Deep Learning and Transfer Learning for Automatic Cell Counting in Microscope Images of Human Cancer Cell Lines
Abstract
:1. Introduction
- A novel dataset containing images and the number of manually counted cells is presented. The images represent a human osteosarcoma cell line (U2OS) and a human leukemia cell line (HL-60). The dataset was collected at the University of Amsterdam, and could be downloaded from: https://doi.org/10.5281/zenodo.4428844 (accessed on 26 May 2021).
- The development of a pipeline for automatically counting cells using a convolutional neural network-based regressor. We indicate a specific architecture of a neural network that in combination with transfer learning allows the achievement of very promising performance.
- We provide baseline results for the newly collected data. Moreover, we present that the proposed approach achieves a human-level performance.
2. Methodology
2.1. Problem Statement
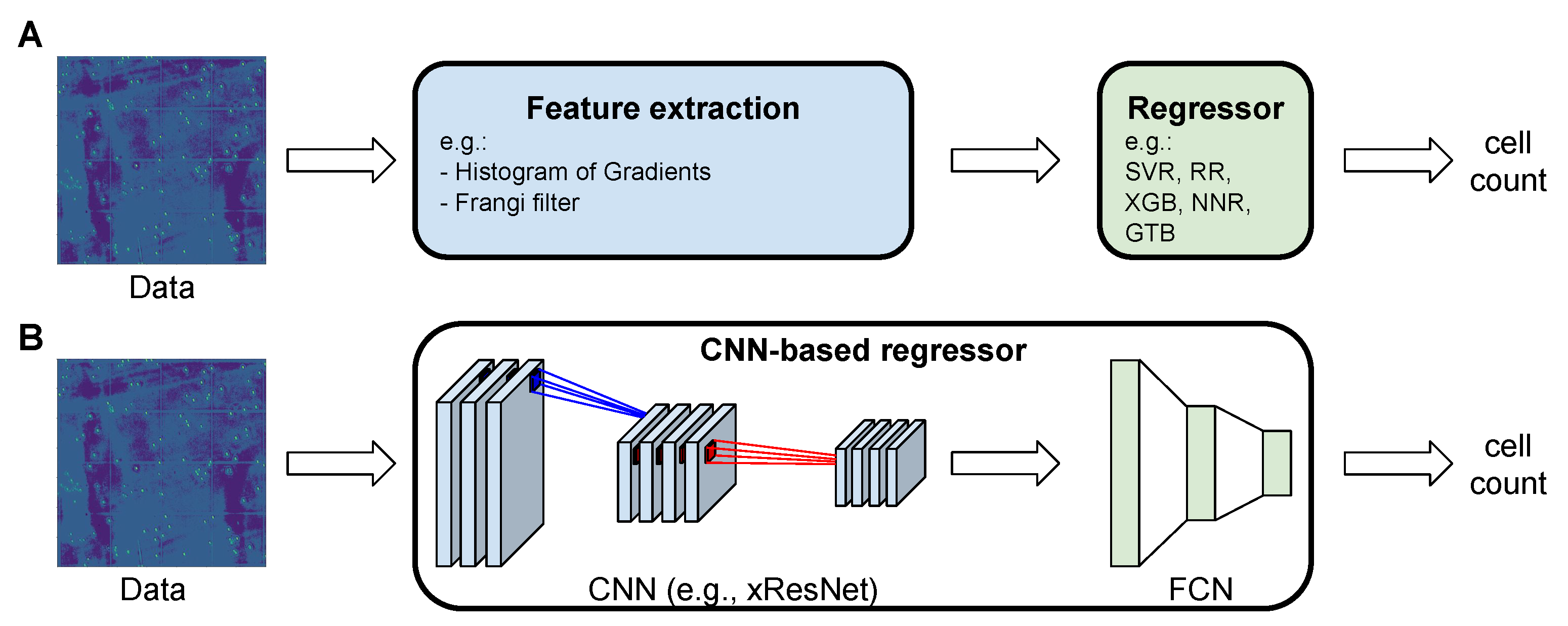
