Identification of Transcription Factors, Biological Pathways, and Diseases as Mediated by N6-methyladenosine Using Tensor Decomposition-Based Unsupervised Feature Extraction
Abstract
1. Introduction
2. Materials and Methods
2.1. m6A, Histone Modification, and Gene Expression
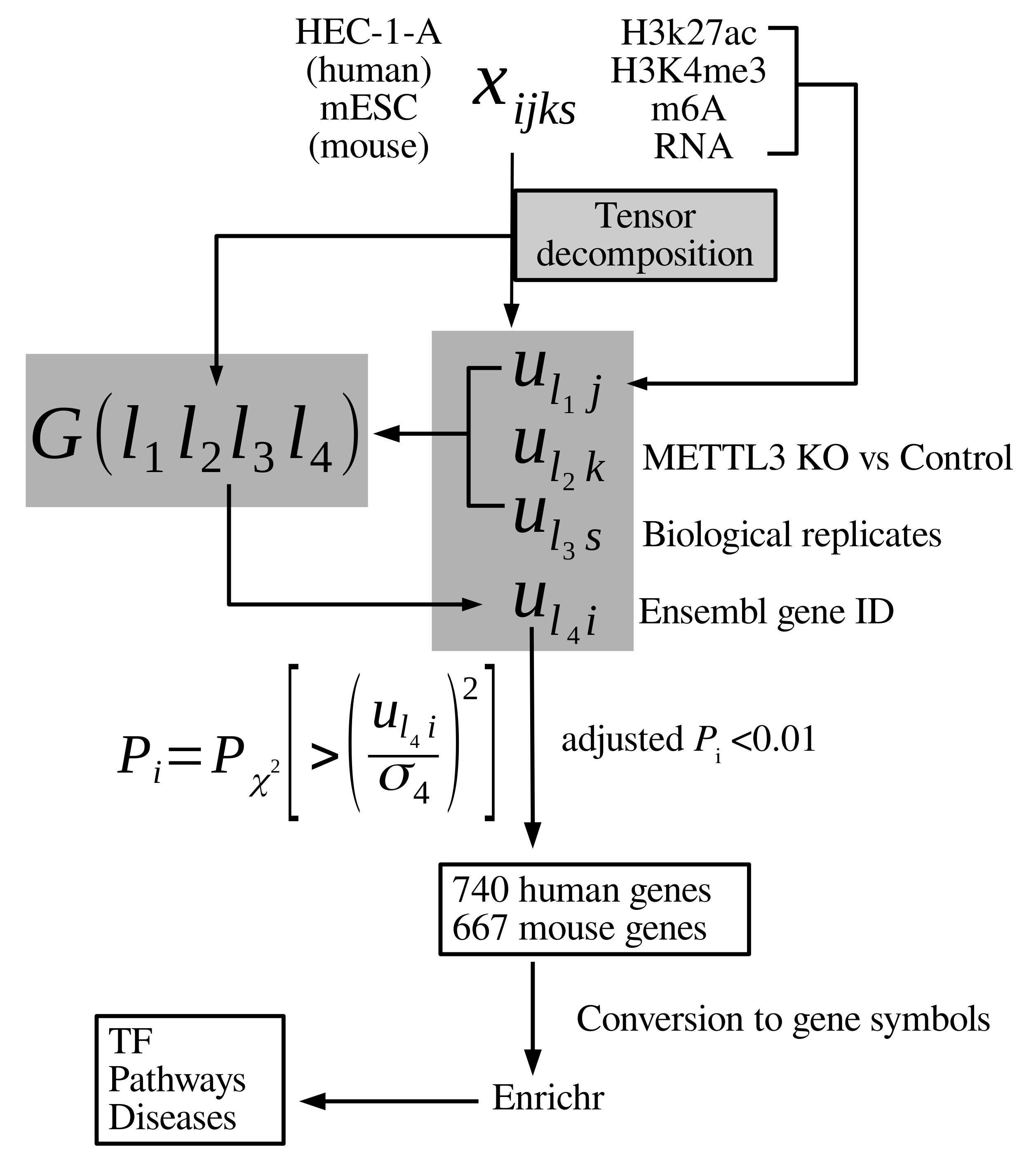
2.2. Tensor
2.3. Tensor Decomposition-Based Unsupervised Feature Extraction
2.4. Enrichment Analysis
3. Results
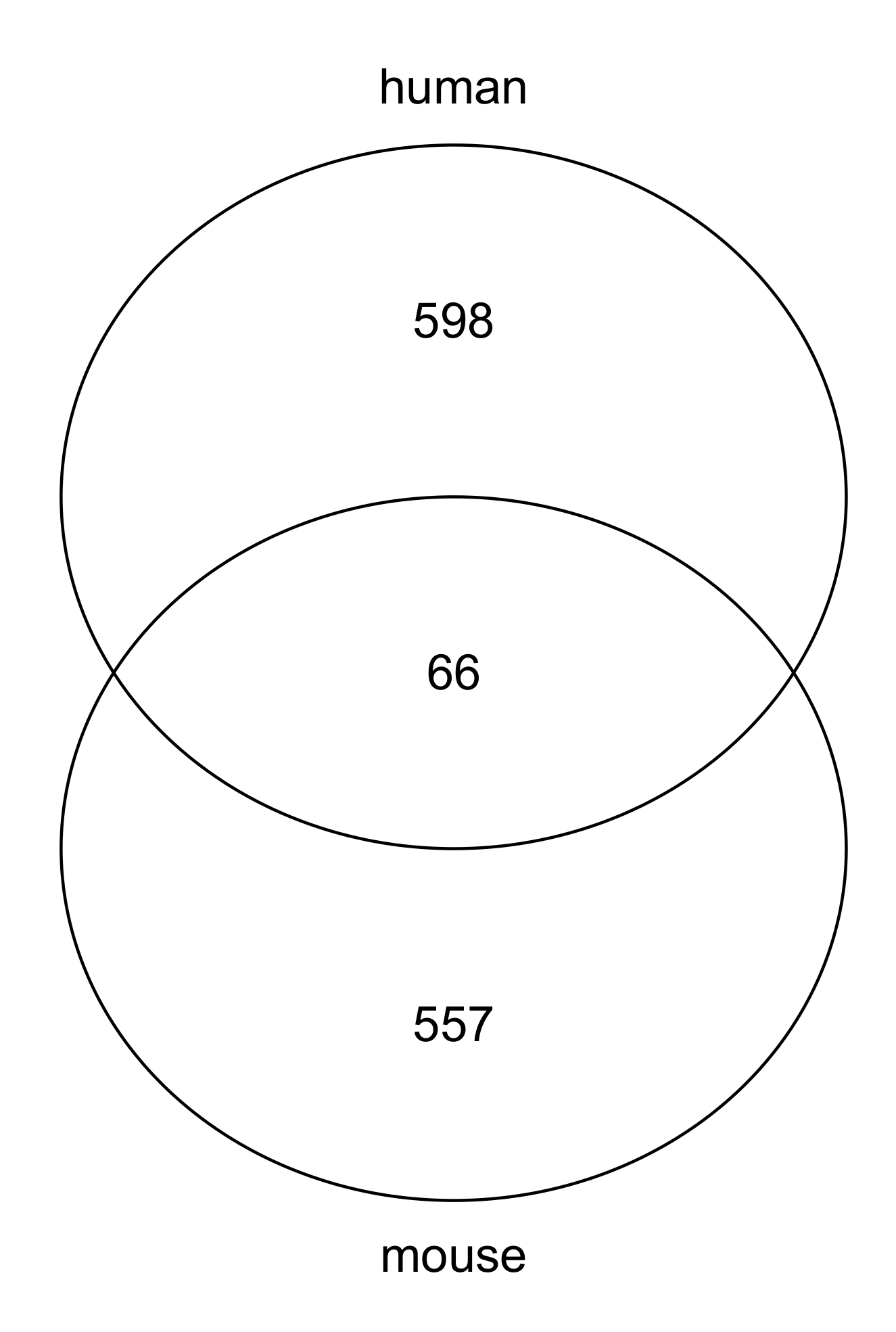
3.1. Gene Selections
- , which is attributed to measurements, represents constant values for histone modifications, m6A, and nascent RNA; it likely exhibits transcription activated by m6A through histone modification.
- (and for mESC), which are both attributed to the distinction between METTL3 KO and control, exhibit the distinction between them; they are more likely related to mA6 mediated effects.
- , which is attributed to two biological replicates, represents constant values between two replicates; it likely exhibits features independent of replicates.
