (compound 3t) was higher than the non-substituted derivative (compound 3a), but the nitro moiety in the ortho position (compound 3r) decreased activity. It suggested that the electron donor nitro group in the meta-para position probably increases and enhances the interactions with the enzyme, maybe establishing π–hole interactions or favoring π-stacking; conversely, when the nitro group is in the ortho position it is possible that it generates a steric hindrance which reduces the interactions with the enzyme.
The AChE and BChE enzyme inhibition activities were found to be very low compared with the IC50 values of the references Tacrine and Donepezil. Compared with Donepezil (IC50: 0.18 µM), only the non-substituted derivative compound 3a showed notable inhibitory activity (IC50: 16.1 µM). These results show that substituents on these compounds inhibit their interaction with the AChE and BChE enzymes.
Several compounds effectively inhibited BACE-1 activity. The IC50 value of the reference compound quercetin was 13.4 µM, and those of 3n, 3e, and 3f were 8.47, 8.63, and 9.92 µM, respectively, which were the lower among the tested compounds. Compound 3n carries 2-F, 3e carries 2-OCH3, and 3f carries 3-OCH3. The presence of fluorine or methoxy in the second position of the compound was observed to increase its interaction with the BACE-1 enzyme. Since the fluorine and methoxy groups attract electrons inductively and donate electrons resonantly, their greater activity in the middle position indicates that the inductive effect is dominant. In general, our compounds showed a moderate inhibitory effect on the MAO-A, MAO-B, and BACE-1 enzymes.
3.4. Reversibility Studies
In the dialysis experiments, the inhibition of MAO-A by 3o was reversed, and its activity increased from 38.7% (the value of AU) to 79.2% (the value of AD) (Figure 2A). The degree of MAO-A recovery after its inhibition by 3o was similar to that observed with the reversible reference inhibitor toloxatone (from 29.7% to 84.5%) and contrary to the values obtained with the irreversible reference inhibitor pargyline (from 25.2% to 26.1%), from which the enzyme activity did not recover. The inhibition of MAO-B by 3s was reversed, and its activity increased from 35.0% (AU) to 84.0% (AD) (Figure 2B). The recovery was also similar to that observed with the reversible reference lazabemide (from 35.3% to 87.9%), whereas the inhibition of MAO-B by the irreversible inhibitor pargyline was not reversed (from 30.6% to 33.8%). These experiments show that 3o and 3s are reversible inhibitors of MAO-A and MAO-B, respectively.

Figure 2.
Reversals of MAO-A inhibition by 3o (A) and MAO-B inhibition by 3s (B) using dialysis experiments.
3.5. Kinetics of MAO-A and MAO-B Inhibition
In kinetic studies of MAO-A inhibition by 3o, Lineweaver–Burk plots and secondary plots showed that 3o competitively inhibited MAO-A (Figure 3A), with a Ki value of 0.35 ± 0.074 µM (Figure 3B). In kinetic studies of MAO-B inhibition by 3s, Lineweaver–Burk plots and secondary plots showed that 3s competitively inhibited MAO-B (Figure 3C), with a Ki value of 1.97 ± 0.65 µM (Figure 3D). These results suggest that 3o and 3s are competitive inhibitors for MAO-A and MAO-B, respectively, and compete with the substrate for the active site.
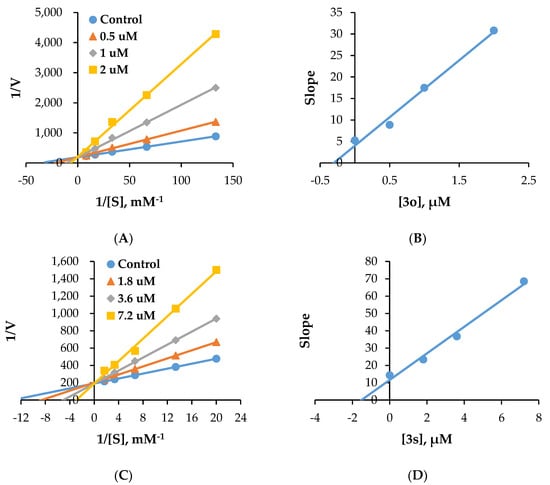
Figure 3.
