Abstract
Artificial Intelligence (AI) technologies have recently been applied to medical imaging for diagnostic support. With respect to fetal ultrasound screening of congenital heart disease (CHD), it is still challenging to achieve consistently accurate diagnoses owing to its manual operation and the technical differences among examiners. Hence, we proposed an architecture of Supervised Object detection with Normal data Only (SONO), based on a convolutional neural network (CNN), to detect cardiac substructures and structural abnormalities in fetal ultrasound videos. We used a barcode-like timeline to visualize the probability of detection and calculated an abnormality score of each video. Performance evaluations of detecting cardiac structural abnormalities utilized videos of sequential cross-sections around a four-chamber view (Heart) and three-vessel trachea view (Vessels). The mean value of abnormality scores in CHD cases was significantly higher than normal cases (p < 0.001). The areas under the receiver operating characteristic curve in Heart and Vessels produced by SONO were 0.787 and 0.891, respectively, higher than the other conventional algorithms. SONO achieves an automatic detection of each cardiac substructure in fetal ultrasound videos, and shows an applicability to detect cardiac structural abnormalities. The barcode-like timeline is informative for examiners to capture the clinical characteristic of each case, and it is also expected to acquire one of the important features in the field of medical AI: the development of “explainable AI.”
1. Introduction
In recent years, deep learning techniques have been developing rapidly, and there is much interest in the adoption of deep learning for medical applications. More than 60 Artificial Intelligence (AI)-equipped medical devices have already been approved by the Food and Drug Administration (FDA) in the United States [1]. Indeed, it has been pointed out that diagnostic systems using deep learning may detect abnormalities and diseases more quickly and accurately than humans can; however, this requires the availability of enough datasets on both normal and abnormal subjects for different diseases [2,3].
It is estimated that congenital heart disease (CHD) exists in approximately 1% of live births, and critical CHD accounts for the largest proportion of infant mortality resulting from birth defects [4,5,6]. In this regard, abnormal cardiac findings on routine prenatal ultrasound screening by mainly obstetricians should trigger a more precise examination as soon as feasible. Proper prenatal diagnosis, allowing for prompt treatment within a week of the birth, is known to markedly improve the prognosis [7]. Fetal ultrasound screening of every pregnancy at risk for CHD is generally recommended at 18 to 22 weeks of gestation worldwide [8,9]. Despite its importance, however, the total prenatal diagnostic rate of 30–50% remains insufficient due to differences in diagnostic skill levels between examiners [8,10,11]. Due to its manual operation, effective fetal cardiac ultrasound screening requires high skill levels and experience among examiners coupled to feedback from fetal or pediatric cardiologists and cardiovascular surgeons. The relatively low incidence of CHD and different levels of medical expertise at hospitals result in inconsistencies. Hence, it is important to develop a system that can always conduct fetal cardiac ultrasound screening with a high skill level.
In the present study, we have used deep learning with relatively small and incomplete datasets of fetal ultrasound videos, to provide diagnostic support for examiners in fetal cardiac ultrasound screening. Each video consisted of the informative sequential cross-sections in our datasets; hence, no high skill levels were required to accurately describe the standardized transverse scanning planes. Generally, experts use their own judgement to determine whether certain cardiac substructures, such as valves and blood vessels, are in the correct anatomical localizations, by comparing normal and abnormal fetal heart images. This process is like the object detection technique, which allows us to distinguish the localizations and classify multiple substructures appearing in videos. Here, we demonstrated a novel deep learning approach for automatic detection of cardiac substructures and its application to detect cardiac structural abnormalities in fetal ultrasound videos.
Related Works
Some supervised deep learning models have been reported for fetal ultrasound images and videos. Temporal HeartNet could automatically predict the visibility, viewing plane, location, and orientation of the heart in fetal ultrasound videos [12]. SonoNet could detect the fetal structures via bounding boxes in fetal ultrasound videos, such as the brain, spine, abdomen, and also the four standardized transverse scanning planes of fetal heart, which were the four-chamber view (4CV), three-vessel view (3VV), right ventricular outflow tract (ROVT), and left ventricular outflow tract (LOVT) [13]. These models focused on plane-based detection of fetal heart and their input data depended on the skill levels of examiners. However, it is still difficult for non-experts to identify the cardiac substructures and describe the scanning planes precisely.
The application of image segmentation methods to fetal ultrasound has been reported. Arnaout et al. used plane-based detection of fetal heart for CHD screening, and performed segmentation of the thorax, heart, spine, and each of the four cardiac chambers using U-net to calculate standard fetal cardiothoracic measurements [14]. We previously employed the time-series information of fetal ultrasound videos in the module that calibrates segmentation results of the ventricular septum [15]. These pixel-by-pixel detection techniques are useful to detect the target with a small shape changing in accordance with the fetal heartbeat.