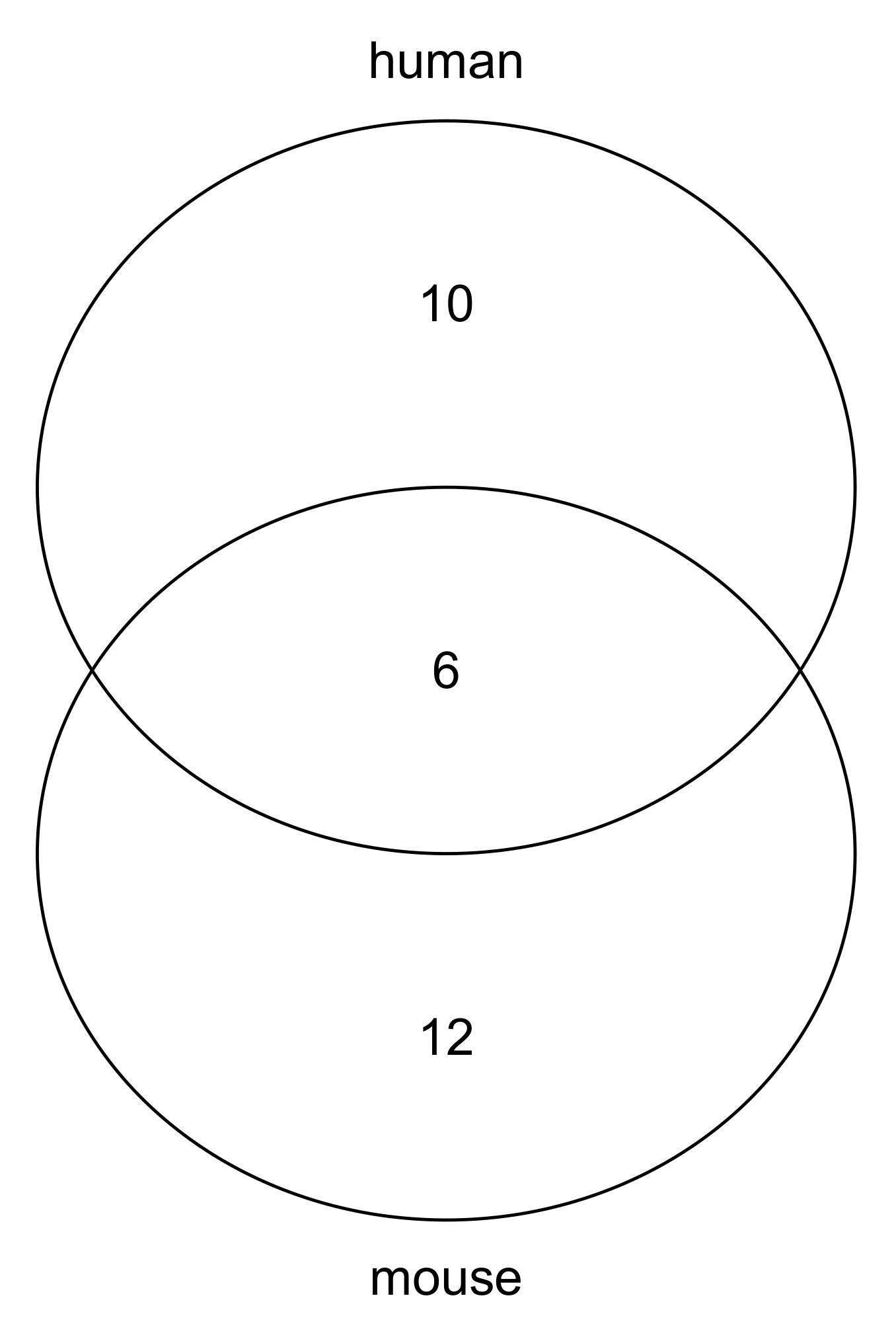
3.2. Transcription Factors Likely to Be Recruited by m6A
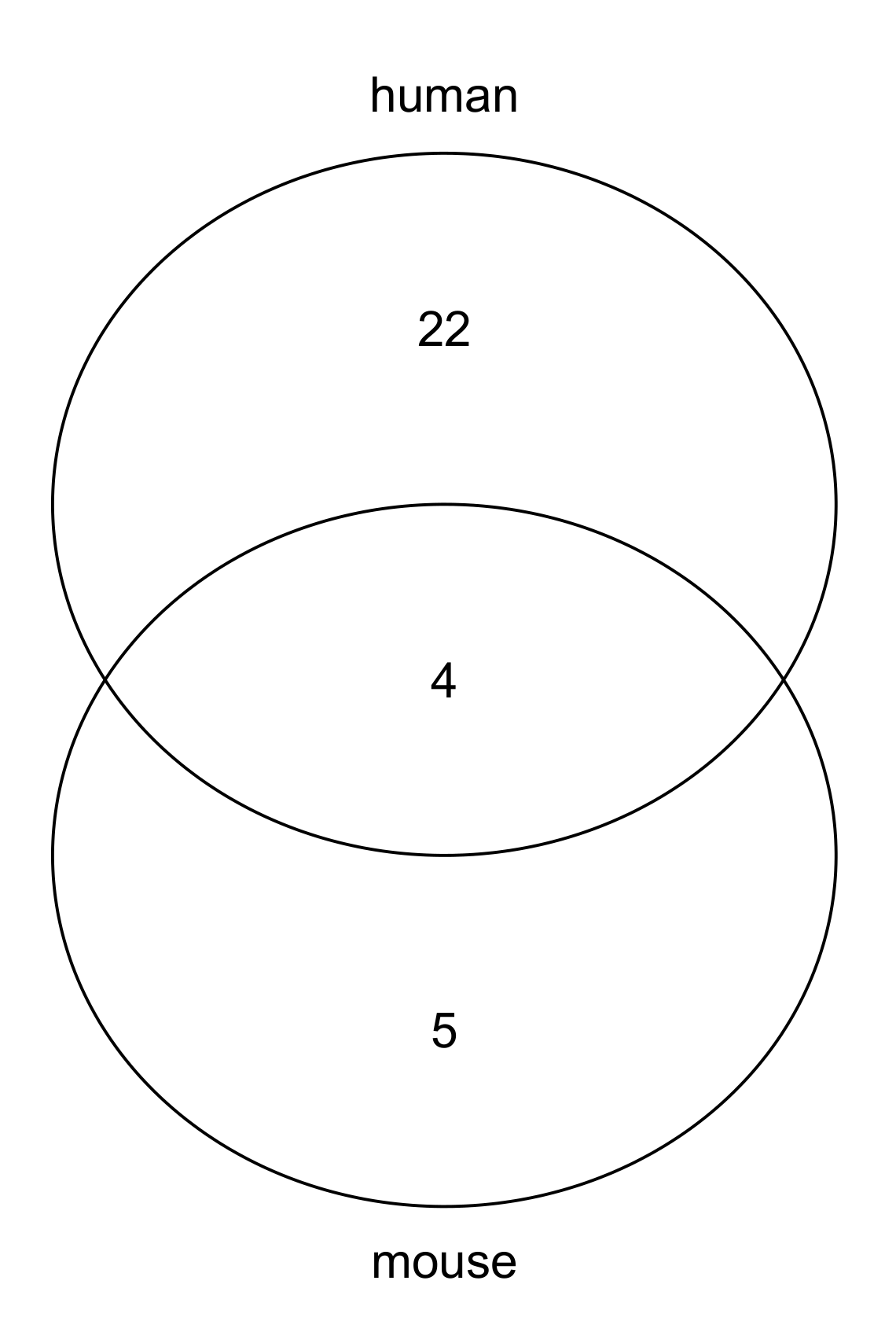
3.3. Biological Processes
3.4. Diseases Mediated by m6A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TD | tensor decomposition |
| TF | transcription factor |
| FE | feature extraction |
| HOSVD | higher order singular value decomposition |
| mESC | mouse embryonic stem cell |
References
- Martin, J. Epitranscriptomics: Mapping methods and beyond. BioTechniques 2018, 65, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2015, 17, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.T.; Tsukahara, T. C-to-U editing and site-directed RNA editing for the correction of genetic mutations. BioSci. Trends 2017, 11, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. Available online: https://rnajournal.cshlp.org/content/3/11/1233 (accessed on 27 December 2020).
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2013, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Fazi, F.; Fatica, A. Interplay Between N6-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front. Cell Dev. Biol. 2019, 7, 116. [Google Scholar] [CrossRef]
- An, S.; Zhang, J.; Wang, Y.; Zhang, Y.; Liu, Q. The Roles of N6-Methyladenosine in Human Diseases. Biochem. Insights 2019, 12. [Google Scholar] [CrossRef]
- Huang, H.; Camats-Perna, J.; Medeiros, R.; Anggono, V.; Widagdo, J. Altered Expression of the m6A Methyltransferase METTL3 in Alzheimer’s Disease. Eneuro 2020. [Google Scholar] [CrossRef]
- Qin, L.; Min, S.; Shu, L.; Pan, H.; Zhong, J.; Guo, J.; Sun, Q.; Yan, X.; Chen, C.; Tang, B.; et al. Genetic analysis of N6-methyladenosine modification genes in Parkinson’s disease. Neurobiol. Aging 2020, 93, 143.e9–143.e13. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18. [Google Scholar] [CrossRef]
- Winkler, R.; Gillis, E.; Lasman, L.; Safra, M.; Geula, S.; Soyris, C.; Nachshon, A.; Tai-Schmiedel, J.; Friedman, N.; Le-Trilling, V.T.K.; et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2018, 20, 173–182. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dou, X.; Chen, C.; Chen, C.; Liu, C.; Xu, M.M.; Zhao, S.; Shen, B.; Gao, Y.; Han, D.; et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 2020, 367, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, J.; Renaud, Y.; Albrecht, S.; Ghavi-Helm, Y.; Roignant, J.Y.; Silies, M.; Junion, G. m6A RNA methylation regulates promoter proximal pausing of RNA Polymerase II. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhou, K.I.; Shi, H.; Lyu, R.; Wylder, A.C.; Matuszek, Ż; Pan, J.N.; He, C.; Parisien, M.; Pan, T. Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol. Cell 2019, 76, 70–81.e9. [Google Scholar] [CrossRef]
- Yang, X.; Liu, Q.L.; Xu, W.; Zhang, Y.C.; Yang, Y.; Ju, L.F.; Chen, J.; Chen, Y.S.; Li, K.; Ren, J.; et al. m6A promotes R-loop formation to facilitate transcription termination. Cell Res. 2019, 29, 1035–1038. [Google Scholar] [CrossRef]