Lineweaver–Burk plots for the inhibition of MAO-A by 3o (A) and MAO-B by 3s (C) and their respective secondary plots (B,D) of the slopes vs. inhibitor concentrations.
3.6. Molecular Docking
In order to investigate the binding mode of compounds 3e, 3f, and 3n towards BACE-1 and 3o and 3s towards MAO-A and MAO-B, molecular docking analyses were performed.
As shown in Figure 4, the binding modes of the three compounds demonstrate that the sulfone moiety and the carbonyl group are crucial for establishing a network of hydrogen bonds with both the main and side chains of T232 (at a distance of 2.2 Å, 2.3 Å and 2.5 Å for 3e, 3f and 3n) and the side chain of S325 (at a distance of 2.2 Å, 2.2 Å and 2.1 Å for 3e, 3f and 3n, respectively) in the S2 pocket. Furthermore, the 2-methoxy-benzyl moiety and the 2-fluorine benzyl moiety of compounds 3e and 3n, respectively, are involved in π–π interactions with Y71 at the S1 pocket. The docking scores were −5.59, −5.90, and −6.88 kcal/mol for 3e, 3f, and 3n, respectively.

Figure 4.
Catalytic site of BACE-1 in complex with compounds 3e (a), 3f (b), and 3n (c), depicted as green, yellow, and blue sticks, respectively. Hydrogen bonds and π–π interactions are indicated by red arrows and green lines, respectively.
As indicated in Figure 5, the para-methyl phenyl ring of compound 3o is trapped in the aromatic cage formed by FAD, Y407, and Y444 of MAO-A and FAD, Y398, and Y435 residues of MAO-B. In addition, compound 3o can establish π-stacking and T-shape π–π contacts with Y407 and F352 of MAO-A, respectively. For MAO-B, π-stacking and T-shape π–π interactions were detected with Y398 and Y326, a MAO-B selective residue. The docking scores were −9.73 and −9.89 kcal/mol in simulations of MAO-A and MAO-B, respectively.
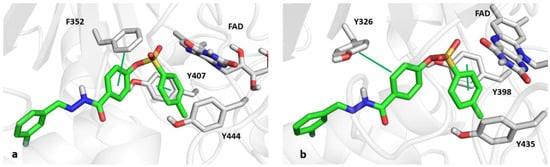
Figure 5.
Panels (a,b) show the catalytic sites of MAO-A and MAO-B, respectively, in complex with compound 3o, depicted as green sticks. The π–π interactions are indicated by green lines.
As shown in Figure 6, the para-methyl ring of compound 3s interacts with the network of aromatic rings of FAD, Y398 and Y435 of MAO-B. Specifically, compound 3s forms π-stacking and T-shape π–π interactions with the Y398 and Y326 of MAO-B. On the other hand, the binding pose of 3s within MAO-A binding pocket resulted to be inverted. Its 3-NO2-benzyl ring was partially trapped in the aromatic cage made of Y407, Y444, and FAD, but stabilized due to the presence of a dipole interaction with F352 residue. The docking scores were −5.67 and −9.88 kcal/mol for MAO-A and MAO-B, respectively.
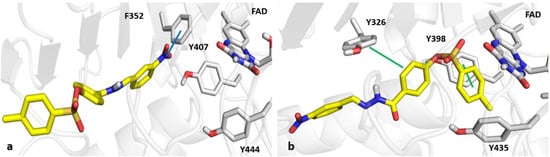
Figure 6.
Panels (a,b) show the catalytic sites of MAO-A and MAO-B, respectively, in complex with compound 3s, depicted as yellow sticks. The π–π and dipole interactions are indicated by green and blue lines, respectively.