2.2. Machine Learning Pipeline
2.2.1. Feature Extractors
HOG
Frangi Filter
2.2.2. Regressors
Support Vector Regression
Gradient Tree Boosting
XGBoost
Ridge Regression
Nearest-Neighbor Regression
2.2.3. Remarks
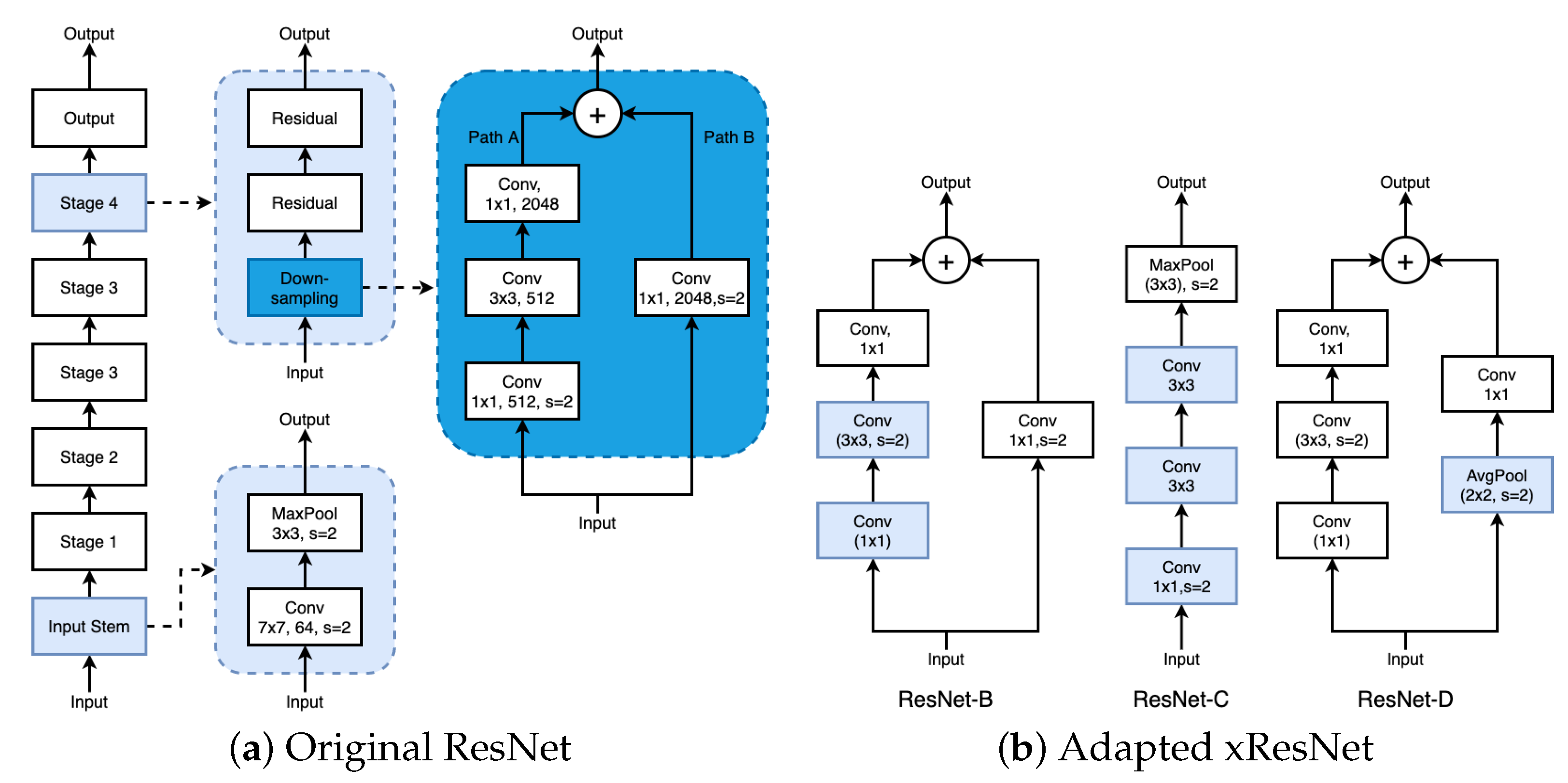
2.3. CNN-Based Regressors
2.3.1. Neural Network Regression
2.3.2. Our CNN-Based Regressor
3. Experiments
3.1. Dataset
- Data Information
- Data Preparation
- The cells tend to have varying circularity ratios, ranging from elongated ellipses to round circles. Therefore, algorithms that rely on the round shape assumption do not hold.
- Some cells have differing levels of staining intensity. Therefore, algorithms that require homogeneous intensity distributions of the object will not perform well.
- Since cells were manually counted using counting chambers, the grid lines are still present in the image. These will cause interference with algorithms that separate foreground from background.