- Taguchi, Y.H. Unsupervised Feature Extraction Applied to Bioinformatics; Springer International Publishing: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [CrossRef]
- Lachmann, A.; Xu, H.; Krishnan, J.; Berger, S.I.; Mazloom, A.R.; Ma’ayan, A. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 2010, 26, 2438–2444. [Google Scholar] [CrossRef] [PubMed]
- Tewari, A.K.; Yardimci, G.; Shibata, Y.; Sheffield, N.C.; Song, L.; Taylor, B.S.; Georgiev, S.G.; Coetzee, G.A.; Ohler, U.; Furey, T.S.; et al. Chromatin accessibility reveals insights into androgen receptor activation and transcriptional specificity. Genome Biol. 2012, 13, R88. [Google Scholar] [CrossRef] [PubMed]
- Magri, L.; Swiss, V.A.; Jablonska, B.; Lei, L.; Pedre, X.; Walsh, M.; Zhang, W.; Gallo, V.; Canoll, P.; Casaccia, P. E2F1 Coregulates Cell Cycle Genes and Chromatin Components during the Transition of Oligodendrocyte Progenitors from Proliferation to Differentiation. J. Neurosci. 2014, 34, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Jeselsohn, R.; Bergholz, J.S.; Pun, M.; Cornwell, M.; Liu, W.; Nardone, A.; Xiao, T.; Li, W.; Qiu, X.; Buchwalter, G.; et al. Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell 2018, 33, 173–186.e5. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Vakoc, C.R.; Ying, L.; Mandat, S.; Wang, H.; Zheng, X.; Blobel, G.A. Exchange of GATA Factors Mediates Transitions in Looped Chromatin Organization at a Developmentally Regulated Gene Locus. Mol. Cell 2008, 29, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Giammartino, D.C.D.; Kloetgen, A.; Polyzos, A.; Liu, Y.; Kim, D.; Murphy, D.; Abuhashem, A.; Cavaliere, P.; Aronson, B.; Shah, V.; et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat. Cell Biol. 2019, 21, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- Nepon-Sixt, B.S.; Bryant, V.L.; Alexandrow, M.G. Myc-driven chromatin accessibility regulates Cdc45 assembly into CMG helicases. Commun. Biol. 2019, 2. [Google Scholar] [CrossRef]
- Pálfy, M.; Schulze, G.; Valen, E.; Vastenhouw, N.L. Chromatin accessibility established by Pou5f3, Sox19b and Nanog primes genes for activity during zebrafish genome activation. PLoS Genet. 2020, 16, 1–25. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Steger, D.J.; Zhuo, D.; Qatanani, M.; Mullican, S.E.; Tuteja, G.; Manduchi, E.; Grant, G.R.; Lazar, M.A. Cell-Specific Determinants of Peroxisome Proliferator-Activated Receptor Function in Adipocytes and Macrophages. Mol. Cell. Biol. 2010, 30, 2078–2089. [Google Scholar] [CrossRef]