For the sake of completeness, all the interactions observed in docking simulations were automatically flagged by GLIDE software, for which the default distance thresholds are equal to 2.8 Å for hydrogen bonds, and to 4.5 Å and 5.5 Å for π-stacking and T-shape contacts, respectively.
3.7. ADME Prediction
In order to evaluate the drug-likeness of the proposed compounds, physiochemical properties were analyzed by using Qikprop tool [39]. The computation of physiochemical properties can help to draw the ADME profile. As shown in Table 2, all the compounds do not show violations of the Lipinki’s rule of five, a well-known parameter for the evaluation of drug-like molecules, and also display a very good value for human oral absorption, a parameter related with potential metabolites, solubility and cell permeability [40].

Table 2.
Physiochemical properties relevant to drug-likeness.
4. Conclusions
In this study, 19 novel N′-(4-/3-/2-/non-substituted benzylidene)-4-[(4-methylphenyl)sulfonyl-oxy]benzohydrazide derivatives were synthesized, and compounds 3o and 3s were found to have the most potent inhibitory activity against MAO-A and MAO-B, respectively, with IC50 values of 1.54 μM for 3o towards MAO-A and 3.64 μM for 3s towards MAO-B, and moderate selectivities. MAO-B inhibitory activity was increased in the order of 3- > 4- > 2-NO2 groups in 3s, 3t, and 3r, respectively. All compounds weakly inhibited AChE and BChE. However, 3e, 3f, and 3n inhibited BACE-1 with IC50 values of 8.63, 9.92, and 8.47 µM, respectively, with higher potencies than the reference compound quercetin. Compounds 3o and 3s were reversible and competitive inhibitors of MAO-A and MAO-B, respectively. It is concluded that 3o and 3s are effective MAO-A and MAO-B inhibitors, respectively, and 3e, 3f, and 3n are effective BACE-1 inhibitors. These results suggest that these compounds can be considered potential agents for the treatment of AD. Computational studies shed light on the interactions of compounds 3e, 3f, and 3n with BACE1 and those of 3s and 3o with MAOs, and these results agree with the experimental data. Compounds 3e, 3f, and 3n can strongly interact with the S1 and S2 binding regions of the BACE-1 catalytic site through a set of both polar and hydrophobic interactions. For compounds 3o and 3s, the networks of aromatic interactions with FAD, Y407, and Y444 of MAO-A and FAD, Y398, and Y435 of MAO-B are crucial for binding at the MAO binding sites. Furthermore, dipole and π–π contacts with the F352 and Y326 residues of MAO-A and MAO-B, respectively, enhance the interaction with binding pockets and can thus explain the IC50 experimental data.
Supplementary Materials
The following is available online at https://www.mdpi.com/article/10.3390/app11135830/s1. Figures S1–S76 for 1H-NMR, 13C-NMR, EST MS, and chromatogram data of the compounds.
