1. Introduction
Cancer is a major disease that causes death worldwide [
1,
2], and current traditional strategies against cancer and other diseases of global importance have certain disadvantages such as a lack of tumor selectivity, nonspecific toxicity, and resistance to multiple drugs [
3]. Challenges in conventional drug delivery continue to include high doses, repeat administration, off-target delivery, and serious side effects. [
4,
5]. Therefore, it is necessary to develop new therapies. Among the approaches used in the development of cancer therapies, great expectations have been generated in systems based on living cells [
6], since they have more complex mechanisms that allow them to cross non-permeable barriers, modulate their microenvironment, and reach specific tissues. Due to these unique characteristics, living cells have been considered as a new class of vectors with high specificity and long persistence [
7]. The term to refer to these living cells that are used as carriers is microbots [
6,
8]. Bacteria-based microbots consist of a drug, plasmid, and protein delivery system conjugated with nanostructures and bacteria as a sensor and an actuator that can point and guide the microbot to its destination through gradients of temperature, pH, oxygen, and different chemoattractants [
5,
9]. Nanostructures are carried either within or on the surface of the bacterial membrane for the delivery of therapeutic loads in the cell [
10]. In addition, this approach takes advantage of the intrinsic characteristics of bacteria themselves, which include chemotaxis, magnetotaxis, galvanotaxis, and phototaxis [
9]. Raj and Das demonstrated the therapeutic effect of microbots to target MCF-7 cancer cells. Those Micobots consisted of a combinatorial approach of the probiotic
Lactobacillus spp. with fluorescent cadmium sulfide nanoparticles [
11]. In 2013, Park et al. demonstrated in an in vivo study that the bacteriobots had chemotactic motility and tumor-targeting ability [
4].
Microbots are one of the most effective vehicles for drug delivery systems [
9] since they possess the invasive property of bacteria and can selectively colonize the hypoxic regions of the tumor [
10].
Tumor hypoxia is one of the most important obstacles to conventional cancer treatments [
12,
13,
14] and, although it is an unfavorable scenario, is used by microbots since its low oxygen microenvironment serves as an ideal habitat [
1]. In this study, we show the use of microbots based on Bifidobacteria and magnetic nanoparticles for gene delivery to a tumor in an in vivo model. For this study, NCs of iron oxide fluorescent magnetic nanoparticles, and chitosan (as a protective agent), a DNA plasmid whose gene expression is controlled by a hypoxic microenvironment, were elaborated. The NCs were attached to the Bifidobacteria membrane to form the microbots, which were characterized by AFM and SEM. Subsequently, microbots were injected intravenously through the tail vein to tumor-bearing mice; after 48 h, the organ or tissue with the expression of the transgene was determined. The transgene is highly expressed in tumor tissue. These microbots are biocompatible and comparatively safe.
2. Materials and Methods
2.1. Plasmid
The plasmid used for the elaboration of NCs contains 6 tandem repeats of the consensus binding site of HIF1α and HIF 2α together (CGTGTACGTG) as a unit of hypoxia-inducible factor (HIF) binding site; the binding sequences also called hypoxia response elements (HRE) are upstream of the promoter region of human thymidine kinase (TK)(sequence ID form GenBank M13643.1). The human TK minimal promoter followed by the 6 HREs were synthesized by Integrated DNA Technologies. Both 6HRE and TK sequences were ligated between the NheI and NcoI sites of the plasmid pHRE-Luc. Six HRE sequences selectively induced luciferase gene expression in response to hypoxia. The 6 HRE sequences and pHRE-Luc vector map can be found in
Supplementary Figures S1 and S2.
2.2. Chemicals
Water-soluble chitosan (20 kDa) was purchased from Coyote Foods (Saltillo, Coah, MX), and pentasodium tripolyphosphate (TPP) was purchased from Sigma Aldrich (St. Louis, MO, USA). They were weighed and resuspended in MilliQ water at concentrations of 2 mg/mL and 0.86 mg/mL, respectively. The pH values of chitosan and TPP were measured and adjusted to pH 5.5 and 3, respectively.
2.3. Fluorescent Magnetic Nanoparticles (FMNPs)
The FMNPs had a size of 75 nm and were composed of an 8 nm core of superparamagnetic iron oxide (SPIO) covered with a biocompatible silica layer and a covalently bound organic fluorophore, whose spectrum is similar to the fluorochrome cyanine 5.5. The FMNPs were purchased from Nvigen (Sunnyvale, CA, USA).
2.4. NCs Production and Characterization
The NCs were developed based on the technique of ionic gelation described by Calvo and Cols (1997), in which the positively charged chitosan amine groups interacted with the negatively charged groups of TPP and the plasmid. Seven hundred and fifty µL of chitosan (1.5 mg) and 750 µL of PBS were added in the tube labeled as positive, while 5000 ng of FMNPs, 50 µg of pHRE-Luc, 187.5 µL of TPP, and 1237.5 µL of PBS were added in the tube labeled as negative. Content of the negative tube was added dropwise to the positive tube under constant shaking; then, they were incubated for 1 h at room temperature with a shaking of 950 rpm. Spontaneous formation of NCs occurred because of the intermolecular bonds between the negative and positive charges. The NCs consisted of the covalent attachment of nanoparticles, chitosan, pHRE-Luc, and TPP.