- Gao, F.; Wei, Z.; An, W.; Wang, K.; Lu, W. The interactomes of POU5F1 and SOX2 enhancers in human embryonic stem cells. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Kim, J.; Xu, S.; Xiong, L.; Yu, L.; Fu, X.; Xu, Y. SALL4 promotes glycolysis and chromatin remodeling via modulating HP1α-Glut1 pathway. Oncogene 2017, 36, 6472–6479. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zeng, C.; Tarasova, N.I.; Chasovskikh, S.; Dritschilo, A.; Timofeeva, O.A. A new role for STAT3 as a regulator of chromatin topology. Transcription 2013, 4, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Sammons, M.A.; Zhu, J.; Drake, A.M.; Berger, S.L. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2014, 25, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Yi, G.; Zhou, H. p63 cooperates with CTCF to modulate chromatin architecture in skin keratinocytes. Epigenet. Chromatin 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Gaspar-Maia, A.; Alajem, A.; Polesso, F.; Sridharan, R.; Mason, M.J.; Heidersbach, A.; Ramalho-Santos, J.; McManus, M.T.; Plath, K.; Meshorer, E.; et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 2009, 460, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Boxer, L.D.; Barajas, B.; Tao, S.; Zhang, J.; Khavari, P.A. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014, 28, 2013–2026. [Google Scholar] [CrossRef]
- Durant, M.; Pugh, B.F. Genome-Wide Relationships between TAF1 and Histone Acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006, 26, 2791–2802. [Google Scholar] [CrossRef]
- Sanij, E.; Diesch, J.; Lesmana, A.; Poortinga, G.; Hein, N.; Lidgerwood, G.; Cameron, D.P.; Ellul, J.; Goodall, G.J.; Wong, L.H.; et al. A novel role for the Pol I transcription factor UBTF in maintaining genome stability through the regulation of highly transcribed Pol II genes. Genome Res. 2015, 25, 201–212. [Google Scholar] [CrossRef]
- Ramos Pittol, J.M.; Oruba, A.; Mittler, G.; Saccani, S.; van Essen, D. Zbtb7a is a transducer for the control of promoter accessibility by NF-kappa B and multiple other transcription factors. PLoS Biol. 2018, 16, 1–33. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2019, 48, D498–D503. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2017, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2020, gkaa970. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Chai, G.; Wu, Y.; Li, J.; Chen, F.; Liu, J.; Luo, G.; Tauler, J.; Du, J.; Lin, S.; et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Jiang, T.; Lei, S.; She, Y.; Shi, H.; Zhou, S.; Ou, J.; Liu, Y. Expression of circular RNAs during C2C12 myoblast differentiation and prediction of coding potential based on the number of open reading frames and N6-methyladenosine motifs. Cell Cycle 2018, 17, 1832–1845. [Google Scholar] [CrossRef]
- Prendergast, G.C.; Gibbs, J.B. Pathways of Ras Function: Connections to the Actin Cytoskeleton; Advances in Cancer Research; Academic Press: Cambridge, MA, USA, 1993; Volume 62, pp. 19–64. [Google Scholar] [CrossRef]
- Wang, J.C.; Lee, J.Y.J.; Christian, S.; Dang-Lawson, M.; Pritchard, C.; Freeman, S.A.; Gold, M.R. The Rap1-cofilin pathway coordinates actin reorganization and MTOC polarization at the B-cell immune synapse. J. Cell Sci. 2017, 130, 1094–1109. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, Y.; Ma, H.; Zeng, R.; Liu, R.; Wang, P.; Jin, X.; Zhao, Y. Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol. Brain 2020, 13. [Google Scholar] [CrossRef]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2019, 21, 36–51. [Google Scholar] [CrossRef]