- Data Augmentation
3.2. Details of the Machine Learning Pipeline
3.3. Details of Our Approach
3.4. Evaluation Metric
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Huang, P.C.; Lin, K.C.; Lin, H.H.; Wang, L.E.; Cheng, C.C.; Chen, T.P.; Chan, Y.K.; Chiang, J.Y. Semi-automatic segmentation and classification of pap smear cells. IEEE J. Biomed. Health Inform. 2013, 18, 94–108. [Google Scholar] [CrossRef]
- Carneiro, G.; Zheng, Y.; Xing, F.; Yang, L. Review of deep learning methods in mammography, cardiovascular, and microscopy image analysis. In Deep Learning and Convolutional Neural Networks for Medical Image Computing; Springer: Berlin/Heidelberg, Germany, 2017; pp. 11–32. [Google Scholar]
- Wainberg, M.; Merico, D.; Delong, A.; Frey, B.J. Deep learning in biomedicine. Nat. Biotechnol. 2018, 36, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, J.; Bise, R. Cell Tracking with Deep Learning for Cell Detection and Motion Estimation in Low-Frame-Rate; Springer: Berlin/Heidelberg, Germany, 2019; pp. 397–405. [Google Scholar]
- Hernandez, D.E.; Chen, S.W.; Hunter, E.E.; Steager, E.B.; Kumar, V. Cell Tracking with Deep Learning and the Viterbi Algorithm. In Proceedings of the 2018 International Conference on Manipulation, Automation and Robotics at Small Scales (MARSS), Nagoya, Japan, 4–8 July 2018; pp. 1–6. [Google Scholar]
- Lugagne, J.B.; Lin, H.; Dunlop, M.J. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol. 2020, 16, e1007673. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.M.; Islam, M.T. Machine learning approach of automatic identification and counting of blood cells. Healthc. Technol. Lett. 2019, 6, 103–108. [Google Scholar] [CrossRef]
- Chandradevan, R.; Aljudi, A.A.; Drumheller, B.R.; Kunananthaseelan, N.; Amgad, M.; Gutman, D.A.; Cooper, L.A.; Jaye, D.L. Machine-based detection and classification for bone marrow aspirate differential counts: Initial development focusing on nonneoplastic cells. Lab. Investig. 2020, 100, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef]
- Khan, A.; Gould, S.; Salzmann, M. Deep Convolutional Neural Networks for Human Embryonic Cell Counting; Springer: Berlin/Heidelberg, Germany, 2016; pp. 339–348. [Google Scholar]
- Anderson, M.; Hinds, P.; Hurditt, S.; Miller, P.; McGrowder, D.; Alexander-Lindo, R. The microbial content of unexpired pasteurized milk from selected supermarkets in a developing country. Asian Pac. J. Trop. Biomed. 2011, 1, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Vieira, F.; Nahas, E. Comparison of microbial numbers in soils by using various culture media and temperatures. Microbiol. Res. 2005, 160, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Gray, T.E.; Thomassen, D.G.; Mass, M.J.; Barrett, J.C. Quantitation of cell proliferation, colony formation, and carcinogen induced cytotoxicity of rat tracheal epithelial cells grown in culture on 3T3 feeder layers. In Vitro 1983, 19, 559–570. [Google Scholar] [CrossRef]
- Kotoura, Y.; Yamamuro, T.; Shikata, J.; Kakutani, Y.; Kitsugi, T.; Tanaka, H. A method for toxicological evaluation of biomaterials based on colony formation of V79 cells. Arch. Orthop. Trauma. Surg. 1985, 104, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hetet, G.; Maurer, A.M.; Chait, Y.; Dhermy, D.; Briere, J. Spontaneous megakaryocyte colony formation in myeloproliferative disorders is not neutralizable by antibodies against IL3, IL6 and GM-CSF. Br. J. Haematol. 1994, 87, 471–476. [Google Scholar] [CrossRef]
