Protein-Engineered Polymers Functionalized with Antimicrobial Peptides for the Development of Active Surfaces
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Genetic Constructions
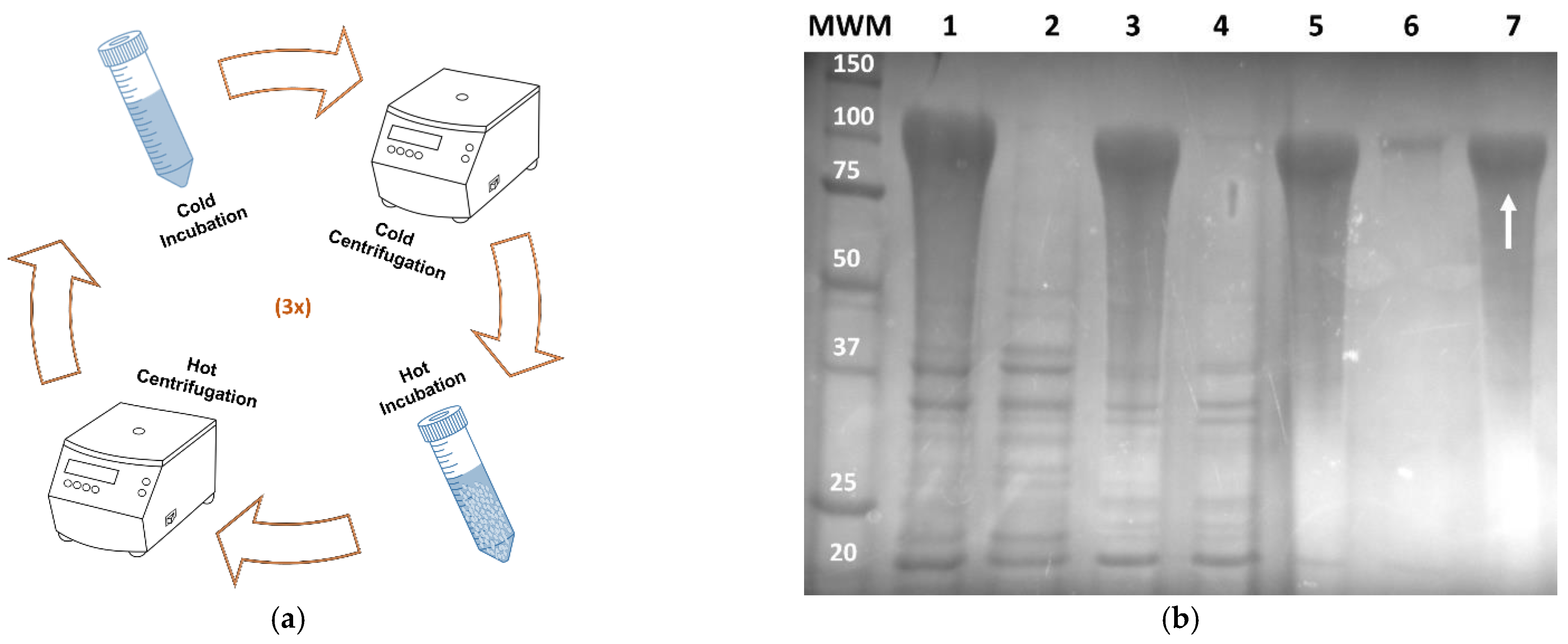
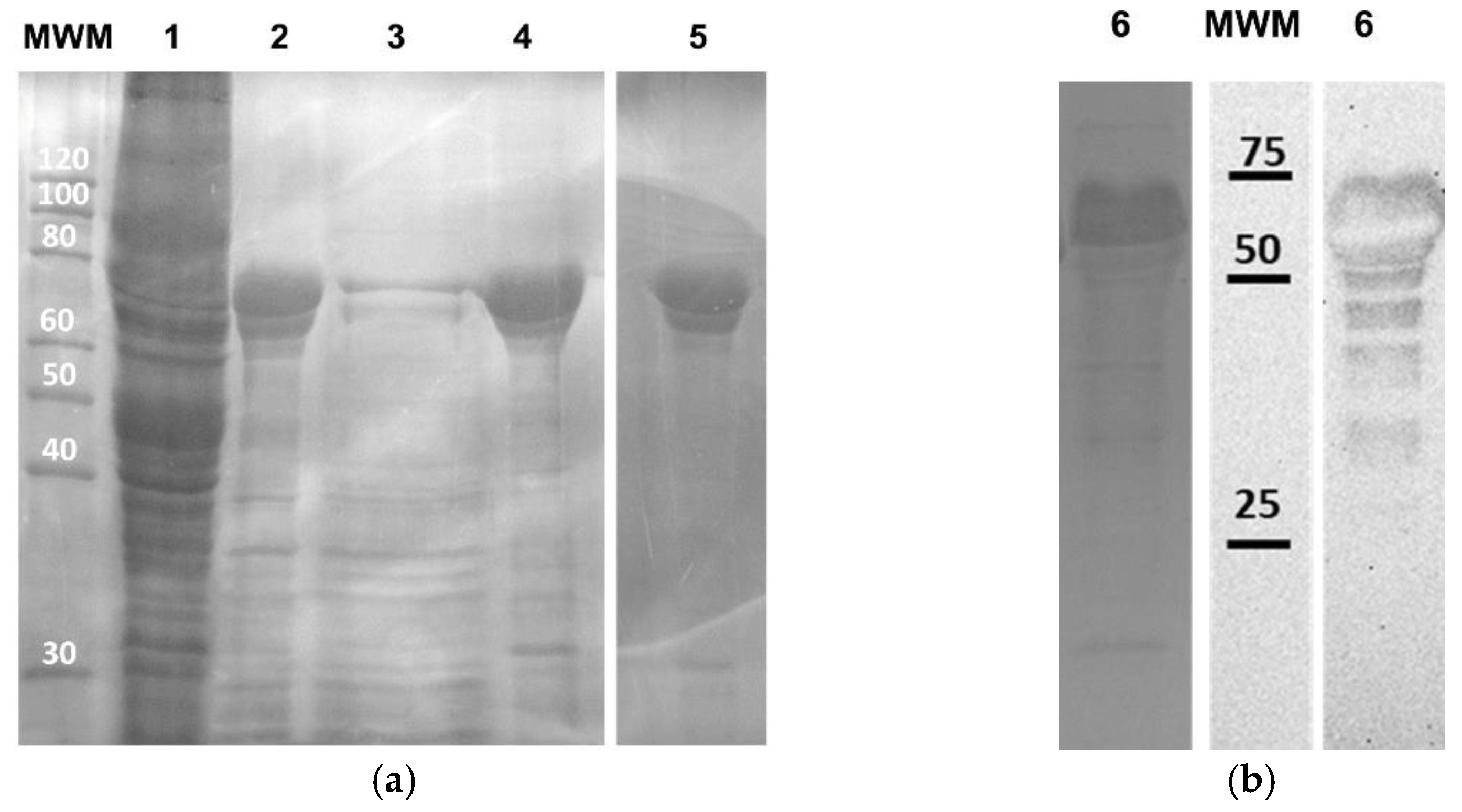
2.2. Protein Production and Purification
2.3. Western Blot Analysis of AMP-SELP
2.4. Preparation of Free-Standing Films
2.5. Structural Characterization of AMP-SELP Films
2.6. Evaluation of Antimicrobial Activity
2.7. Statistical Analysis
3. Results and Discussion
3.1. Production and Purification of Recombinant AMP-A200 and AMP-SELP
3.2. Secondary Structure Analysis of AMP-SELP Films
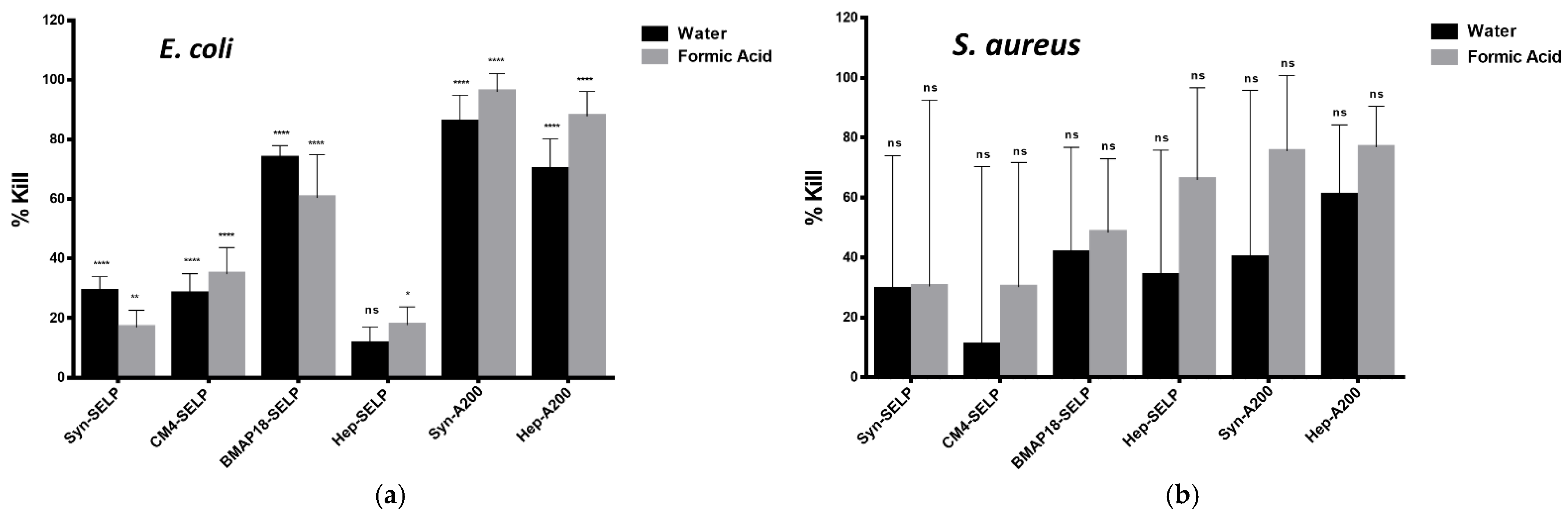
3.3. Antimicrobial Activity of AMP-ELR and AMP-SELP Films
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Y.J.; Holmberg, A.L.; Olsen, B.D. Artificially engineered protein polymers. Annu. Rev. Chem. Biomol. Eng. 2017, 8, 549–575. [Google Scholar] [CrossRef]
- Abascal, N.C.; Regan, L. The past, present and future of protein-based materials. R. Soc. Open Biol. 2018, 8, 180113. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.-G.; Pan, F.; Xia, X.-X. Synthetic biology for protein-based materials. Curr. Opin. Biotechnol. 2020, 65, 197–204. [Google Scholar] [CrossRef]
- Rabotyagova, O.S.; Cebe, P.; Kaplan, D.L. Protein-based block copolymers. Biomacromolecules 2011, 12, 269–289. [Google Scholar] [CrossRef]
- Machado, R.; Da Costa, A.; Sencadas, V.; Pereira, A.M.; Collins, T.; Rodríguez-Cabello, J.C.; Lanceros-Méndez, S.; Casal, M. Exploring the properties of genetically engineered silk-elastin-like protein films. Macromol. Biosci. 2015, 15, 1698–1709. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.; Azevedo-Silva, J.; Correia, C.; Collins, T.; Arias, F.J.; Rodriguez-Cabello, J.C.; Casal, M. High level expression and facile purification of recombinant silk-elastin-like polymers in auto induction shake flask cultures. AMB Express 2013, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Acosta, S.; Ibañez-Fonseca, A.; Aparicio, C.; Rodríguez-Cabello, J.C. Antibiofilm coatings based on protein-engineered polymers and antimicrobial peptides for preventing implant-associated infections. Biomater. Sci. 2020, 8, 2866–2877. [Google Scholar] [CrossRef]