- Widagdo, J.; Anggono, V. The m6A-epitranscriptomic signature in neurobiology: From neurodevelopment to brain plasticity. J. Neurochem. 2018, 147, 137–152. [Google Scholar] [CrossRef]
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Röh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403.e9. [Google Scholar] [CrossRef]
- Ueberham, U.; Arendt, T. The Role of Smad Proteins for Development, Differentiation and Dedifferentiation of Neurons. In Trends in Cell Signaling Pathways in Neuronal Fate Decision; InTech: London, UK, 2013. [Google Scholar] [CrossRef]
- Luo, L. RHO GTPASES in neuronal morphogenesis. Nat. Rev. Neurosci. 2000, 1, 173–180. [Google Scholar] [CrossRef]
- Li, M.; Armelloni, S.; Ikehata, M.; Corbelli, A.; Pesaresi, M.; Calvaresi, N.; Giardino, L.; Mattinzoli, D.; Nisticò, F.; Andreoni, S.; et al. Nephrin expression in the adult rodent central nervous system and its interaction with glutamate receptors. J. Pathol. 2011, 225, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Sebastian-delaCruz, M.; Olazagoitia-Garmendia, A.; Gonzalez-Moro, I.; Santin, I.; Garcia-Etxebarria, K.; Castellanos-Rubio, A. Implication of m6A mRNA Methylation in Susceptibility to Inflammatory Bowel Disease. Epigenomes 2020, 4, 16. [Google Scholar] [CrossRef]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Guan, R.; Hong, W.; Huang, B.; Liu, P.; Guo, X.; Hu, S.; Yu, M.; Hou, B. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann. Transl. Med. 2020, 8, 387. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.W.; Imam, H.; Khan, M.; Siddiqui, A. N6-Methyladenosine Modification of Hepatitis B and C Viral RNAs Attenuates Host Innate Immunity via RIG-I Signaling. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Ji, G.; Huang, C.; He, S.; Gong, Y.; Song, G.; Li, X.; Zhou, L. Comprehensive analysis of m6A regulators prognostic value in prostate cancer. Aging 2020, 12, 14863–14884. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, X.; Chen, N.; Lv, X.; Ge, X.; Lu, K.; Zhou, X.; Yang, J.; Han, Y.; Hu, S.; et al. N6-Methyladenosine RNA Methylation Regulators Contribute to Malignant Progression and Survival Prediction in Chronic Lymphocytic Leukemia. Blood 2019, 134, 1738. [Google Scholar] [CrossRef]






| GEO ID | Treatments | Observed | Replicates |
|---|---|---|---|
| GSM4174073 | Control | H3K4me3 | r1 |
| GSM4174074 | Control | H3K4me3 | r2 |
| GSM4174075 | KD | H3K4me3 | r1 |
| GSM4174076 | KD | H3K4me3 | r2 |
| GSM4174077 | Control | H3K27ac | r1 |
| GSM4174078 | Control | H3K27ac | r2 |
| GSM4174079 | KD | H3K27ac | r1 |
| GSM4174080 | KD | H3K27ac | r2 |
| GSM4174099 | Control | MeRIP | r1 |
| GSM4174100 | Control | MeRIP | r2 |
| GSM4174101 | KD | MeRIP | r1 |
| GSM4174102 | KD | MeRIP | r2 |
| GSM4174167 | Control | nuclearRNA | r1 |