- Krastev, D.B.; Slabicki, M.; Paszkowski-Rogacz, M.; Hubner, N.C.; Junqueira, M.; Shevchenko, A.; Mann, M.; Neugebauer, K.M.; Buchholz, F. A systematic RNAi synthetic interaction screen reveals a link between p53 and snoRNP assembly. Nat. Cell Biol. 2011, 13, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Qian, X.; Si, H.; Xu, G.; Han, R.; Ni, Y. Significantly improved solvent tolerance of Escherichia coli by global transcription machinery engineering. Microb. Cell Fact. 2015, 14, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hébert, E.M.; Debouttière, P.J.; Lepage, M.; Sanche, L.; Hunting, D.J. Preferential tumour accumulation of gold nanoparticles, visualised by Magnetic Resonance Imaging: Radiosensitisation studies in vivo and in vitro. Int. J. Radiat. Biol. 2010, 86, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Nishio, K.; Kato, H.; Shinohara, N.; Nakamura, A.; Fujita, K.; Kinugasa, S.; Endoh, S.; Yamamoto, K.; Yamamoto, O.; et al. In vitro evaluation of cellular responses induced by stable fullerene C60 medium dispersion. J. Biochem. 2010, 148, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Sanders, B.G.; Kline, K. Tocotrienols induce apoptosis in breast cancer cell lines via an endoplasmic reticulum stress-dependent increase in extrinsic death receptor signaling. Breast Cancer Res. Treat. 2010, 124, 361–375. [Google Scholar] [CrossRef]
- Azari, H.; Louis, S.A.; Sharififar, S.; Vedam-Mai, V.; Reynolds, B.A. Neural-colony forming cell assay: An assay to discriminate bona fide neural stem cells from neural progenitor cells. JOVE (J. Vis. Exp.) 2011, e2639. [Google Scholar] [CrossRef]
- Galli, R. The neurosphere assay applied to neural stem cells and cancer stem cells. In Target Identification and Validation in Drug Discovery; Springer: Berlin/Heidelberg, Germany, 2013; pp. 267–277. [Google Scholar]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, M. Hemocytometer Protocol. Available online: https://www.hemocytometer.org/hemocytometer-protocol/ (accessed on 25 May 2021).
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 886–893. [Google Scholar]
- Kong, H.; Akakin, H.C.; Sarma, S.E. A generalized Laplacian of Gaussian filter for blob detection and its applications. IEEE Trans. Cybern. 2013, 43, 1719–1733. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikäinen, M.; Harwood, D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 1–74. [Google Scholar] [CrossRef] [PubMed]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. arXiv 2015, arXiv:1411.4038. [Google Scholar]
- Shrestha, A.; Mahmood, A. Review of deep learning algorithms and architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks; Springer: Berlin/Heidelberg, Germany, 2013; pp. 411–418. [Google Scholar]
- Liu, F.; Yang, L. A novel cell detection method using deep convolutional neural network and maximum-weight independent set. In Deep Learning and Convolutional Neural Networks for Medical Image Computing; Springer: Berlin/Heidelberg, Germany, 2017; pp. 63–72. [Google Scholar]
- Xie, Y.; Xing, F.; Kong, X.; Su, H.; Yang, L. Beyond classification: Structured regression for robust cell detection using convolutional neural network. Med. Image Comput. Comput. Assist. Interv. 2015, 9351, 358–365. [Google Scholar] [PubMed] [Green Version]
- Akram, S.U.; Kannala, J.; Eklund, L.; Heikkilä, J. Cell segmentation proposal network for microscopy image analysis. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 21–29. [Google Scholar]