- da Costa, A.; Pereira, A.M.; Sampaio, P.; Rodríguez-Cabello, J.C.; Gomes, A.C.; Casal, M.; Machado, R. Protein-Based Films Functionalized with a Truncated Antimicrobial Peptide Sequence Display Broad Antimicrobial Activity. ACS Biomater. Sci. Eng. 2021, 7, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.C.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Antimicrobial functionalized genetically engineered spider silk. Biomaterials 2011, 32, 4255–4266. [Google Scholar] [CrossRef]
- Franco, A.R.; Fernandes, E.M.; Rodrigues, M.T.; Rodrigues, F.J.; Gomes, M.E.; Leonor, I.B.; Kaplan, D.L.; Reis, R.L. Antimicrobial coating of spider silk to prevent bacterial attachment on silk surgical sutures. Acta Biomater. 2019, 99, 236–246. [Google Scholar] [CrossRef]
- Acosta, S.; Quintanilla, L.; Alonso, M.; Aparicio, C.; Rodríguez-Cabello, J.C. Recombinant AMP/Polypeptide Self-Assembled Monolayers with Synergistic Antimicrobial Properties for Bacterial Strains of Medical Relevance. ACS Biomater. Sci. Eng. 2019, 5, 4708–4716. [Google Scholar] [CrossRef]
- da Costa, A.; Machado, R.; Ribeiro, A.; Collins, T.; Thiagarajan, V.; Neves-Petersen, M.T.; Rodríguez-Cabello, J.C.; Gomes, A.C.; Casal, M. Development of elastin-like recombinamer films with antimicrobial activity. Biomacromolecules 2015, 16, 625–635. [Google Scholar] [CrossRef]
- da Costa, A.; Pereira, A.M.; Gomes, A.C.; Rodríguez-Cabello, J.C.; Sencadas, V.; Casal, M.; Machado, R. Single step fabrication of antimicrobial fibre mats from a bioengineered protein-based polymer. Biomed. Mater. 2017, 12, 045011. [Google Scholar] [CrossRef]
- Yigit, S.; Dinjaski, N.; Kaplan, D.L. Fibrous proteins: At the crossroads of genetic engineering and biotechnological applications. Biotechnol. Bioeng. 2016, 113, 913–929. [Google Scholar] [CrossRef]
- Ibáñez-Fonseca, A.; Flora, T.; Acosta, S.; Rodríguez-Cabello, J.C. Trends in the design and use of elastin-like recombinamers as biomaterials. Matrix Biol. 2019, 84, 111–126. [Google Scholar] [CrossRef]
- Girotti, A.; Fernández-Colino, A.; López, I.M.; Rodríguez-Cabello, J.C.; Arias, F.J. Elastin-like recombinamers: Biosynthetic strategies and biotechnological applications. Biotechnol. J. 2011, 6, 1174–1186. [Google Scholar] [CrossRef]
