Distribution of Growth-Inhibiting Bacteria against the Toxic Dinoflagellate Alexandrium catenella (Group I) in Akkeshi-Ko Estuary and Akkeshi Bay, Hokkaido, Japan
Abstract
1. Introduction
2. Materials and Methods
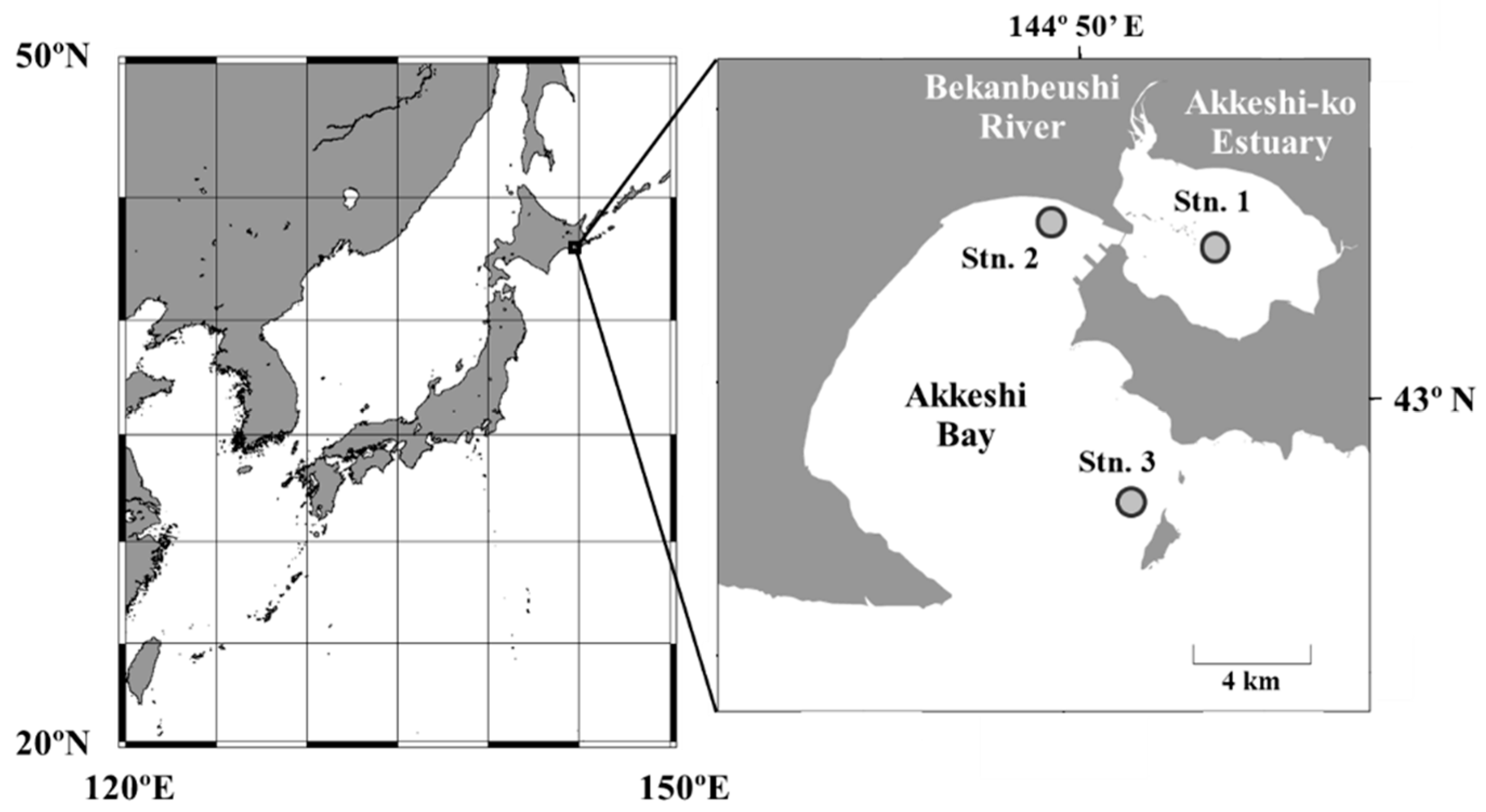