- Shelhamer, E.; Rakelly, K.; Hoffman, J.; Darrell, T. Clockwork convnets for video semantic segmentation. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 852–868. [Google Scholar]
- Xie, W.; Noble, J.A.; Zisserman, A. Microscopy cell counting and detection with fully convolutional regression networks. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2018, 6, 283–292. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Support vector regression. In Efficient Learning Machines; Springer: Berlin/Heidelberg, Germany, 2015; pp. 67–80. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Altman, N.S. An introduction to kernel and nearest-neighbor nonparametric regression. Am. Stat. 1992, 46, 175–185. [Google Scholar]
- Sun, C.; Wang, D.; Lu, H.; Yang, M.H. Learning spatial-aware regressions for visual tracking. arXiv 2018, arXiv:1706.07457. [Google Scholar]
- Hernández, C.X.; Sultan, M.M.; Pande, V.S. Using deep learning for segmentation and counting within microscopy data. arXiv 2018, arXiv:1802.10548. [Google Scholar]
- Marana, A.N.; Velastin, S.; Costa, L.; Lotufo, R. Estimation of crowd density using image processing. In Proceedings of the IEEE Colloquium on Image Processing for Security Applications (Digest No: 1997/074), London, UK, 10 March 1997. [Google Scholar]
- Kong, D.; Gray, D.; Tao, H. A viewpoint invariant approach for crowd counting. In Proceedings of the 18th International Conference on Pattern Recognition (ICPR’06), Hong Kong, China, 20–24 August 2006; Volume 3, pp. 1187–1190. [Google Scholar]
- He, T.; Zhang, Z.; Zhang, H.; Zhang, Z.; Xie, J.; Li, M. Bag of tricks for image classification with convolutional neural networks. arXiv 2019, arXiv:1812.01187. [Google Scholar]
- Howard, J.; Gugger, S. Fastai: A layered API for deep learning. Information 2020, 11, 108. [Google Scholar] [CrossRef] [Green Version]
- Nixon, M.; Aguado, A. Feature Extraction and Image Processing for Computer Vision; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Frangi, A.F.; Niessen, W.J.; Vincken, K.L.; Viergever, M.A. Multiscale vessel enhancement filtering. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention, Cambridge, MA, USA, 11–13 October 1998; pp. 130–137. [Google Scholar]
- Longo, A.; Morscher, S.; Najafababdi, J.M.; Jüstel, D.; Zakian, C.; Ntziachristos, V. Assessment of hessian-based Frangi vesselness filter in optoacoustic imaging. Photoacoustics 2020, 20, 100200. [Google Scholar] [CrossRef]
- Shahzad, A.; Goh, C.; Saad, N.; Walter, N.; Malik, A.S.; Meriaudeau, F. Subcutaneous veins detection and backprojection method using Frangi vesselness filter. In Proceedings of the 2015 IEEE Symposium on Computer Applications & Industrial Electronics (ISCAIE), Langkawi, Malaysia, 12–14 April 2015; pp. 65–68. [Google Scholar]
- Drucker, H.; Burges, C.J.; Kaufman, L.; Smola, A.; Vapnik, V. Support vector regression machines. Adv. Neural Inf. Process. Syst. 1996, 9, 155–161. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. arXiv 2016, arXiv:1603.02754. [Google Scholar]
- Hoerl, A.E.; Kennard, R.W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 1970, 12, 55–67. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Vapnik, V.N. The Nature of Statistical Learning; Springer: Berlin/Heidelberg, Germany, 1995. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Mutlag, W.K.; Ali, S.K.; Aydam, Z.M.; Taher, B.H. Feature Extraction Methods: A Review. J. Phys. Conf. Ser. IOP Publ. 2020, 1591, 012028. [Google Scholar] [CrossRef]