Author Contributions
Conceptualization: H.E.S., İ.S.D. and H.K.; synthesis: H.E.S., İ.S.D., S.Y., A.B.A., A.K. and M.A.A.; biological activity: J.M.O. and G.S.J.; docking analysis: N.G. and O.N.; original draft writing: J.M.O., İ.S.D., H.E.S. and B.M.; review and editing: H.K., İ.S.D. and B.M.; supervision: H.K.; funding acquisition: H.K. and A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (NRF-2019R1A2C1088967), Republic of Korea, and by Scientific Research Projects Coordination Unit of Karadeniz Technical University (Project Number: FLO-2019-7883), Turkey.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Authors acknowledge Taif University Researchers Supporting Project number (TURSP-2020/68), Taif University, Taif, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cao, Z.; Song, Q.; Yu, G.; Liu, Z.; Cong, S.; Tan, Z.; Deng, Y. Novel 3-benzylidene/benzylphthalide Mannich base derivatives as potential multifunctional agents for the treatment of Alzheimer’s disease. Bioorg. Med. Chem. 2021, 35, 116074. [Google Scholar] [CrossRef]
- George, G.T. Cholinesterase inhibitors for the treatment of Alzheimer’s disease: Getting on and staying on. Curr. Ther. Res. 2003, 64, 216–235. [Google Scholar]
- Herholz, K.; Bauer, B.; Wienhard, K.; Kracht, L.; Mielke, R.; Lenz, O.; Strotmann, T.; Heiss, H.D. In-vivo measurements of regional acetylcholine esterase activity in degenerative dementia: Comparison with blood flow and glucose metabolism. J. Neural. Transm. 2000, 107, 1457–1468. [Google Scholar] [CrossRef]
- Dorababu, A. Synthesis, pharmacological evaluation and structure-activity relationship of recently discovered enzyme antagonist azoles. Heliyon 2020, 6, e03656. [Google Scholar] [CrossRef]
- Lane, R.M.; Potkin, S.G.; Enz, A. Targeting acetylcholinesterase and butyrylcholinesterase in dementia. Int. J. Neuropsychopharmacol. 2006, 9, 101–124. [Google Scholar] [CrossRef]
- Ege, T.; Şelimen, H. Monoamine oxidase inhibitory effects of medicinal plants in management of Alzheimer’s disease. JOTCSA 2021, 8, 239–248. [Google Scholar]
- Sadaoui, N.; Bec, N.; Barragan-Montero, V.; Kadri, N.; Cuisinier, F.; Larroque, C.; Arab, K.; Khettal, B. The essential oil of algerian ammodaucus leucotrichus Coss. & Dur. and its effect on the cholinesterase and monoamine oxidase activities. Fitoterapia 2018, 130, 1–5. [Google Scholar]
- Ghosh, A.K.; Osswald, H.L. BACE1 (beta-secretase) inhibitors for the treatment of Alzheimer’s disease. Chem. Soc. Rev. 2014, 43, 6765–6813. [Google Scholar] [CrossRef] [Green Version]
- Aalto, T.R.; Firman, M.C.; Rigler, N.E. p-Hydroxybenzoic acid esters as preservatives: I. Uses, antibacterial and antifungal studies, properties and determination. J. Am. Pharm. Assoc. Sci. Ed. 1953, 42, 449–457. [Google Scholar] [CrossRef]
- Steinberg, D.C. 2005 preservatives use: Frequency report and registration. Cosmet. Toilet. 2006, 121, 65–69. [Google Scholar]
- Wang, Z.; Kang, D.; Chen, M.; Wu, G.; Feng, D.; Zhao, T.; Zhou, N.; Huo, Z.; Jing, L.; Zuo, X.; et al. Design, synthesis, and antiviral evaluation of novel hydrazone-substituted thiophene[3,2-d]pyrimidine derivatives as potent human immunodeficiency virus-1 inhibitors. Chem. Biol. Drug Des. 2018, 92, 2009–2021. [Google Scholar] [CrossRef]