Size and zeta potential of NCs were determined by dynamic light scattering (DLS) by the Zetasizer apparatus (ZS90, Malvern), and Zetasizer Software version 7.11 was used. Refractive index (RI) and absorption used for NCs measurements were 1.330 and 0.001, respectively (chitosan values), while for the dispersant (water) the following values were used: RI of 1.330, a viscosity of 0.8872 cP, and a dielectric constant of 78.5. The ZS90 equipment has a red laser at 4 mW 632.8 nm. Results obtained from the three measurements are reported in a size and zeta-potential graph.
2.5. Microbot Production and Characterization
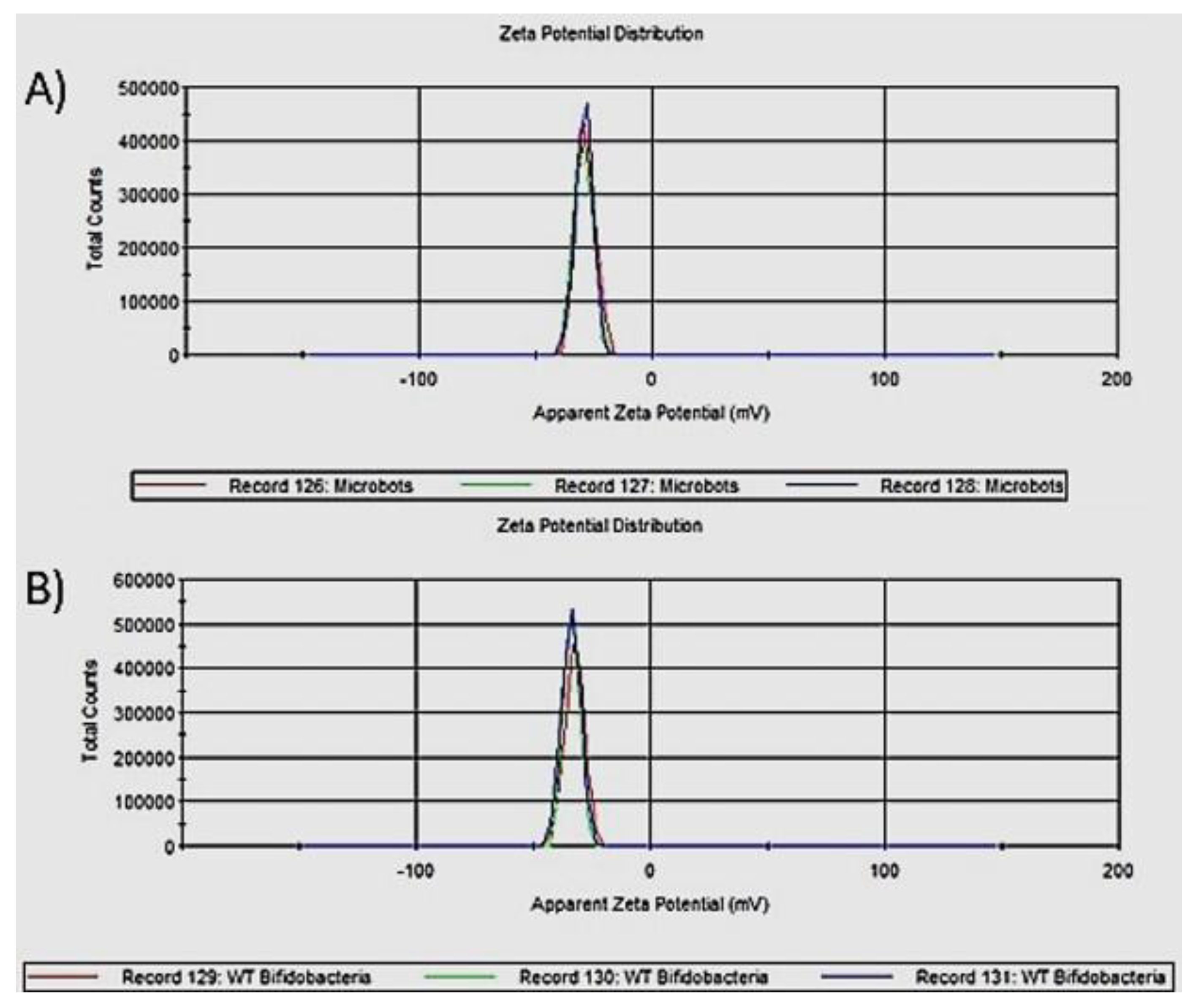
For the development of the microbots, the combination of Bifidobacteria of the strain Bifidobacterium breve donated by the Instituto de Productos Lácteos de Asturias (IPLA) and NCs was used (the NCs were prepared as previously explained). Seven hundred and fifty µL of chitosan (1.5 mg) were added in the tube labeled as positive, and 60 μL of NCs, 1 × 109 Bifidobacteria (resuspended in 100 μL of saline solution), 93.75 μL of TPP, and 93.75 μL of genipin 0.1 M (acting as cross-linking agents) were added in the tube labeled as negative. Content of the negative tube was added dropwise to the positive tube under constant shaking; then, they were incubated at 4 °C for 1 h with a shaking of 950 rpm. After that, they were passed through membrane filters of 1 µm to discard free NCs not adhered to microbot membrane. The surface charge of the microbots and Bifidobacterium breve wild-type were evaluated by dynamic light scattering (DLS) by the Zetasizer apparatus (ZS90, Malvern) and Zetasizer Software version 7.11. Refractive index (RI) and absorption used for measurements were 1.450 and 0.001, respectively (protein values), while for the dispersant (water) the following values were used: RI of 1.330, a viscosity of 0.8872 cP, and a dielectric constant of 78.5. Results obtained from the three measurements are reported in a zeta potential graph. For AFM measurements, 5 µL of microbots and Bifidobacterium breve wild-type were placed on a slide and allowed to dry for a few minutes. After that, the reading method was semi-contact topography, using an RTESPA probe (Bruker, Massachusetts) with the cantilever Olympus AC240TS antimony doped silicon with a length of 125 μm and a resonance frequency of 140 kHz. Measurements were performed using a model NTEGRA atomic force microscope (NT-MDT). The WSxM 5.0 Program was used for image analysis. SEM analysis was performed on JEOL Model JSM-6390LV at 15 KV to obtain finer details of microbots. Twenty microliters of microbots and 20 µL of the 1% fixative solution (0.1 M phosphate buffer and 25% glutaraldehyde) were added on a coverslip and allowed to fix for 30 min at 4 °C. Remaining liquid was removed and dried to visualize the sample in SEM.