- Urry, D.W.; Hayes, L.C.; Gowda, D.C.; Harris, C.M.; Harris, R.D. Reduction-driven polypeptide folding by the ΔTt mechanism. Biochem. Biophys. Res. Commun. 1992, 188, 611–617. [Google Scholar] [CrossRef]
- Rodriguez-Cabello, J.C. Smart elastin-like polymers. In Biomaterials. Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 2004. [Google Scholar] [CrossRef]
- Megeed, Z.; Cappello, J.; Ghandehari, H. Genetically engineered silk-elastinlike protein polymers for controlled drug delivery. Adv. Drug Deliv. Rev. 2002, 54, 1075–1091. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Genet. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2009, 1788, 1687–1692. [Google Scholar] [CrossRef]
- Lai, Y.; Gallo, R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial peptides: Features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Gao, T.; Zhang, N.; He, J.; Wu, F. Covalent immobilization of DJK-5 peptide on porous titanium for enhanced antibacterial effects and restrained inflammatory osteoclastogenesis. Colloids Surf. B Biointerfaces 2021, 202, 111697. [Google Scholar] [CrossRef]
- Song, D.W.; Kim, S.H.; Kim, H.H.; Lee, K.H.; Ki, C.S.; Park, Y.H. Multi-biofunction of antimicrobial peptide-immobilized silk fibroin nanofiber membrane: Implications for wound healing. Acta Biomater. 2016, 39, 146–155. [Google Scholar] [CrossRef]
- Boix-Lemonche, G.; Guillem-Marti, J.; Lekka, M.; D’Este, F.; Guida, F.; Manero, J.M.; Skerlavaj, B. Membrane perturbation, altered morphology and killing of Staphylococcus epidermidis upon contact with a cytocompatible peptide-based antibacterial surface. Colloids Surf. B Biointerfaces 2021, 203, 111745. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.G.; Chen, X.; Astleford-Hopper, K.; He, J.; Mullikin, A.F.; Mansky, K.C.; Aparicio, C. Antimicrobial and enzyme-responsive multi-peptide surfaces for bone-anchored devices. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 125, 112108. [Google Scholar] [CrossRef]
- Onaizi, S.A.; Leong, S.S. Tethering antimicrobial peptides: Current status and potential challenges. Biotechnol. Adv. 2011, 29, 67–74. [Google Scholar] [CrossRef]
- Bagheri, M.; Beyermann, M.; Dathe, M. Immobilization Reduces the Activity of Surface-Bound Cationic Antimicrobial Peptides with No Influence upon the Activity Spectrum. Antimicrob. Agents Chemother. 2009, 53, 1132. [Google Scholar] [CrossRef]