- Aslan, H.G.; Özcan, S.; Karacan, N. The antibacterial activity of some sulfonamides and sulfonyl hydrazones, and 2D-QSAR study of a Series of sulfonyl hydrazones. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012, 98, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kadyan, K.; Duhan, M.; Sindhu, J.; Singh, V.; Saharan, B.S. Design, synthesis, conformational and molecular docking study of some novel acyl hydrazone based molecular hybrids as antimalarial and antimicrobial agents. Chem. Cent. J. 2017, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Parlar, S.; Erzurumlu, Y.; Ilhan, R.; Kirmizibayrak, P.B.; Alptüzün, V.; Erciyas, E. Synthesis and evaluation of pyridinium-hydrazone derivatives as potential antitumoral agents. Chem. Biol. Drug Des. 2018, 92, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Rollas, S.; Küçükgüzel, S.G. Biological activities of hydrazone derivatives. Molecules 2007, 12, 1910–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popiołek, Ł. Hydrazide-hydrazones as potential antimicrobial agents: Overview of the literature since 2010. Med. Chem. Res. 2017, 26, 287–301. [Google Scholar] [CrossRef] [Green Version]
- Angelova, V.T.; Valcheva, V.; Pencheva, T.; Voynikov, Y.; Vassilev, N.; Mihaylova, R.; Momekov, G.; Shivachev, B. Synthesis, antimycobacterial activity and docking study of 2-aroyl-[1]benzopyrano[4,3-c]pyrazol-4(1H)-one derivatives and related hydrazide-hydrazones. Bioorg. Med. Chem. Lett. 2017, 27, 2996–3002. [Google Scholar] [CrossRef]
- Özdemir, U.O.; Arslan, F.; Hamurcu, F. Synthesis, characterization, antibacterial activities and carbonic anhydrase enzyme inhibitor effects of new arylsulfonylhydrazone and their Ni(II), Co(II) complexes. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2010, 75, 121–126. [Google Scholar] [CrossRef]
- Backes, G.L.; Neumann, D.M.; Jursic, B.S. Synthesis and antifungal activity of substituted salicyl aldehyde hydrazones, hydrazides and sulfohydrazides. Bioorg. Med. Chem. 2014, 22, 4629–4636. [Google Scholar] [CrossRef]
- Tatar, E.; Şenkardeş, S.; Sellitepe, H.E.; Küçükgüzel, Ş.G.; Karaoğlu, Ş.A.; Bozdeveci, A.; Clercq, E.D.; Pannecouque, C.; Hadda, T.B.; Kucukguzel, I. Synthesis, and prediction of molecular properties and antimicrobial activity of some acylhydrazones derived from N-(arylsulfonyl)methionine. Turkish. J. Chem. 2016, 40, 510–534. [Google Scholar] [CrossRef]
- Kadawla, M.; Sinha, J.; Verma, P.K. Microwave assisted synthesis of novel 3-mercapto-4,5-disubstituted 1,2,4-triazole derivatives and evaluation of antimicrobial, antitubercular activity. Pharma. Innov. J. 2018, 7, 41–47. [Google Scholar]
- Nurkenova, O.A.; Satpaevaa, Z.B.; Schepetkin, I.A.; Khlebnikov, A.I.; Turdybekov, K.M.; Seilkhanov, T.M.; Fazylo, S.D. Synthesis and biological activity of hydrazones of o- and p-hydroxybenzoic acids. Spatial structure of 5-bromo-2-hydroxybenzylidene-4-hydroxybenzohydrazide. Russ. J. Gen. Chem. 2017, 87, 2299–2306. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Jalla, A.; Shama, A.A.; Stafford, J.M.; Cao, B. Chromatography-free and eco-friendly synthesis of aryl tosylates and mesylates. Synthesis 2015, 47, 2578–2585. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.M.; Jang, H.J.; Kim, W.J.; Kang, M.G.; Baek, S.C.; Lee, J.P.; Park, D.; Oh, S.R.; Kim, H. Calycosin and 8-O-methylretusin isolated from Maackia amurensis as potent and selective reversible inhibitors of human monoamine oxidase-B. Int. J. Biol. Macromol. 2020, 151, 441–448. [Google Scholar] [CrossRef]