2.6. In Vivo Studies
To generate tumor-bearing mice, tumor implantation was carried out in C57BL/6 mice using the B16F10 line. In vivo, the intradermal implant of B16-F10 cells in C57BL/6 mice results in an aggressively growing solid tumor. The mouse melanoma cells line B16F10 were grown in DMEM medium supplemented with 10% fetal bovine serum and 1% antifungal antibiotic at 37 °C in 5% CO2. Female C57BL/6 mice (6–8 weeks old) from Harlan Laboratories (Indianapolis, IN, USA) were injected with 1 × 105 B16F10 cells subcutaneously into the thigh right; after 15–20 days, a solid tumor growth was observed. After tumor-cell implantation, the mice were divided into four groups of three mice per group (n = 3) and treatments (microbots) were injected IV into each mouse through the tail vein. A neodymium magnet with a 3/8” diameter and intensity of 3T was placed in the tumor area for 1 h to target microbots to the tumor; 48 h later all mice were sacrificed. Subsequently for the autopsy, the organs (spleen, heart, liver, kidney, lung, and tumor) were removed. A part was plated in agar cMRS to determine the biodistribution and the other part to analyze by fluorescence and luminescence. This study was approved by the Comité de Ética de Investigación de Bienestar Animal of Biological Sciences Faculty (CEIBA) of the Autonomous University of Nuevo Leon (UANL). All animal-handling procedures were performed according to the Mexican Official Standard NOM-062-ZOO-1999, technical specifications for the production care, and use of laboratory animals.
2.7. Fluorescence and Luminescence Analysis
NCs loaded into the membrane of the microbot carry an organic fluorophore with which the location of the nanoparticles can be determined. Fifty microliters of the supernatant of the lysate of each organ were taken and mixed with 100 μL of physiological solution, and then a150 μL sample was placed in a black 96 well flat bottom plate from Costar Corning (Corning, NY, USA). The fluorescence spectrum was analyzed by a Varioskan Lux from ThermoFisher Scientific (Waltham, MA, USA). The emission and excitation lengths used were those established by the manufacturer: 673 nm and 695 nm, respectively. To analyze the luciferase activity, the ONE-Glo Luciferase assay system kit from Promega (Madison, WI, USA) was used according to the manufacturer’s recommendations. A total of 150 μL of the lysate was used and analyzed by a Varioskan Lux luminometer from ThermoFisher Scientific.
The fluorescence and luminescence results were normalized with respect to their protein concentrations. The data are indicated as the relative fluorescence units (RFU/mg of total protein) and relative light units (RLU/mg of total protein).