- Hasan, J.; Crawford, R.J.; Ivanova, E.P. Antibacterial surfaces: The quest for a new generation of biomaterials. Trends Biotechnol. 2013, 31, 295–304. [Google Scholar] [CrossRef]
- Alves, D.; Olívia Pereira, M. Mini-review: Antimicrobial peptides and enzymes as promising candidates to functionalize biomaterial surfaces. Biofouling 2014, 30, 483–499. [Google Scholar] [CrossRef]
- Kazemzadeh-Narbat, M.; Cheng, H.; Chabok, R.; Alvarez, M.M.; de la Fuente-Nunez, C.; Phillips, K.S.; Khademhosseini, A. Strategies for antimicrobial peptide coatings on medical devices: A review and regulatory science perspective. Crit. Rev. Biotechnol. 2021, 41, 94–120. [Google Scholar] [CrossRef] [PubMed]
- Chouirfa, H.; Bouloussa, H.; Migonney, V.; Falentin-Daudre, C. Review of titanium surface modification techniques and coatings for antibacterial applications. Acta Biomater. 2019, 83, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Ahmadabadi, H.Y.; Yu, K.; Kizhakkedathu, J.N. Surface modification approaches for prevention of implant associated infections. Colloids Surf. B Biointerfaces 2020, 193, 111116. [Google Scholar] [CrossRef]
- Atefyekta, S.; Pihl, M.; Lindsay, C.; Heilshorn, S.C.; Andersson, M. Antibiofilm elastin-like polypeptide coatings: Functionality, stability, and selectivity. Acta Biomater. 2019, 83, 245–256. [Google Scholar] [CrossRef]
- Tiller, J.C.; Liao, C.-J.; Lewis, K.; Klibanov, A.M. Designing surfaces that kill bacteria on contact. Proc. Natl. Acad. Sci. USA 2001, 98, 5981–5985. [Google Scholar] [CrossRef]
- Lewis, K.; Klibanov, A.M. Surpassing nature: Rational design of sterile-surface materials. Trends Biotechnol. 2005, 23, 343–348. [Google Scholar] [CrossRef]
- da Costa, A.; Pereira, A.M.; Gomes, A.C.; Rodriguez-Cabello, J.C.; Casal, M.; Machado, R. Production of bioactive hepcidin by recombinant DNA tagging with an elastin-like recombinamer. New Biotechnol. 2018, 46, 45–53. [Google Scholar] [CrossRef]
- Machado, R.; Ribeiro, A.J.; Padrão, J.; Silva, D.; Nobre, A.; Teixeira, J.; Arias, F.; Cunha, A.M.; Rodríguez-Cabello, J.C.; Casal, M. Exploiting the sequence of naturally occurring elastin: Construction, production and characterization of a recombinant thermoplastic protein-based polymer. J. Nano Res. 2009, 6, 133–145. [Google Scholar] [CrossRef]