In fetal ultrasound, deep learning-based detection of cardiac abnormalities is still challenging because CHD is relatively rare and noisy acoustic shadows affect ultrasound images, making it a daunting task to prepare complete training datasets [16]. To overcome these issues, we have to consider an applied method for detection of cardiac structural abnormalities using small and incomplete datasets.
2. Materials and Methods
2.1. Data Preparation
A total of 363 pregnant women having a fetus with a normal heart or CHD underwent fetal cardiac ultrasound screening at 18–34 weeks. Patients were examined in the four Showa University Hospitals (Tokyo and Yokohama, Japan). All women were enrolled in research protocols approved by the Institutional Review Board of RIKEN, Fujitsu Ltd., Showa University, and the National Cancer Center (approval ID: Wako1 29-4). All methods were performed in accordance with the Ethical Guidelines for Medical and Health Research Involving Human Subjects, and with regard to the handling of data, we followed Data Handling Guidelines for the Medical AI project. Not only expert sonographers, but also obstetricians with at least three years of experience, obtained fetal ultrasound videos under the guidance of experts. A total of 772 screening videos were acquired using commercially available ultrasonography machines (Voluson® E8 or E10, GE Healthcare, Chicago, IL, USA) equipped with an abdominal 2–6 MHz transducer in accordance with the guidelines [17,18]. A cardiac preset was used, and images were magnified until the chest fills at least one-half to two-thirds of the screen. Each video consisted of the sequential cross-sections from the level of the stomach, through the heart, to the vascular arches, mainly in apical view. All data consisted of 349 normal cases and 14 CHD cases, and were randomly assigned for deep learning, as shown in Figure 1. The characteristics of the CHD cases are listed in Supplementary Table S1.
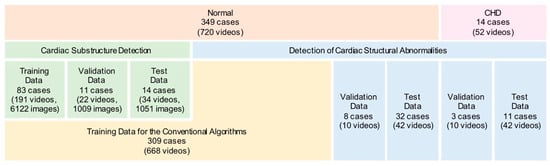
Figure 1.
Data preparation for deep learning.
2.2. Cardiac Substructure Detection
In the present study, we propose a novel architecture of Supervised Object detection with Normal data Only (SONO) to detect fetal cardiac substructures and structural abnormalities, as shown in Figure 2. The experimental flow charts also show our key-feature methods (Supplementary Figure S1). Using the checkpoints in the standardized screening for CHD, the expert annotated the correct positions of 18 different anatomical substructures with bounding boxes in 8182 frames from 247 normal fetal ultrasound videos, including a crux, ventricular septum, right atrium, tricuspid valve, right ventricle, left atrium, mitral valve, left ventricle, pulmonary artery, ascending aorta, superior vena cava, descending aorta, stomach, spine, umbilical vein, inferior vena cava, pulmonary vein, and ductus arteriosus. The selected substructures are shown in Figure 3. The performance of our SONO, based on a convolutional neural network (CNN) for real-time object detection, YOLOv2 [19], was evaluated using the annotated dataset which was randomly assigned into 191 videos for training, 22 videos for validation, and 34 videos for test data. The implementation details and training details of the CNN are shown in Appendix A. This CNN can predict the localization and classification of each substructure simultaneously, measuring the intersection over union (IoU) of the ground truth and the predicted box, and the conditional probability, given that there was an object. It defined that a substructure was detected somewhere in the same frame of the ground truth in 0 IoU. To evaluate the detection accuracy, the mean average precision (mAP) was calculated in IoU > 0 [20].
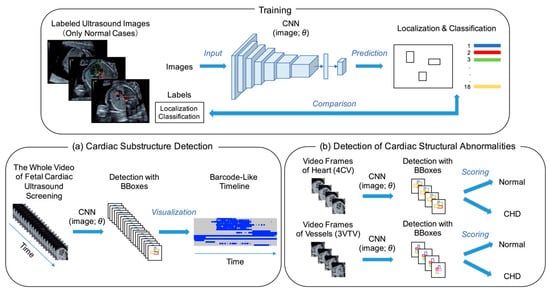
Figure 2.
Architecture of supervised object detection with normal data only (SONO) for detection of (a) fetal cardiac substructures and (b) structural abnormalities. A convolutional neural network (CNN) was trained with labeled ultrasound images and performed each detection with bounding boxes (BBoxes). θ: parameter; 4CV: four-chamber view; 3VTV: three-vessel trachea view.