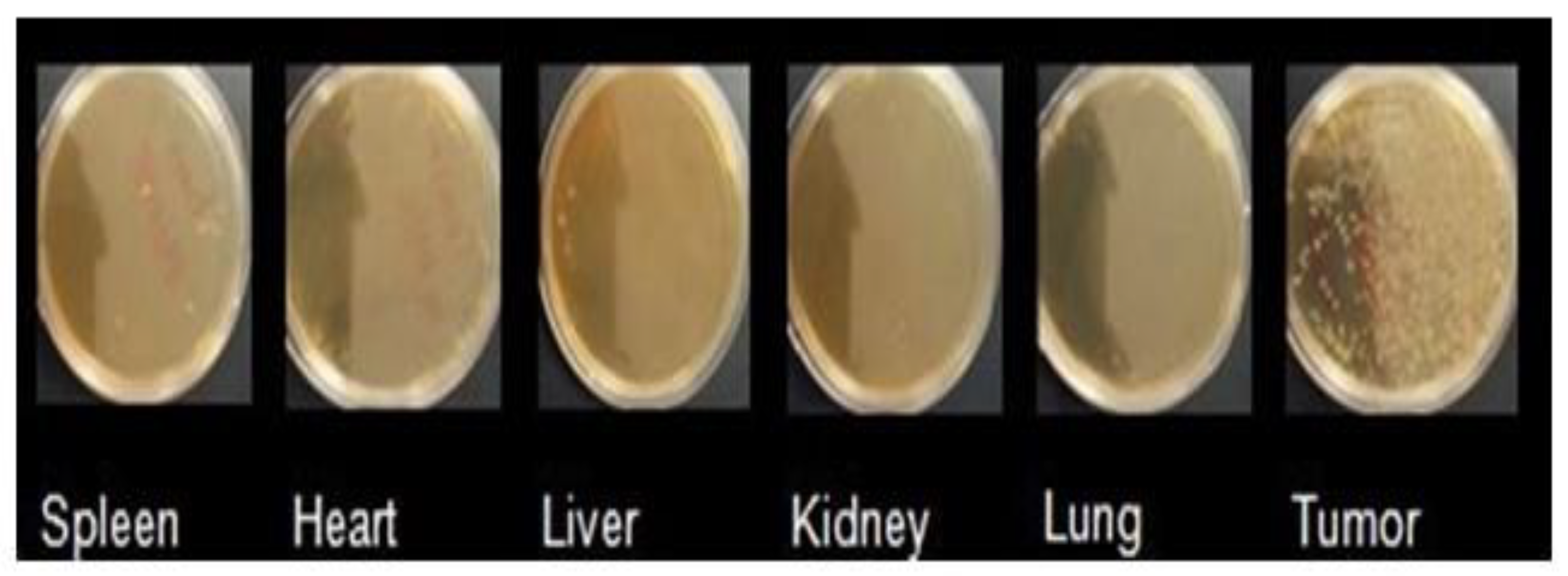
2.8. Microbot Biodistribution
Organs of mice sacrificed at 48 h were macerated and plated in agar cMRS with antibiotic. Agar cMRS plates were incubated at 37 °C under anaerobic conditions for 24 h.
2.9. Statistical Analysis
All data are presented as the mean ± standard error of the mean (SEM). Statistical and graphic analysis was carried out using Graph Pad Prism version 6.0 software. The normality of the data was verified, and subsequently, a post ANOVA test was performed for the multiple comparison of means by the Tukey test. Significance was defined as P,0.05 (* p < 0.05, ** p < 0.01, and *** p < 0.001).
4. Discussion
Current strategies to fight cancer usually present certain disadvantages since they are not selective for cancer cells and affect normal tissues. That is why we are looking for new alternatives [
3,
15]. In cancer, the use of microbots based on anaerobic bacteria as specific transporters targeted to tumor tissues has been explored since they have more complex mechanisms that allow them to reach specific tissues, cross nonpermeable barriers easily, and modulate their microenvironment [
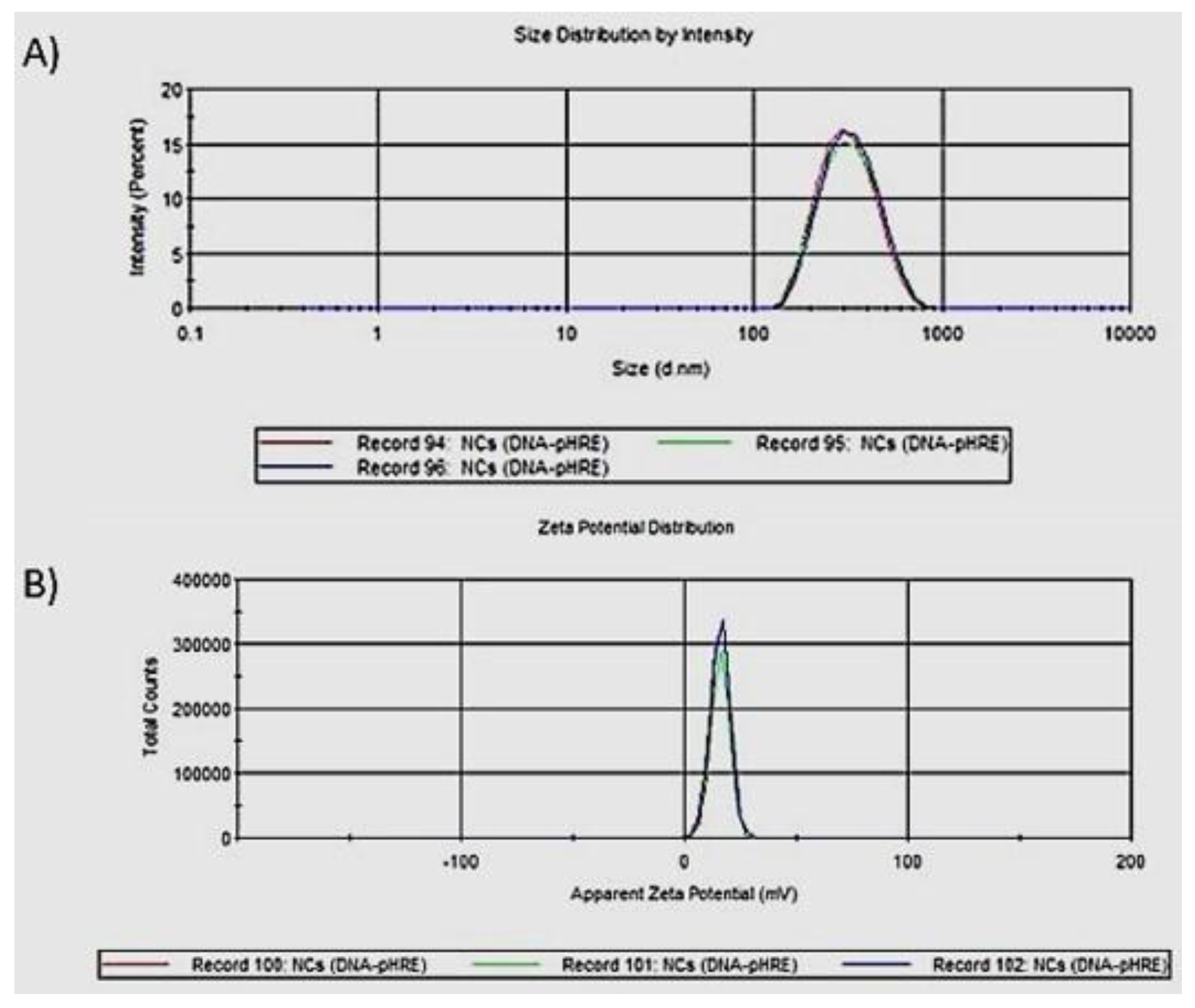