| GSM4174168 | Control | nuclearRNA | r2 |
| GSM4174169 | KD | nuclearRNA | r1 |
| GSM4174170 | KD | nuclearRNA | r2 |
| GEO ID | Treatments | Observed | Replicates |
|---|---|---|---|
| GSM3912479 | Control | H3K27ac | r1 |
| GSM3912480 | Control | H3K27ac | r2 |
| GSM3912481 | Control | H3K4me3 | r1 |
| GSM3912482 | Control | H3K4me3 | r2 |
| GSM3912485 | KO1 | H3K27ac | r1 |
| GSM3912486 | KO1 | H3K27ac | r2 |
| GSM3912487 | KO1 | H3K4me3 | r1 |
| GSM3912488 | KO1 | H3K4me3 | r2 |
| GSM3912491 | KO2 | H3K27ac | r1 |
| GSM3912492 | KO2 | H3K27ac | r2 |
| GSM3912493 | KO2 | H3K4me3 | r1 |
| GSM3912494 | KO2 | H3K4me3 | r2 |
| GSM3912555 | Control | nuclearRNA | r1 |
| GSM3912556 | Control | nuclearRNA | r2 |
| GSM3912557 | KO1 | nuclearRNA | r1 |
| GSM3912558 | KO1 | nuclearRNA | r2 |
| GSM3912559 | KO2 | nuclearRNA | r1 |
| GSM3912560 | KO2 | nuclearRNA | r2 |
| GSM3912800 | Control | MeRIP | r1 |
| GSM3912801 | Control | MeRIP | r2 |
| GSM3912802 | KO1 | MeRIP | r1 |
| GSM3912803 | KO1 | MeRIP | r2 |
| GSM3912804 | KO2 | MeRIP | r1 |
| GSM3912805 | KO2 | MeRIP | r2 |
| Human | Mouse | ||
|---|---|---|---|
| 1 | −3.8580489 | 11.451996 | 0.3547811 |
| 2 | −8.3420060 | −3.105822 | 0.5361026 |
| 3 | 1.2758702 | −1.979357 | −15.3679662 |
| 4 | 0.4298687 | 21.579441 | 2.8415422 |
| 5 | −72.7824354 | −82.092367 | −31.1421313 |
| 6 | 10.1455134 | 1.676667 | 19.6058307 |
| 7 | −10.1008315 | −7.248264 | 66.0183422 |
| 8 | −10.3176091 | 5.825304 | 4.3903138 |
| 9 | −1.4442934 | 38.314394 | −23.3314778 |
| 10 | 0.3916004 | 6.841401 | 7.6772055 |
| Human | |||
|---|---|---|---|
| Not Selected | Selected | ||
| mouse | not selected | 15704 | 496 |
| selected | 499 | 63 | |
| CHEA | AR E2F1 ESR1 GATA2 KLF4 MYC NANOG NFE2L2 PPARG POU5F1 SMAD4 SALL4 SOX2 STAT3 TP53 TP63 |
| ENCODE | CHD1 CTCF PBX3 RCOR1 RELA TAF1 TCF3 UBTF YY1 ZBTB7A ZNF384 |
| Pathway Name | Found | Ratio | p-Value | FDR |
|---|---|---|---|---|
| Transcriptional regulation of pluripotent stem cells | 13/45 | |||
| Developmental Biology | 23/1241 | |||
| Generic Transcription Pathway | 24/1554 | |||
| POU5F1 (OCT4), SOX2, NANOG activate genes related to proliferation | 6/21 | |||
| Gene expression (Transcription) | 25/1851 | |||
| RNA Polymerase II Transcription | 24/1693 | |||
| Interleukin-4 and Interleukin-13 signaling | 9/211 | |||
| ESR-mediated signaling | 9/256 | |||
| Estrogen-dependent gene expression | 7/154 | |||
| Transcriptional regulation of granulopoiesis | 5/71 | |||
| Signaling by Nuclear Receptors | 9/385 | |||
| POU5F1 (OCT4), SOX2, NANOG repress genes related to differentiation | 3/10 | |||
| Binding of TCF/LEF:CTNNB1 to target gene promoters | 3/10 | |||
| Activation of PUMA and translocation to mitochondria | 3/10 | |||
| RUNX3 regulates WNT signaling | 3/10 | |||
| Repression of WNT target genes | 3/16 | |||
| Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors | 4/52 | |||
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | 3/20 | |||