- Machado, R.; Bessa, P.C.; Reis, R.L.; Rodriguez-Cabello, J.C.; Casal, M. Elastin-based nanoparticles for delivery of bone morphogenetic proteins. In Nanoparticles in Biology and Medicine. Methods in Molecular Biology (Methods and Protocols); Humana Press: Totowa, NJ, USA, 2012. [Google Scholar] [CrossRef]
- Freire, D.O.; da Cunha, N.B.; Leite, M.L.; Kostopoulos, A.G.; da Silva, S.N.; de Souza, A.C.; Nolasco, D.O.; Franco, O.L.; Mortari, M.R.; Dias, S.C. Wasp venom peptide, synoeca-MP, from Synoeca surinama shows antimicrobial activity against human and animal pathogenic microorganisms. Pept. Sci. 2019, 112, e24141. [Google Scholar] [CrossRef]
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar] [CrossRef]
- Krause, A.; Neitz, S.; Mägert, H.-J.; Schulz, A.; Forssmann, W.-G.; Schulz-Knappe, P.; Adermann, K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000, 480, 147–150. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Min, C.; Sang, M.; Han, Y.Y.; Ma, X.; Xue, X.Q.; Zhang, S.Q. A cationic amphiphilic peptide ABP-CM4 exhibits selective cytotoxicity against leukemia cells. Peptides 2010, 31, 1504–1510. [Google Scholar] [CrossRef]
- Li, B.-C.; Zhang, S.-Q.; Dan, W.-B.; Chen, Y.-Q.; Cao, P. Expression in Escherichia coli and purification of bioactive antibacterial peptide ABP-CM4 from the Chinese silk worm, Bombyx mori. Biotechnol. Lett. 2007, 29, 1031–1036. [Google Scholar] [CrossRef]
- Mardirossian, M.; Pompilio, A.; Crocetta, V.; De Nicola, S.; Guida, F.; Degasperi, M.; Gennaro, R.; Di Bonaventura, G.; Scocchi, M. In vitro and in vivo evaluation of BMAP-derived peptides for the treatment of cystic fibrosis-related pulmonary infections. Amino Acids 2016, 48, 2253–2260. [Google Scholar] [CrossRef]
- Lee, E.K.; Kim, Y.-C.; Nan, Y.H.; Shin, S.Y. Cell selectivity, mechanism of action and LPS-neutralizing activity of bovine myeloid antimicrobial peptide-18 (BMAP-18) and its analogs. Peptides 2011, 32, 1123–1130. [Google Scholar] [CrossRef]
- Lombardi, L.; Maisetta, G.; Batoni, G.; Tavanti, A. Insights into the antimicrobial properties of hepcidins: Advantages and drawbacks as potential therapeutic agents. Molecules 2015, 20, 6319–6341. [Google Scholar] [CrossRef]