7],. In this study, we developed microbots based on Bifidobacteria and fluorescent magnetic nanoparticles coated with chitosan and pHRE-Luc to be used as gene carriers. We showed the simultaneous use of Bifidobacterium with NCs (microbots) to deliver pDNA in an in vivo model and the transgene was highly expressed in tumor tissue compared with other organs. The microbot does not express the transgene (bioluminescent gene); rather, it acts as a delivery vector that transfects the tumor cells and transfers the transgene for expression by the mammalian cells. The bioluminescent gene is loaded on the NCs, which are carried on the bacterial surface. Herein, we take advantage of the anaerobic bacterial habit to combine with functional NPs and the hypoxic promoter, which induce high gene reporter expression in response to hypoxia and improve the efficiency of treatment in the tumor hypoxic environment. These microbots are biocompatible and comparatively safe. NCs that were a size of 200–300 nm were obtained (
Figure 1A); this size is ideal for transfections in tumor therapy since they can extravasate to the tumor area [
16]. Zeta potential of +16 mV was obtained (
Figure 1B), and a positive potential represents an advantage in targeted delivery therapies and transfection efficiency since the cell membrane is negatively charged, so cationic NCs can have a strong electrostatic interaction with the cell, which results in rapid endocytosis and allows for efficient transfection [