- İnik, Ö.; Ceyhan, A.; Balcıoğlu, E.; Ülker, E. A new method for automatic counting of ovarian follicles on whole slide histological images based on convolutional neural network. Comput. Biol. Med. 2019, 112, 103350. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Kavukcuoglu, K.; Farabet, C. Convolutional networks and applications in vision. In Proceedings of the 2010 IEEE International Symposium on Circuits and Systems, Paris, France, 30 May–2 June 2010; pp. 253–256. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. arXiv 2016, arXiv:1512.03385. [Google Scholar]
- Lauvrak, S.; Munthe, E.; Kresse, S.; Stratford, E.; Namløs, H.; Meza-Zepeda, L.; Myklebost, O. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer 2013, 109, 2228–2236. [Google Scholar] [CrossRef] [PubMed]
- Weglarz-Tomczak, E.; Rijlaarsdam, D.J.; Tomczak, J.M.; Brul, S. GEM-based metabolic profiling for Human Bone Osteosarcoma under different glucose and glutamine availability. Int. J. Mol. Sci. 2021, 22, 1470. [Google Scholar] [CrossRef]
- Birnie, G. The HL60 cell line: A model system for studying human myeloid cell differentiation. Br. J. Cancer Suppl. 1988, 9, 41. [Google Scholar]
- Ilse, M.; Tomczak, J.M.; Forré, P. Designing Data Augmentation for Simulating Interventions. arXiv 2020, arXiv:2005.01856. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Sgdr: Stochastic gradient descent with warm restarts. arXiv 2016, arXiv:1608.03983. [Google Scholar]
- Smith, L.N. A disciplined approach to neural network hyper-parameters: Part 1—Learning rate, batch size, momentum, and weight decay. arXiv 2018, arXiv:1803.09820. [Google Scholar]
- Oruganti, T.; Laufer, J.G.; Treeby, B.E. Vessel filtering of photoacoustic images. In Proceedings of the Photons Plus Ultrasound: Imaging and Sensing 2013, San Francisco, CA, USA, 3–5 February 2013; Volume 8581, p. 85811W. [Google Scholar]
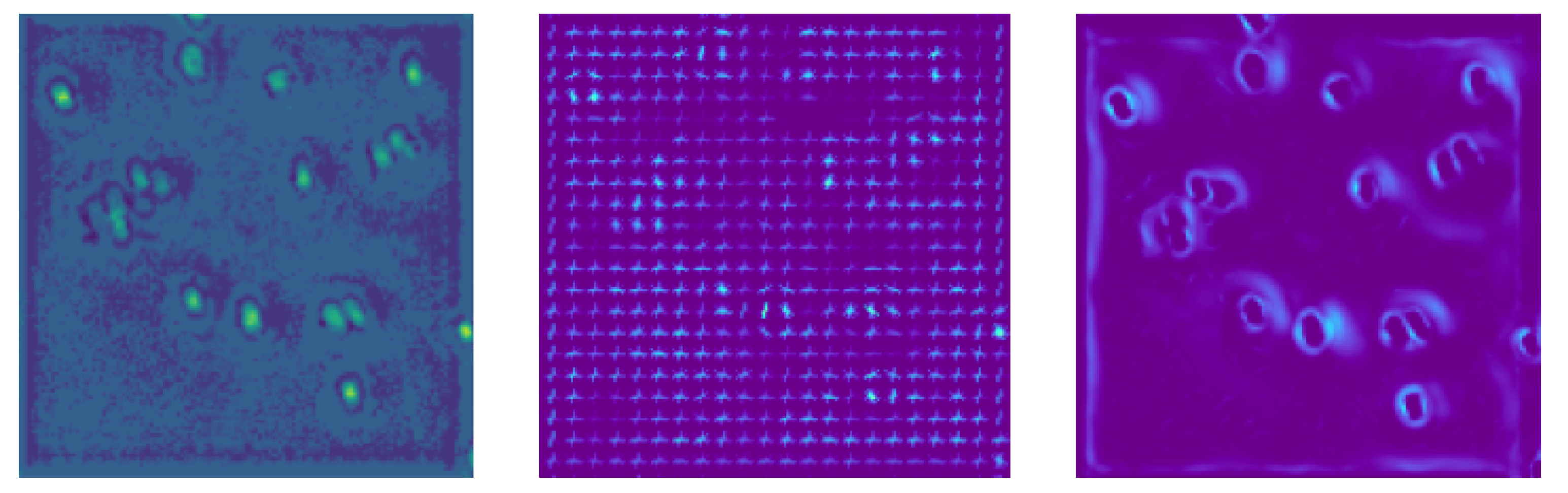





| MAE ± std | IMG | HOG | Frangi |
|---|---|---|---|
| SVR | 88 ± 65 | 94 ± 69 | 86 ± 64 |
| RR | 88 ± 71 | 41 ± 37 | 45 ± 52 |
| NNR | 107 ± 101 | 98 ± 96 | 111 ± 101 |
| XGB | 86 ± 81 | 85 ± 79 | 81 ± 79 |
| GTB | 75 ± 58 | 82 ± 55 | 67 ± 51 |
| Our w/o TL | 32 ± 33 | x | x |
| Our w/TL | 12 ± 15 | x | x |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lavitt, F.; Rijlaarsdam, D.J.; van der Linden, D.; Weglarz-Tomczak, E.; Tomczak, J.M. Deep Learning and Transfer Learning for Automatic Cell Counting in Microscope Images of Human Cancer Cell Lines. Appl. Sci. 2021, 11, 4912. https://doi.org/10.3390/app11114912
Lavitt F, Rijlaarsdam DJ, van der Linden D, Weglarz-Tomczak E, Tomczak JM. Deep Learning and Transfer Learning for Automatic Cell Counting in Microscope Images of Human Cancer Cell Lines. Applied Sciences. 2021; 11(11):4912. https://doi.org/10.3390/app11114912
Chicago/Turabian StyleLavitt, Falko, Demi J. Rijlaarsdam, Dennet van der Linden, Ewelina Weglarz-Tomczak, and Jakub M. Tomczak. 2021. "Deep Learning and Transfer Learning for Automatic Cell Counting in Microscope Images of Human Cancer Cell Lines" Applied Sciences 11, no. 11: 4912. https://doi.org/10.3390/app11114912
APA StyleLavitt, F., Rijlaarsdam, D. J., van der Linden, D., Weglarz-Tomczak, E., & Tomczak, J. M. (2021). Deep Learning and Transfer Learning for Automatic Cell Counting in Microscope Images of Human Cancer Cell Lines. Applied Sciences, 11(11), 4912. https://doi.org/10.3390/app11114912