- Lee, J.P.; Kang, M.G.; Lee, J.Y.; Oh, J.M.; Baek, S.C.; Leem, H.H.; Park, D.; Cho, M.L.; Kim, H. Potent inhibition of acetylcholinesterase by sargachromanol I from Sargassum siliquastrum and by selected natural compounds. Bioorg. Chem. 2019, 103043. [Google Scholar] [CrossRef]
- Heo, J.H.; Eom, B.H.; Ryu, H.W.; Kang, M.G.; Park, J.E.; Kim, D.Y.; Kim, J.H.; Park, D.; Oh, S.R.; Kim, H. Acetylcholinesterase and butyrylcholinesterase inhibitory activities of khellactone coumarin derivatives isolated from Peucedanum japonicum Thurnberg. Sci. Rep. 2020, 10, 21695. [Google Scholar] [CrossRef]
- Jeong, G.S.; Kang, M.G.; Han, S.A.; Noh, J.I.; Park, J.E.; Nam, S.J.; Park, D.; Yee, S.T.; Kim, H. Selective inhibition of human monoamine oxidase B by 5-hydroxy-2-methyl-chroman-4-one isolated from an endogenous lichen fungus Daldinia fissa. J. Fungi. 2021, 7, 84. [Google Scholar] [CrossRef]
- Oh, J.M.; Kang, M.G.; Hong, A.; Park, J.E.; Kim, S.H.; Lee, J.P.; Baek, S.C.; Park, D.; Nam, S.J.; Cho, M.L.; et al. Potent and selective inhibition of human monoamine oxidase-B by 4-dimethylaminochalcone and selected chalcone derivatives. Int. J. Biol. Macromol. 2019, 137, 426–432. [Google Scholar] [CrossRef]
- Baek, S.C.; Lee, H.W.; Ryu, H.W.; Kang, M.G.; Park, D.; Kim, S.H.; Cho, M.L.; Oh, S.R.; Kim, H. Selective inhibition of monoamine oxidase A by hispidol. Bioorg. Med. Chem. Lett. 2018, 15, 584–588. [Google Scholar] [CrossRef]
- Son, S.Y.; Ma, J.; Kondou, Y.; Yoshimura, M.; Yamashita, E.; Tsukihara, T. Structure of human monoamine oxidase A at 2.2-a resolution: The control of opening the entry for substrates/inhibitors. Proc. Natl. Acad. Sci. USA 2008, 105, 5739–5744. [Google Scholar] [CrossRef] [Green Version]
- Binda, C.; Wang, J.; Pisani, L.; Caccia, C.; Carotti, A.; Salvati, P.; Edmondson, D.E.; Mattevi, A. Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: Safinamide and coumarin analogs. J. Med. Chem. 2007, 50, 5848–5852. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, M.J.; Greenblatt, H.; Chen, W.; Paz, A.; Dym, O.; Peleg, Y.; Chen, T.; Shen, X.; He, J.; et al. Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations. Acta. Crystallogr. D. Biol. Crystallogr. 2012, 68, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger. Release 2020-4: Glide; Schrödinger, LLC: New York, NY, USA, 2020. [Google Scholar]
- Schrödinger. Release 2020-4: LigPrep; Schrödinger, LLC: New York, NY, USA, 2020. [Google Scholar]
- Schrödinger. Release 2020-4: Protein Preparation Wizard; Epik, Schrödinger, LLC: New York, NY, USA, 2016; Impact, Schrödinger, LLC: New York, NY, 2016; Prime, Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- Khattab, S.N.; Abdel Moneim, S.A.; Bekhit, A.A.; El Massry, A.M.; Hassan, S.Y.; El-Faham, A.; Ali Ahmed, H.E.; Amer, A. Exploring new selective 3-benzylquinoxaline-based MAO-A inhibitors: Design, synthesis, biological evaluation and docking studies. Eur. J. Med. Chem. 2015, 93, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.; Montiel, E.; Ulrich, H.; Paz, C. Selective secretase targeting for Alzheimer’s disease therapy. J. Alzheimers Dis. 2021. [Google Scholar] [CrossRef]
- Mathew, B.; Oh, J.M.; Baty, R.S.; Batiha, G.E.; Parambi, D.G.T.; Gambacorta, N.; Nicolotti, O.; Kim, H. Piperazine-substituted chalcones: A new class of MAO-B, AChE, and BACE-1 inhibitors for the treatment of neurological disorders. Environ. Sci. Pollut. Res. 2021. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-2: QikProp; Schrödinger, LLC: New York, NY, USA, 2021.
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).