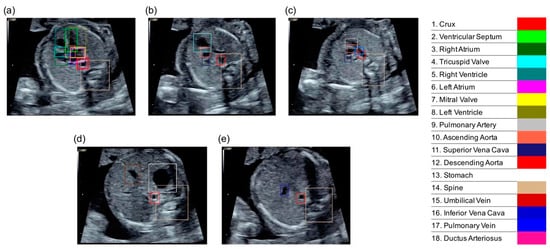
Figure 3.
The correct localizations of 18 different anatomical substructures were annotated with bounding boxes. (a) 4CV; (b) 3VV (three-vessel view); (c) 3VTV; (d,e) abdomen view.
2.3. Visualization of the Detection Result
The detection probability of each substructure was measured and described in a barcode-like timeline to visualize its progress along with the sweep scanning. The vertical axis represented the 18 selected substructures, and the horizontal axis represented the examination timeline in a rightward direction, which followed the probe scanning in the order of the abdomen, heart structure, outflow tracts, and vessels. A probability ≥0.01 was set as well-detected and shown as a blue bar, and <0.01 as non-detected and a gray bar. The whole cardiac ultrasound screening video of a case of tetralogy of Fallot (TOF), one of the most common CHDs, was used as test data. The resulting colored barcode-like timeline was examined and compared with that from normal fetal heart videos.
2.4. Performance Evaluations of Detecting Cardiac Structural Abnormalities
To conduct performance evaluations of detecting cardiac structural abnormalities, we used 104 sets of the sequential 20 video frames of cross-sections around a 4CV (Heart) and around a three-vessel trachea view (3VTV) (Vessels), acquired from 40 normal and 14 CHD cases. They were randomly assigned into 10 videos for validation and 42 videos for testing in both the normal and CHD datasets. In SONO, the abnormality score was calculated using the total number of well-detected substructures among the eight selected substructures (crux, ventricular septum, right atrium, tricuspid valve, right ventricle, left atrium, mitral valve, and left ventricle) for Heart, and the four selected substructures (pulmonary artery, ascending aorta, superior vena cava, and ductus arteriosus) for Vessels in each set. These selected substructures were important checkpoints for fetal cardiac ultrasound screening; in particular, the guidelines recommended that the evaluation should be done around each standardized transverse scanning plane [17,18]. We defined,
The abnormality score ranged from 0 to 1 where t represented the frame number and T represented the maximum number. In this study, we focused on the abovementioned 20 video frames (T = 20) and calculated the abnormality scores in Heart and Vessels. h(t) and v(t) represented the total number of substructures with the probability ≥ 0.01 in each frame. Then, we compared the accuracy of detecting cardiac structural abnormalities in SONO, using 191 videos of 83 normal cases for training, with other conventional anomaly detection algorithms for general images, such as a typical convolutional autoencoder (ConvAE) for a frame (ConvAE-1frame), ConvAE [21], AE + global feature [22], and anomaly detection with generative adversarial networks (AnoGAN) [23], for which the training data consisted of 668 videos of 309 normal cases. ConvAE and AE + global feature directly applied to video analyses; however, the other methods were originally intended for still image analyses. The reconstruction errors for each individual frame were used. We assumed that the minimum value of reconstruction errors reflected the background noise and calculated an abnormality score for each method using the range between the maximum and minimum value of reconstruction errors. The code has been uploaded to GitHub (https://github.com/rafcc/2020-prenatal-sono).
2.5. Statistical Analysis
Dependent continuous variables were compared using nonparametric tests (Mann–Whitney U test). All statistical tests were two-tailed and a p value < 0.05 was considered statistically significant. To evaluate the performance of detecting cardiac structural abnormalities in SONO and the other algorithms, a receiver operating characteristic (ROC) analysis was performed and the area under the ROC curve (AUC) produced by each algorithm was compared in Heart and Vessels.
3. Results
3.1. Average Precisions of Cardiac Substructure Detection
Table 1 shows that our SONO achieved a mAP of 0.70 in test data and suppressed over-fitting in the training data through validation. According to the average precision (AP) for each substructure, a crux, ventricular septum, both sides of the ventricle, and atrium were all well-detected. The outflow tracts, pulmonary artery, and ascending aorta were all detected with enough precision. In contrast, the detection performance of the tricuspid valve, mitral valve, inferior vena cava, pulmonary vein, and ductus arteriosus was still poor.

Table 1.
Average precisions (AP) of cardiac substructure detection and its mean value (mAP) were demonstrated.