| Signaling by Interleukins | 10/639 | |||
| Transcriptional regulation by RUNX3 | 5/118 | |||
| TFAP2 (AP-2) family regulates transcription of growth factors and their receptors | 3/21 | |||
| Intrinsic Pathway for Apoptosis | 4/61 |
| "Regulation of actin cytoskeleton"; "Adherens junction"; "Focal adhesion"; "Rap1 signaling pathway"; "Ras signaling pathway"; "Proteoglycans in cancer" |
| "Developmental Biology Homo sapiens R-HSA-1266738"; "Axon guidance Homo sapiens R-HSA-422475"; "Downregulation of SMAD2/3:SMAD4 transcriptional activity Homo sapiens R-HSA-2173795"; "L1CAM interactions Homo sapiens R-HSA-373760"; "Nephrin interactions Homo sapiens R-HSA-373753"; "Signaling by Rho GTPases Homo sapiens R-HSA-194315" |
| "NEURONAL EPITHELIUM"; "PLACENTA (BULK)"; "ASTROCYTE"; "FIBROBLAST" |
| Disease Perturbations from GEO down |
| "Crohn’s disease DOID-8778 human GSE6731 sample 757"; "Ulcerative Colitis C0009324 human GSE6731 sample 249"; "Ulcerative colitis DOID-8577 human GSE6731 sample 759"; "Ulcerative colitis DOID-8577 human GSE6731 sample 760"; "Hepatitis C DOID-1883 human GSE20948 sample 599"; "Diabetic Nephropathy C0011881 human GSE1009 sample 223"; "Cardiac Hypertrophy C1383860 rat GSE1055 sample 354"; "Prostate cancer DOID-10283 human GSE3868 sample 639"; "Hepatitis C DOID-1883 human GSE20948 sample 597"; "Cancer of prostate C0376358 human GSE3868 sample 135" |
| Disease Perturbations from GEO up |
| "Pancreatitis DOID-4989 mouse GSE3644 sample 513 Asthma"; "Allergic C0155877 human GSE3004 sample 360"; "Acute pancreatitis C0001339 mouse GSE3644 sample 376"; "Chronic lymphocytic leukemia DOID-1040 human GSE6691 sample 786" |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taguchi, Y.-h.; Dharshini, S.A.P.; Gromiha, M.M. Identification of Transcription Factors, Biological Pathways, and Diseases as Mediated by N6-methyladenosine Using Tensor Decomposition-Based Unsupervised Feature Extraction. Appl. Sci. 2021, 11, 213. https://doi.org/10.3390/app11010213
Taguchi Y-h, Dharshini SAP, Gromiha MM. Identification of Transcription Factors, Biological Pathways, and Diseases as Mediated by N6-methyladenosine Using Tensor Decomposition-Based Unsupervised Feature Extraction. Applied Sciences. 2021; 11(1):213. https://doi.org/10.3390/app11010213
Chicago/Turabian StyleTaguchi, Y-h., S. Akila Parvathy Dharshini, and M. Michael Gromiha. 2021. "Identification of Transcription Factors, Biological Pathways, and Diseases as Mediated by N6-methyladenosine Using Tensor Decomposition-Based Unsupervised Feature Extraction" Applied Sciences 11, no. 1: 213. https://doi.org/10.3390/app11010213
APA StyleTaguchi, Y.-h., Dharshini, S. A. P., & Gromiha, M. M. (2021). Identification of Transcription Factors, Biological Pathways, and Diseases as Mediated by N6-methyladenosine Using Tensor Decomposition-Based Unsupervised Feature Extraction. Applied Sciences, 11(1), 213. https://doi.org/10.3390/app11010213