17]. An excessive positive charge can cause cytotoxicity, nonspecific interactions, and blood aggregation [
18]. The magnetic NPs used for the elaboration of NCs in this study are composed of an SPIO core, which is a material used in clinical magnetic resonance diagnoses. The NPs shell was made of silica, a biocompatible material component similar to those used in daily supplements of calcium, and the fluorescence was given by a covalently bound organic fluorophore that has been approved by the Food and Drug Administration (FDA) [
19]. To protect the pDNA, chitosan was used, which is a biocompatible and nontoxic biopolymer, whose use has been approved by the FDA and has already been used in various food and medical areas [
20].
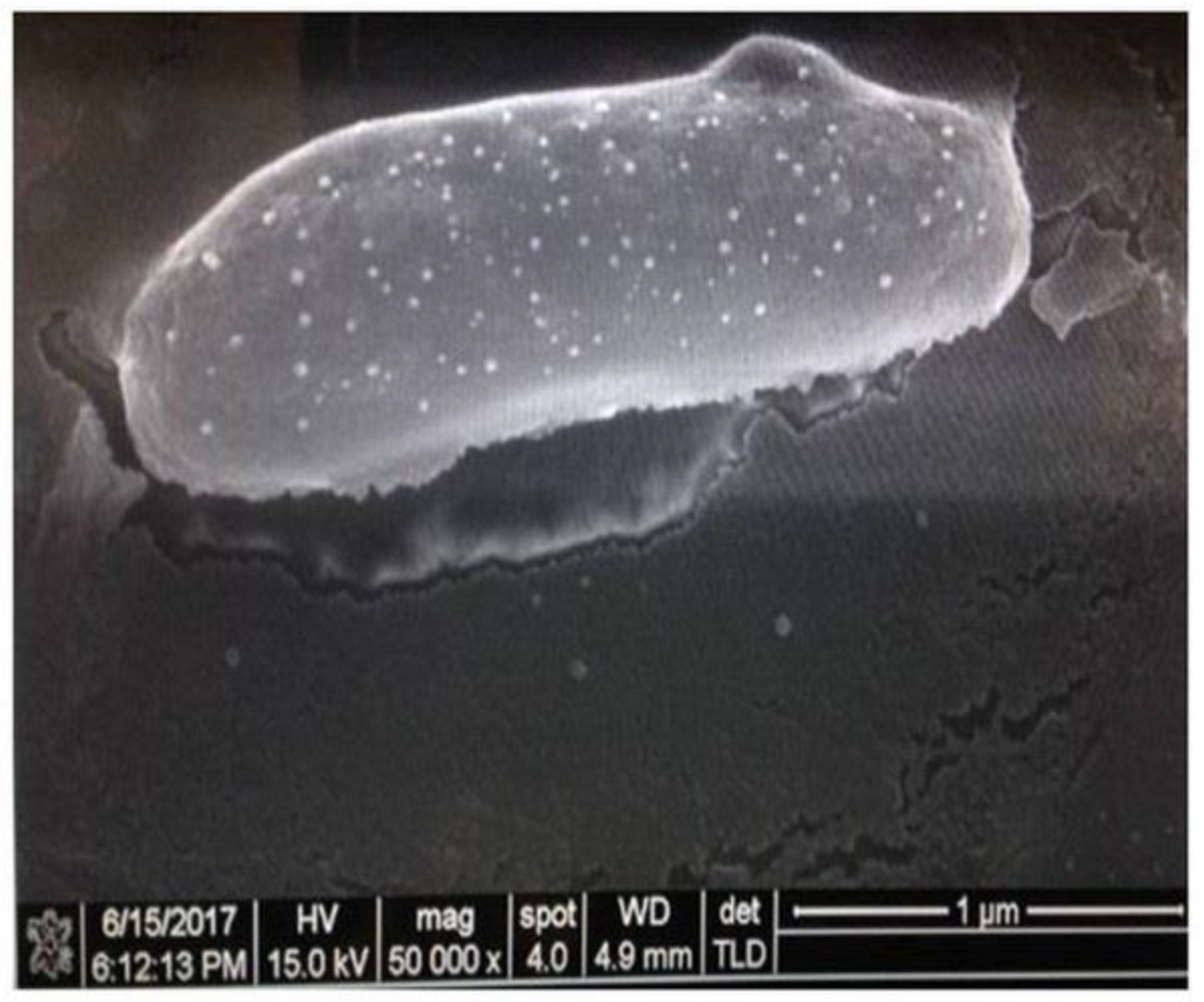
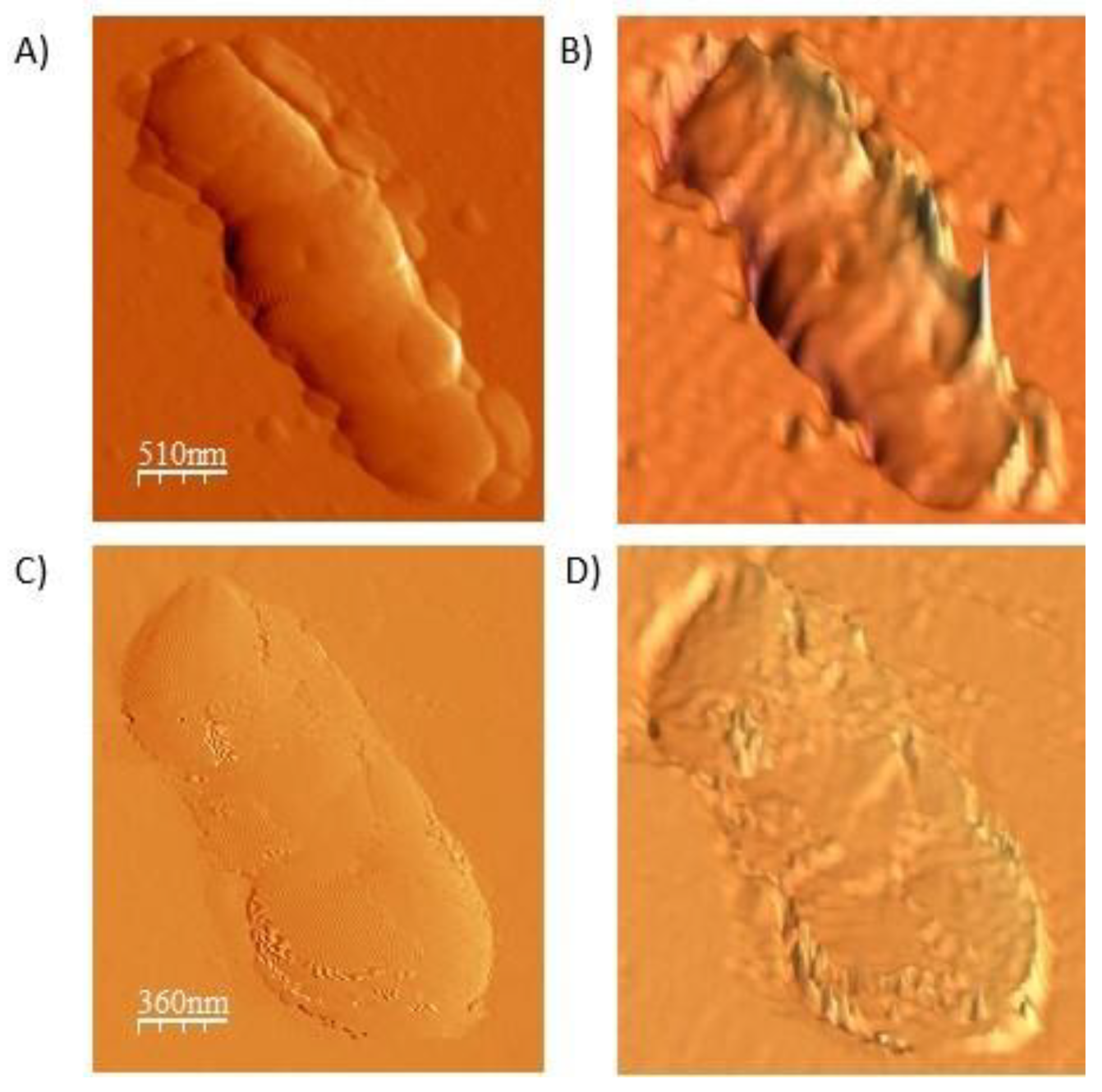
For the determination of the shape and morphology of the microbots, AFM was used, and the results confirmed that the NCs uniformly adhered to the bacterium’s surface (
Figure 3). In addition, this result was validated by SEM, and microbots with a diameter between 1–2 µm were observed (
Figure 2).