3.2. Barcode-Like Timeline
The whole examination time was 10–15 s per video, which consisted of approximately 300–600 sequential ultrasound frames. With the exception of the screening videos with the probe shake and sweep iteration by each examiner, the representative barcode-like timelines of normal cases were clearly distinguished between three parts consisting of the abdomen, heart structure, and outflow tract/blood vessels. In normal cases, the diagnostic components of a 4CV and 3VTV were well-detected and located in their correct anatomical positions; the other substructures were also well-detected along with their correct scanning timing (Figure 4a). On the other hand, in the TOF case, the detection probabilities of the heart structures around the 4CV and 3VTV were poor. The probabilities raw data and the whole examination timeline is shown in Supplementary Table S2. In particular, a pulmonary artery was not clearly detected, which was an obvious difference from the normal cases in the timelines (Figure 4b). The TOF consists of four features of the heart and its blood vessels: ventricular septal defect (VSD), pulmonary stenosis, aortic override, and right ventricular hypertrophy. A narrowing of the pulmonary artery induces a morphological change in outflow tracts and around the 3VTV. Through SONO, undetectable substructures indicated the possibility of their pathological findings.
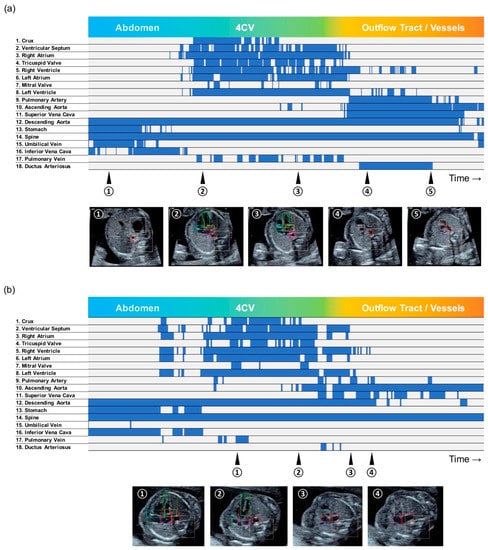
Figure 4.
Barcode-like timeline in (a) a normal case, and (b) a tetralogy of Fallot (TOF) case. The vertical axis represented the 18 selected substructures, and the horizontal axis represented the examination timeline in a rightward direction. A probability ≥0.01 was set as well-detected and shown as a blue bar, and <0.01 as non-detected and a gray bar.
3.3. Detection of Cardiac Structural Abnormalities
To make a validation and test dataset of CHD for detection of cardiac structural abnormalities, we collected the ultrasound screening videos obtained from 14 CHD cases. We defined the abnormality score of each video through a calculation using the probability of the selected cardiac substructures for Heart and Vessels. The mean value of abnormality scores in CHD cases (Heart = 0.251, Vessels = 0.418) was significantly higher than normal cases (Heart = 0.087, Vessels = 0.083; p < 0.001), as shown in Supplementary Figure S2. These results indicated that this abnormality score was suitable to use to distinguish morphological anomalies from a normal fetal heart and vessels.
Furthermore, the ROC analyses were used to assess the performance of detecting cardiac structural abnormalities in Heart and Vessels, and Figure 5 shows our SONO compared to other conventional algorithms. The AUCs produced by SONO were 0.787 in Heart and 0.891 in Vessels. The AUCs produced by ConvAE-1frame, ConvAE, AE + global feature, and AnoGAN in Heart/Vessels were 0.747/0.706, 0.517/0.542, 0.656/0.673, and 0.656/0.651, respectively (Table 2). Therefore, SONO demonstrated superior performance to any other conventional ones in this comparison analysis, and detected the abnormalities more accurately in Vessels than Heart.
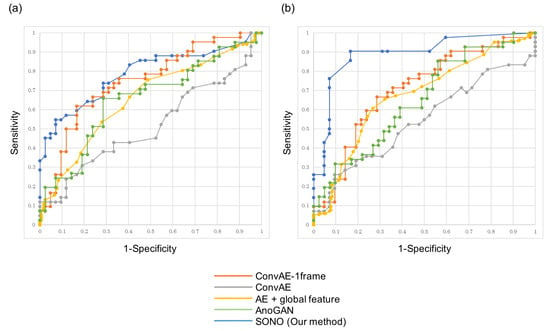
Figure 5.
Receiver operating characteristic (ROC) curves showing performance comparison of SONO and the four conventional anomaly detection algorithms in detection of cardiac structural abnormalities in (a) Heart and (b) Vessels.

Table 2.