- Machado, R.; Da Costa, A.; Sencadas, V.; Garcia-Arévalo, C.; Costa, C.M.; Padrao, J.; Gomes, A.; Lanceros-Méndez, S.; Rodríguez-Cabello, J.C.; Casal, M. Electrospun silk-elastin-like fibre mats for tissue engineering applications. Biomed. Mater. 2013, 8, 065009. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.R.; Palma Kimmerling, E.; Silva, C.; Rodrigues, F.J.; Leonor, I.B.; Reis, R.L.; Kaplan, D.L. Silk-based antimicrobial polymers as a new platform to design drug-free materials to impede microbial infections. Macromol. Biosci. 2018, 18, 1800262. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.; Machado, R.; da Costa, A.; Ribeiro, A.; Collins, T.; Gomes, A.C.; Leonor, I.B.; Kaplan, D.L.; Reis, R.L.; Casal, M. Silk-based biomaterials functionalized with fibronectin type II promotes cell adhesion. Acta Biomater. 2017, 47, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Teng, W.; Cappello, J.; Wu, X. Recombinant silk-elastinlike protein polymer displays elasticity comparable to elastin. Biomacromolecules 2009, 10, 3028–3036. [Google Scholar] [CrossRef]
- Lyons, R.E.; Lesieur, E.; Kim, M.; Wong, D.C.; Huson, M.G.; Nairn, K.M.; Brownlee, A.G.; Pearson, R.D.; Elvin, C.M. Design and facile production of recombinant resilin-like polypeptides: Gene construction and a rapid protein purification method. Protein Eng. Des. Sel. 2007, 20, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Bandekar, J. Amide modes and protein conformation. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 1992, 1120, 123–143. [Google Scholar] [CrossRef]
- Surewicz, W.K.; Mantsch, H.H.; Chapman, D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: A critical assessment. Biochemistry 1993, 32, 389–394. [Google Scholar] [CrossRef]
- Hu, X.; Kaplan, D.; Cebe, P. Determining Beta-Sheet Crystallinity in Fibrous Proteins by Thermal Analysis and Infrared Spectroscopy. Macromolecules 2006, 39, 6161–6170. [Google Scholar] [CrossRef]
- Arrondo, J.L.R.; Goñi, F.M. Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog. Biophys. Mol. Biol. 1999, 72, 367–405. [Google Scholar] [CrossRef]
- Susi, H.; Michael Byler, D. Protein structure by Fourier transform infrared spectroscopy: Second derivative spectra. Biochem. Biophys. Res. Commun. 1983, 115, 391–397. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Rauscher, S.; Pomès, R. The liquid structure of elastin. Elife 2017, 6, e26526. [Google Scholar] [CrossRef]