The in vivo distributions of microbots and reporter gene expression was determined in a tumor-bearing mouse model two days after IV administration by luminescence and fluorescence.
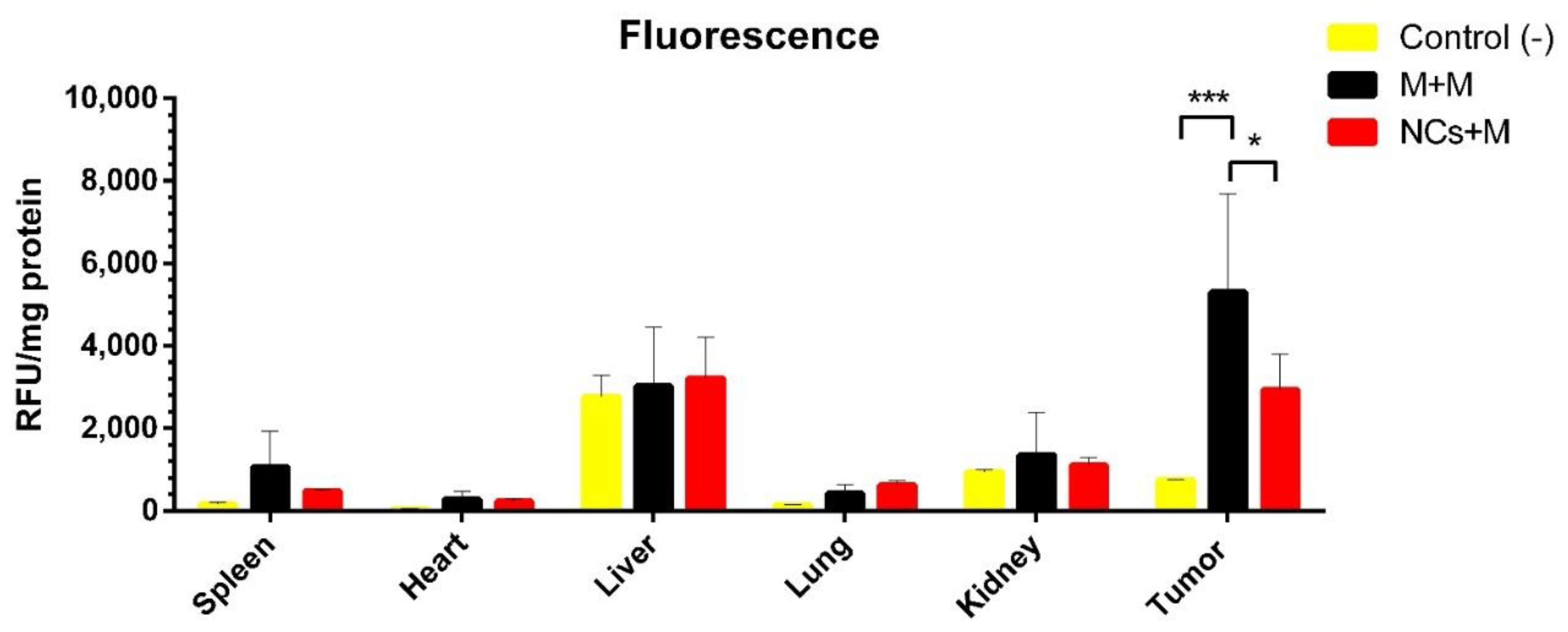
Greater red fluorescence was obtained in the tumor tissue than in the other organs (spleen, heart, liver, lung, kidney), indicating the presence of microbots in the tumor area. In addition, when comparing the specific transport efficiency of the two systems, r the microbots were better than the NCs alone group (
Figure 5). The fluorescence obtained from the tumor tissue does not compare with that of the other organs; however, minimum fluorescence was observed in the liver and kidney since they play an important role in the excretion of nanoparticles and substances [
21,
22]. Multiple reports have indicated that tumor blood vessels have structural abnormalities and are interrupted by circular fenestrations with a size of 400 nm to 1–2 microns which allows microbots to be extravasated to the tumor cells [
12,
14,
23] since microbots can colonize hypoxic areas of the tumor due to their complex mechanisms [
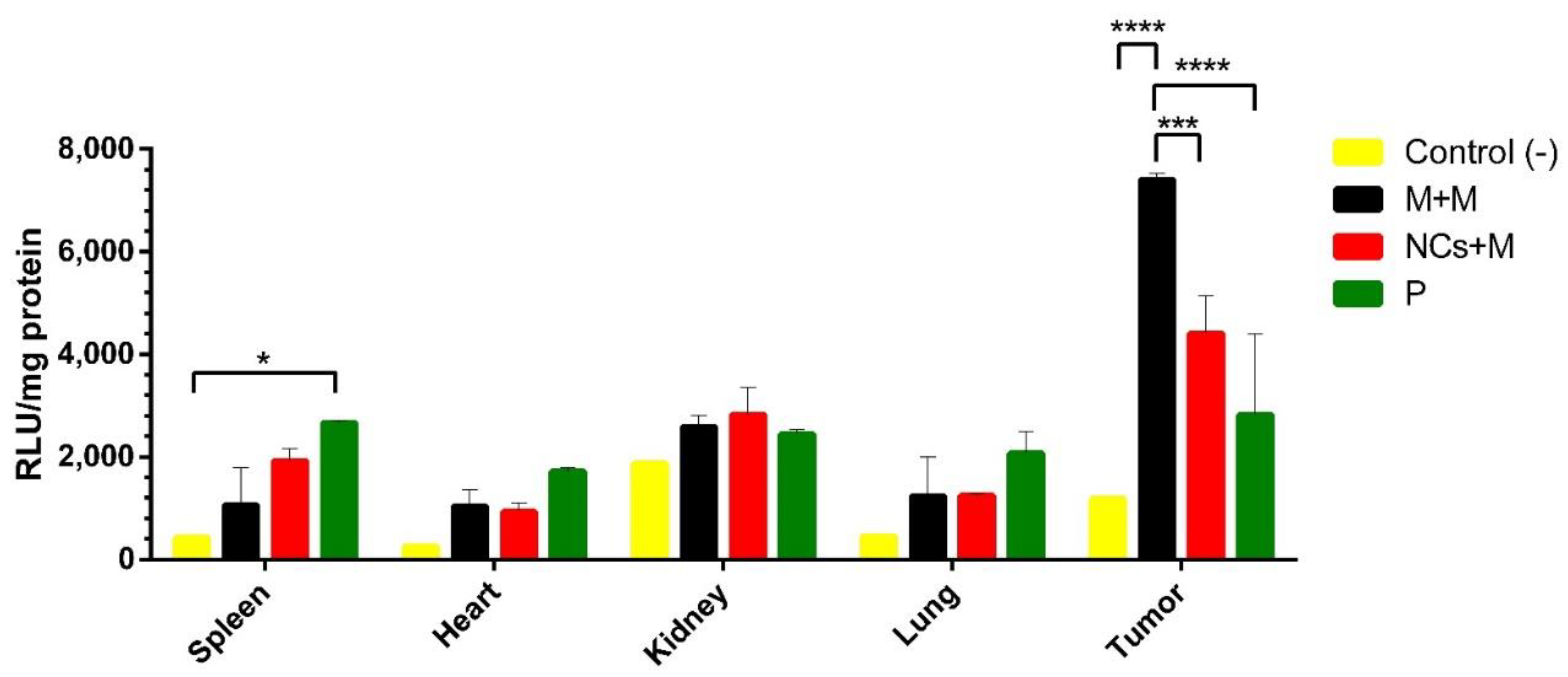
7]. Once in the tumor area, the NCs adhered to the bacterial membrane were released (due to the low pH in the tumor area) and were endocytosed by the tumor cells for the gene expression of the pHRE-Luc vector. Luciferase expression was measured 48 h after IV injection of the microbots that contained 50 µg of the pHRE-Luc vector. We found higher luciferase expression in the tumor tissue (
Figure 6); this is attributed to the hypoxic promoter and the presence of hypoxic response elements that enhance the expression of an adjacent gene (luciferase) under hypoxic conditions [
24,
25].