The areas under the receiver operating characteristic curves (AUCs) for SONO and the other algorithms in Heart and Vessels.
3.4. Graphical User Interface
We integrated abovementioned technologies and proposed a graphical user interface (GUI) for clinical implementation, as shown in Supplementary Videos S1 and S2. The cardiac substructure detection and its probability measurement took place at a real-time speed. The colored bounding boxes automatically indicated where different substructures are supposed to be located in fetal ultrasound videos. The detection probabilities of cardiac substructures in each frame were measured and real-timely demonstrated in the upper right table. Along with the sweep scanning, the abnormality scores were calculated and its transitive graph were displayed at the bottom right of the screen. The heart and vessels areas were colored and emphasized. Furthermore, after the examination was finished and the report button was clicked, another window was opened in the same screen. It displayed a barcode-like timeline of the whole examination and the mean value of abnormality scores in the heart and vessels. In the TOF case, the lines of abnormality score dramatically increased in the graph, and the report window displayed a different timeline from normal cases and high abnormality scores.
4. Discussion
Fetal cardiac ultrasound assessments of an affected pregnancy should be performed sufficiently early to provide time for a proper treatment if needed. The importance of fetal cardiac ultrasound screening, incorporating multiple views of the heart and blood vessels, has been advocated to improve the prenatal detection rate for CHD [8]. Recent advances in computer processing and transducer technology have also expanded the capacity of fetal ultrasound to include a wide variety of new modalities and sophisticated measures for cardiac structure and function. Nevertheless, the detection rate remains inaccurate and dependent on the type of ultrasound practice and experience of the examiners [24,25]. Previous experience with CHDs and exposure to practical advice and feedback from experts, cardiologists, and cardiovascular surgeons are necessary to become a well-qualified examiner. The manual operation adds to the practical difficulties of normalizing the sweep scanning techniques and the resulting images. The research and development of the modalities with fixed patient or subject and constant measurement time, including computed tomography (CT), magnetic resonance imaging (MRI), X-ray, and pathological images, have led to advances in high quality controls [26,27]. However, the characteristic issues in ultrasound described above have slowed the progress of research, and there have been few publications and products associated with deep learning-based analyses of ultrasound images compared to other modalities [28,29,30]. Some models to support CHD screening by detecting the standardized transverse scanning planes have been reported, but the robustness of their input data needs to be considered [12,13,14].
We investigated deep learning using relatively small and incomplete datasets. The low incidence rate of CHD limited our ability to collect large volumes of relevant ultrasound images or videos for deep learning training. On the other hand, most pregnant women have a singleton fetus with a normal heart, among which there is little structural atypia. Therefore, we developed a novel application of object detection supervised from the dataset of normal cases only, to detect fetal cardiac substructures and structural abnormalities in fetal cardiac ultrasound screening. We analyzed fetal ultrasound videos, which consisted of the informative sequential cross-sections in an examiner-independent manner. For quality control, a high quality expert assisted in addressing the technical variety of annotation of the 18 different anatomical substructures. Our proposed SONO achieved a high detection ability, whereas the detail of their AP distribution implied that there were the detectable and undetectable substructures. Relatively small substructures such as a tricuspid valve, mitral valve, pulmonary vein, and ductus arteriosus were undetectable.
We converted the video data into a barcode-like timeline. Enhancing the perspicuity of the whole examination, the barcode-like timeline made it easy to identify which substructures affected the diagnosis and hence, shorten the confirmation time. The examination results were standardized regardless of the technical levels of examiners, using automatic cardiac substructure detection. Our analyses comparing normal and some CHD cases showed that this timeline correctly captured their clinical characteristics. The important findings were that a pulmonary artery was not detected as normal in TOF, which reflects its narrowing. In CHD cases, we could see the probability transition and identify the critical differences from normal cases. While previous methods have tried to hide the detection variability in video sequences, this study showed the variability in video object detection as useful information for examiners. The barcode-like timeline is useful in terms of explainability, and can be highlighted as one of the features of “explainable AI.”
To assess detection ability of cardiac structural abnormalities, we focused on the sequential 20 video frames of cross-sections around the 4CVs and 3VTVs. Through the ROC analysis, SONO performed better than the four conventional anomaly detection algorithms in both test datasets. In addition, SONO used one-third of the videos of the other algorithms in the training dataset, thereby reducing the cost and effort of data collection. Furthermore, the detection accuracy of outflow tracts and vessels was higher than the other heart structures in SONO. The conventional algorithms, ConvAE and AE + global feature, were engineering advanced and adapted to high quality images photographed with a security camera; however, their domain specific abilities of anomaly detection were insufficient for the low-resolution ultrasound videos. AnoGAN, originally intended for still ultrasound images, and the versatile algorithm ConvAE-1frame were inferior to SONO regarding fetal ultrasound videos.