- Quintanilla-Sierra, L.; García-Arévalo, C.; Rodriguez-Cabello, J. Self-assembly in elastin-like recombinamers: A mechanism to mimic natural complexity. Mater. Today Bio 2019, 2, 100007. [Google Scholar] [CrossRef]
- Wilson, D.; Valluzzi, R.; Kaplan, D. Conformational transitions in model silk peptides. Biophys. J. 2000, 78, 2690–2701. [Google Scholar] [CrossRef]
- Rabotyagova, O.S.; Cebe, P.; Kaplan, D.L. Role of polyalanine domains in β-sheet formation in spider silk block copolymers. Macromol. Biosci. 2009, 10, 49–59. [Google Scholar] [CrossRef]
- Haaber, J.; Cohn, M.T.; Frees, D.; Andersen, T.J.; Ingmer, H. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS ONE 2012, 7, e41075. [Google Scholar] [CrossRef]
- Skerlavaj, B.; Gennaro, R.; Bagella, L.; Merluzzi, L.; Risso, A.; Zanetti, M. Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J. Biol. Chem. 1996, 271, 28375–28381. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Zhang, J.; Xu, X.Z.; Han, Y.Y.; Cui, X.W.; Chen, Y.Q.; Zhang, S.Q. The antibacterial peptide ABP-CM4: The current state of its production and applications. Amino Acids 2012, 42, 2393–2402. [Google Scholar] [CrossRef]
- Jordan, J.B.; Poppe, L.; Haniu, M.; Arvedson, T.; Syed, R.; Li, V.; Kohno, H.; Kim, H.; Schnier, P.D.; Harvey, T.S.; et al. Hepcidin revisited, disulfide connectivity, dynamics, and structure. J. Biol. Chem. 2009, 284, 24155–24167. [Google Scholar] [CrossRef]
- Taraballi, F.; Natalello, A.; Campione, M.; Villa, O.; Doglia, S.M.; Paleari, A.; Gelain, F. Glycine-spacers influence functional motifs exposure and self-assembling propensity of functionalized substrates tailored for neural stem cell cultures. Front. Neuroeng. 2010, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- van Rosmalen, M.; Krom, M.; Merkx, M. Tuning the flexibility of glycine-serine linkers to allow rational design of multidomain proteins. Biochemistry 2017, 56, 6565–6574. [Google Scholar] [CrossRef]
- Gabriel, M.; Nazmi, K.; Veerman, E.C.; Nieuw Amerongen, A.V.; Zentner, A. Preparation of LL-37-grafted titanium surfaces with bactericidal activity. Bioconjugate Chem. 2006, 17, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Kaplan, D.L. 2.212—Silk Biomaterials. In Comprehensive Biomaterials; Ducheyne, P., Ed.; Elsevier: Oxford, UK, 2011. [Google Scholar]