Also, the microbots were compared with two other gene delivery systems: NCs alone and the naked plasmid, and the microbots showed significant differences in gene delivery efficiency.
Bifidobacterium breve was used as a vehicle to load NCs and transport them into tumor tissues because it is a non-pathogenic, non-immunogenic bacterium; it is anaerobic and can colonize hypoxic tissues rather than normal tissues and organs. Also, its bacterial membrane rich in peptidoglycans allows chemical modifications to load NCs on its surface [
26].
Microbots proved to be a very specific and efficient gene delivery system since the expression of the delivered gene is specific to tumor tissues due to the following reasons: (1) the ability of the microbots to only colonize in tumors and not in other tissues since the bacterium only survives under the ideal conditions, such as hypoxia; (2) the magnetic NCs attached to the microbot membrane are directed by a magnet to the tumor tissue, improving specific transport efficiency; (3) chitosan was used to protect DNA from degradation; and (4) the low levels of oxygen in the solid tumors microenvironment generate the accumulation of HIFs, which can enhance the expression of our pHRE-Luc vector. However, in tissues with normal oxygen, this level accumulation of HIFs does not occur (compared to hypoxic tissues). All the components are biocompatible and safe, and the therapeutic use of Bifidobacteria in humans is in the clinical phase. Our results show that the microbots are a highly specific and efficient gene delivery system and can be used to carry and express genes in specific tissues (tumor tissues). This system can be used for drugs and therapeutic gene delivery due to its specificity, therefore reducing collateral damage to normal cells.