Limitations
There are several limitations in this study. First, owing to the relatively low incidence of CHD, we used the small volume of CHD data from limited institutions. Our training data consisted of only normal cases; however, further CHD data collection is needed as test data for the validity and reliability evaluation of detecting cardiac structural abnormalities, by cooperating with other hospitals throughout Japan or globally. Second, our fetal ultrasound videos were obtained using the same type of ultrasonography machine. In terms of the robustness, we have to verify whether SONO works in a different equipment and setting. Third, SONO consisted of mainly apical view data and could not handle any kind of fetal presentations. Inputting further non-apical view datasets to the CNN might resolve this limitation. Finally, it was still hard for SONO to capture the isomerism, complete transposition of large vessels, and the subtle changes of the cardiac substructures, such as a ventricular hypertrophy, ventricular septal defect, and valve abnormalities. Therefore, we have to consider add-on technologies including image segmentation, for further accurate detection of these findings.
5. Conclusions
This study demonstrated that our proposed SONO can detect cardiac substructures and indicate structural abnormalities in fetal ultrasound videos. The barcode-like timeline is a useful diagram to capture the whole examination process and characteristics of each cardiac substructure. SONO and the barcode-like timeline require further examinations for clinical implementation; however, these technologies have the potential to be practically used as the operation guidance and clinical report to support examiners in fetal cardiac ultrasound screening.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/11/1/371/s1, Figure S1: Experimental flow charts, Figure S2: Abnormality scores in the Heart and Vessels, Table S1: Characteristics of the 14 cases with congenital heart disease, Table S2: Raw data of the detection probabilities of 18 cardiac substructures along with the whole examination timeline, Video S1: Graphical user interface in a normal case, Video S2: Graphical user interface in a TOF case.
Author Contributions
Conceptualization, M.K., A.S. (Akira Sakai) and R.K.; methodology, M.K., A.S. (Akira Sakai) and R.K.; software, M.K., A.S. (Akira Sakai) and H.H.; validation, M.K., A.S. (Akira Sakai) and R.K.; investigation, M.K. and A.S. (Akira Sakai); resources, R.K., R.M., T.A. and A.S. (Akihiko Sekizawa); data curation, M.K., A.S. (Akira Sakai) and R.K.; writing—original draft preparation, M.K., A.S. (Akira Sakai) and R.K.; writing—review and editing, R.M., S.Y., K.S., A.D., H.M., H.H., T.A., K.A., S.K., A.S. (Akihiko Sekizawa) and R.H.; supervision, M.K. and R.H.; project administration, M.K., A.S. (Akira Sakai) and R.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the subsidy for Advanced Integrated Intelligence Platform (MEXT), and the commissioned projects income for RIKEN AIP-FUJITSU Collaboration Center.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of RIKEN, Fujitsu Ltd., Showa University, and the National Cancer Center (approval ID: Wako1 29-4).
Informed Consent Statement
This research protocol was approved by the medical ethics committees of the four collaborating research facilities, and data collection was conducted in an opt-out manner.
Data Availability Statement
Data sharing is not applicable owing to the patient privacy rights. The source code of the method proposed in this study is available on GitHub at https://github.com/rafcc/2020-prenatal-sono.
Acknowledgments
The authors are grateful to Hisayuki Sano, Hiroyuki Yoshida, and all members of Hamamoto laboratory for their helpful discussion and support. We also thank the medical doctors in the Showa University Hospitals for data collection.
Conflicts of Interest
R.H. has received the joint research grant from Fujitsu Ltd. The other authors declare no conflict of interest.
Appendix A
This appendix describes implementation details, preprocessing, and training of YOLOv2. In this study, we followed except slight modification of training parameters for our data [19]. The implementation of YOLOv2 in this paper is available from (https://github.com/pjreddie/darknet), which is the original code of YOLOv2 developed by the authors of [19]. YOLOv2 is implemented using C language with a Python wrapper. About the network configuration and pre-training process, we totally followed the paper [19]. YOLOv2 employs the darknet-19 network, which is a convolutional network with a leaky ReLU activation function; the detailed configuration is described in the main text of [19]. In the pre-training process, we pre-trained the darknet-19 network using ImageNet, and adopted it as the backbone network of YOLOv2 by changing the input resolution 224 × 224 pixels to 416 × 416 pixels according to the description in [19].