| Sample | Water | Formic Acid | ||
|---|---|---|---|---|
| Untreated | MetOH-Treated | Untreated | MetOH-Treated | |
| Hep-SELP | 45.62% | 54.91% | 44.90% | 48.48% |
| Syn-SELP | 38.40% | 54.56% | 48.70% | 51.86% |
| CM4-SELP | 40.86% | 49.75% | 44.42% | 55.68% |
| BMAP18-SELP | 37.62% | 55.02% | 45.95% | 52.7% |
| Sample | % Kill (E. coli) | % Kill (S. aureus) | Reference |
|---|---|---|---|
| Hep-A200 (a) | 69.9 ± 10.2 | 60.8 ± 23.5 | This work |
| Hep-A200 (b) | 87.7 ± 8.3 | 76.7 ± 13.7 | This work |
| Syn-A200 (a) | 85.9 ± 9.0 | 75.4 ± 25.4 | This work |
| Syn-A200 (b) | 95.9 ± 6.2 | 85.6 ± 8.2 | This work |
| CM4-A200 (b) | 86.5 ± 0.7 | 69.7 ± 4.5 | [12] |
| BMAP18-A200 (b) | 100.0 | 98.8 ± 2.0 | [8] |
| Hep-SELP (a) | 11.4 ± 5.5 | 33.9 ± 41.9 | This work |
| Hep-SELP (b) | 17.7 ± 6.1 | 65.9 ± 30.8 | This work |
| Syn-SELP (a) | 28.9 ± 4.9 | 29.3 ± 44.7 | This work |
| Syn-SELP (b) | 16.9 ± 5.8 | 30.3 ± 62.2 | This work |
| CM4-SELP (a) | 28.1 ± 6.8 | 10.7 ± 59.6 | This work |
| CM4-SELP (b) | 34.8 ± 8.9 | 30.1 ± 4.8 | This work |
| BMAP18-SELP (a) | 73.7 ± 4.2 | 41.5 ± 35.3 | This work |
| BMAP18-SELP (b) | 60.4 ± 14.4 | 48.4 ± 24.5 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, A.M.; Gomes, D.; da Costa, A.; Dias, S.C.; Casal, M.; Machado, R. Protein-Engineered Polymers Functionalized with Antimicrobial Peptides for the Development of Active Surfaces. Appl. Sci. 2021, 11, 5352. https://doi.org/10.3390/app11125352
Pereira AM, Gomes D, da Costa A, Dias SC, Casal M, Machado R. Protein-Engineered Polymers Functionalized with Antimicrobial Peptides for the Development of Active Surfaces. Applied Sciences. 2021; 11(12):5352. https://doi.org/10.3390/app11125352
Chicago/Turabian StylePereira, Ana Margarida, Diana Gomes, André da Costa, Simoni Campos Dias, Margarida Casal, and Raul Machado. 2021. "Protein-Engineered Polymers Functionalized with Antimicrobial Peptides for the Development of Active Surfaces" Applied Sciences 11, no. 12: 5352. https://doi.org/10.3390/app11125352
APA StylePereira, A. M., Gomes, D., da Costa, A., Dias, S. C., Casal, M., & Machado, R. (2021). Protein-Engineered Polymers Functionalized with Antimicrobial Peptides for the Development of Active Surfaces. Applied Sciences, 11(12), 5352. https://doi.org/10.3390/app11125352