The pre-trained YOLOv2 was used to train the fetal cardiac substructures as follows. The stochastic gradient descent method with the Nesterov momentum was adopted for optimization. The learning rate was set to 0.001, momentum factor to 0.9, and weight decay to 0.0005. The mini-batch size was set to 64. The maximum number of iterations (i.e., the number of processed mini-batches) was set to 80,200. The learning rate was multiplied by a factor of 0.1 for 40,000 iterations and 60,000 iterations. Models were saved for every 1000 iterations, and the model with the highest mAP of cardiac substructure detection for the validation data was selected. As for the preprocessing step, the input images were resized to 416 × 416 pixels.
About the software version, YOLOv2 was compiled using GCC 4.8.5. The Python version was 3.6, and other libraries used in this study were scikit-learn 0.19.1, OpenCV-Python 3.4.0.12, and NumPy 1.14.1. About the hardware, we employed the computer server, which has Intel(R) Xeon(R) CPU E5-2690 v4 at 2.60 GHz, GeForce GTX 1080 Ti.
References
- Hamamoto, R.; Suvarna, K.; Yamada, M.; Kobayashi, K.; Shinkai, N.; Miyake, M.; Takahashi, M.; Jinnai, S.; Shimoyama, R.; Sakai, A.; et al. Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers 2020, 12, 3532. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Petrini, J.R.; Broussard, C.S.; Gilboa, S.M.; Lee, K.A.; Oster, M.; Honein, M.A. Racial differences by gestational age in neonatal deaths attributable to congenital heart defects—United States, 2003–2006. MMWR Morb. Mortal. Wkly. Rep. 2010, 59, 1208–1211. [Google Scholar]
- Wren, C.; Richmond, S.; Donaldson, L. Temporal variability in birth prevalence of cardiovascular malformations. Heart 2000, 83, 414–419. [Google Scholar] [CrossRef]
- Meberg, A.; Otterstad, J.E.; Froland, G.; Lindberg, H.; Sorland, S.J. Outcome of congenital heart defects—A population-based study. Acta Paediatr. 2000, 89, 1344–1351. [Google Scholar] [CrossRef]
- Holland, B.J.; Myers, J.A.; Woods, C.R., Jr. Prenatal diagnosis of critical congenital heart disease reduces risk of death from cardiovascular compromise prior to planned neonatal cardiac surgery: A meta-analysis. Ultrasound Obstet. Gynecol. 2015, 45, 631–638. [Google Scholar] [CrossRef]
- Donofrio, M.T.; Moon-Grady, A.J.; Hornberger, L.K.; Copel, J.A.; Sklansky, M.S.; Abuhamad, A.; Cuneo, B.F.; Huhta, J.C.; Jonas, R.A.; Krishnan, A.; et al. Diagnosis and treatment of fetal cardiac disease: A scientific statement from the American Heart Association. Circulation 2014, 129, 2183–2242. [Google Scholar] [CrossRef]
- American Institute of Ultrasound in Medicine. AIUM practice guideline for the performance of fetal echocardiography. J. Ultrasound Med. 2013, 32, 1067–1082. [Google Scholar] [CrossRef]
- Tegnander, E.; Williams, W.; Johansen, O.J.; Blaas, H.G.; Eik-Nes, S.H. Prenatal detection of heart defects in a non-selected population of 30,149 fetuses—Detection rates and outcome. Ultrasound Obstet. Gynecol. 2006, 27, 252–265. [Google Scholar] [CrossRef]
- Cuneo, B.F.; Curran, L.F.; Davis, N.; Elrad, H. Trends in prenatal diagnosis of critical cardiac defects in an integrated obstetric and pediatric cardiac imaging center. J. Perinatol. 2004, 24, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Bridge, C.P.; Noble, J.A.; Zisserman, A. Temporal HeartNet: Towards human-level automatic analysis of fetal cardiac screening video. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Philadelphia, PA, USA, 2017; pp. 341–349. [Google Scholar]
- Baumgartner, C.F.; Kamnitsas, K.; Matthew, J.; Fletcher, T.P.; Smith, S.; Koch, L.M.; Kainz, B.; Rueckert, D. SonoNet: Real-Time Detection and Localisation of Fetal Standard Scan Planes in Freehand Ultrasound. IEEE Trans. Med. Imaging 2017, 36, 2204–2215. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, R.; Curran, L.; Zhao, Y.; Levine, J.; Chinn, E.; Moon-Grady, A. Expert-level prenatal detection of complex congenital heart disease from screening ultrasound using deep learning. medRxiv 2020. [Google Scholar] [CrossRef]
- Dozen, A.; Komatsu, M.; Sakai, A.; Komatsu, R.; Shozu, K.; Machino, H.; Yasutomi, S.; Arakaki, T.; Asada, K.; Kaneko, S.; et al. Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information. Biomolecules 2020, 10, 1526. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Sinclair, M.; Zimmer, V.; Hou, B.; Rajchl, M.; Toussaint, N.; Oktay, O.; Schlemper, J.; Gomez, A.; Housden, J.; et al. Weakly Supervised Estimation of Shadow Confidence Maps in Fetal Ultrasound Imaging. IEEE Trans. Med. Imaging 2019, 38, 2755–2767. [Google Scholar] [CrossRef] [PubMed]
- American College of Obstetricians & Gynecologists. ACOG Practice Bulletin No. 101: Ultrasonography in pregnancy. Obstet. Gynecol. 2009, 113, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.S.; Allan, L.D.; Chaoui, R.; Copel, J.A.; DeVore, G.R.; Hecher, K.; Lee, W.; Munoz, H.; Paladini, D.; Tutschek, B.; et al. ISUOG Practice Guidelines (updated): Sonographic screening examination of the fetal heart. Ultrasound Obstet. Gynecol. 2013, 41, 348–359. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7263–7271. [Google Scholar]
- Everingham, M.; Eslami, S.A.; Van Gool, L.; Williams, C.K.; Winn, J.; Zisserman, A. The pascal visual object classes challenge: A retrospective. Int. J. Comput. Vis. 2015, 111, 98–136. [Google Scholar] [CrossRef]
- Hasan, M.; Choi, J.; Neumann, J.; Roy-Chowdhury, A.K.; Davis, L.S. Learning temporal regularity in video sequences. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 733–742. [Google Scholar]
- Narasimhan, M.G.; Kamath, S. Dynamic video anomaly detection and localization using sparse denoising autoencoders. Multimed. Tools Appl. 2018, 77, 13173–13195. [Google Scholar] [CrossRef]
- Schlegl, T.; Seeböck, P.; Waldstein, S.M.; Schmidt-Erfurth, U.; Langs, G. Unsupervised anomaly detection with generative adversarial networks to guide marker discovery. In International Conference on Information Processing in Medical Imaging; Springer: Philadelphia, PA, USA, 2017; pp. 146–157. [Google Scholar]
- Friedberg, M.K.; Silverman, N.H.; Moon-Grady, A.J.; Tong, E.; Nourse, J.; Sorenson, B.; Lee, J.; Hornberger, L.K. Prenatal detection of congenital heart disease. J. Pediatr. 2009, 155, 26–31. [Google Scholar] [CrossRef]
- Ogge, G.; Gaglioti, P.; Maccanti, S.; Faggiano, F.; Todros, T. Prenatal screening for congenital heart disease with four-chamber and outflow-tract views: A multicenter study. Ultrasound Obstet. Gynecol. 2006, 28, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Tsuzuki, T.; Akatsuka, J.; Ueki, M.; Morikawa, H.; Numata, Y.; Takahara, T.; Tsuyuki, T.; Tsutsumi, K.; Nakazawa, R.; et al. Automated acquisition of explainable knowledge from unannotated histopathology images. Nat. Commun. 2019, 10, 5642. [Google Scholar] [CrossRef] [PubMed]
- Toba, S.; Mitani, Y.; Yodoya, N.; Ohashi, H.; Sawada, H.; Hayakawa, H.; Hirayama, M.; Futsuki, A.; Yamamoto, N.; Ito, H.; et al. Prediction of Pulmonary to Systemic Flow Ratio in Patients With Congenital Heart Disease Using Deep Learning-Based Analysis of Chest Radiographs. JAMA Cardiol. 2020, 5, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Codari, M.; Sardanelli, F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur. Radiol. Exp. 2018, 2, 35. [Google Scholar] [CrossRef]
- Barinov, L.; Jairaj, A.; Becker, M.; Seymour, S.; Lee, E.; Schram, A.; Lane, E.; Goldszal, A.; Quigley, D.; Paster, L. Impact of Data Presentation on Physician Performance Utilizing Artificial Intelligence-Based Computer-Aided Diagnosis and Decision Support Systems. J. Digit. Imaging 2019, 32, 408–416. [Google Scholar] [CrossRef]
- Kusunose, K.; Abe, T.; Haga, A.; Fukuda, D.; Yamada, H.; Harada, M.; Sata, M. A Deep Learning Approach for Assessment of Regional Wall Motion Abnormality From Echocardiographic Images. JACC Cardiol. Imaging 2020, 13, 374–381. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).