2.1. Sampling
2.2. Observation of Phytoplankton
2.3. Enumeration and Cultivation of the Bacteria in the Seawater Samples
2.4. Enumeration and Cultivation of the Bacteria on the Leaves of Seagrasses
2.5. Direct Count of the Bacteria
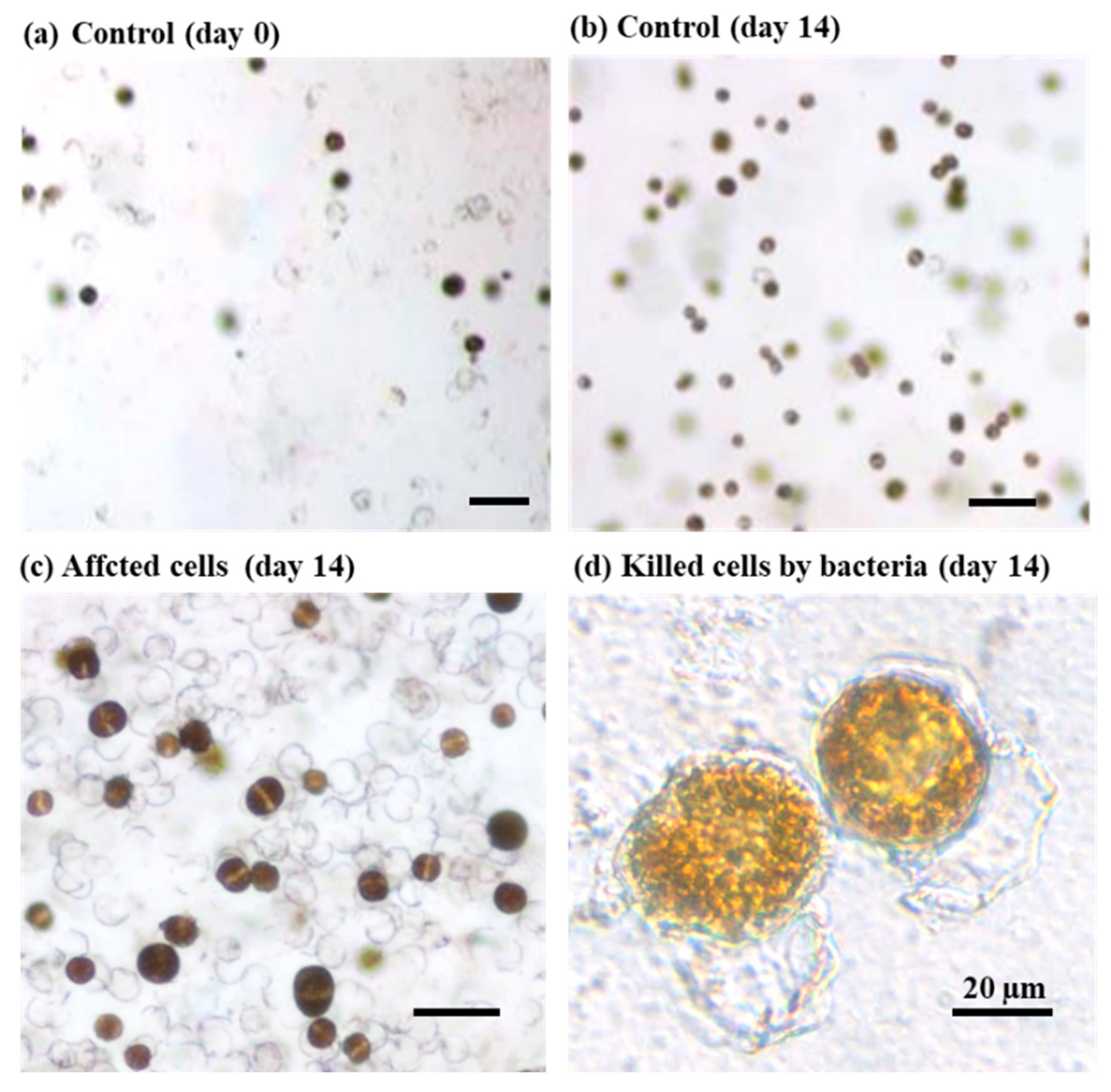
2.6. Screening of the Growth-Inhibiting Bacteria against A. catenella (Group I)
2.7. 16S Ribosomal RNA Gene Sequencing of Bacteria
3. Results
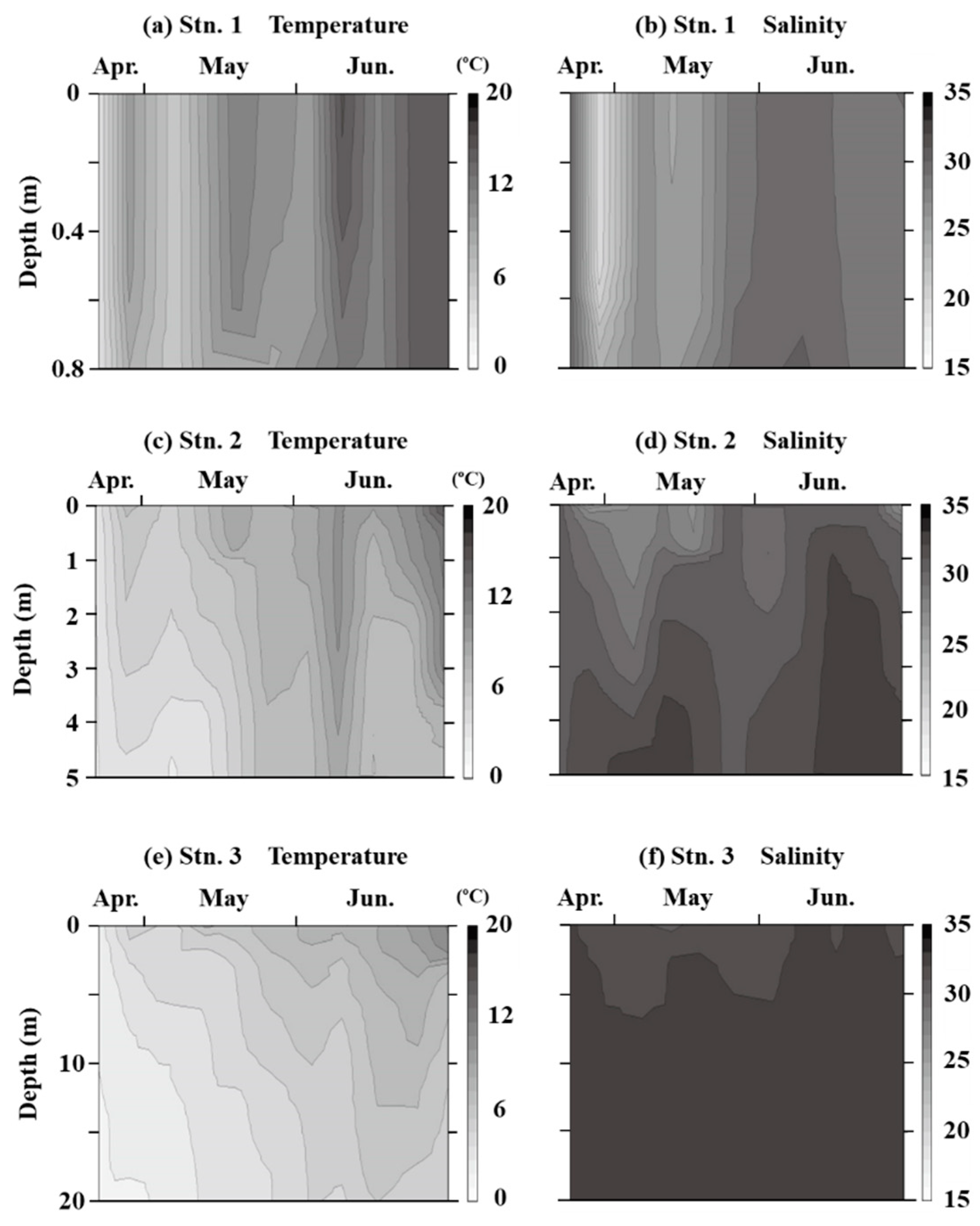
3.1. Water Environments and Seagrass Bed
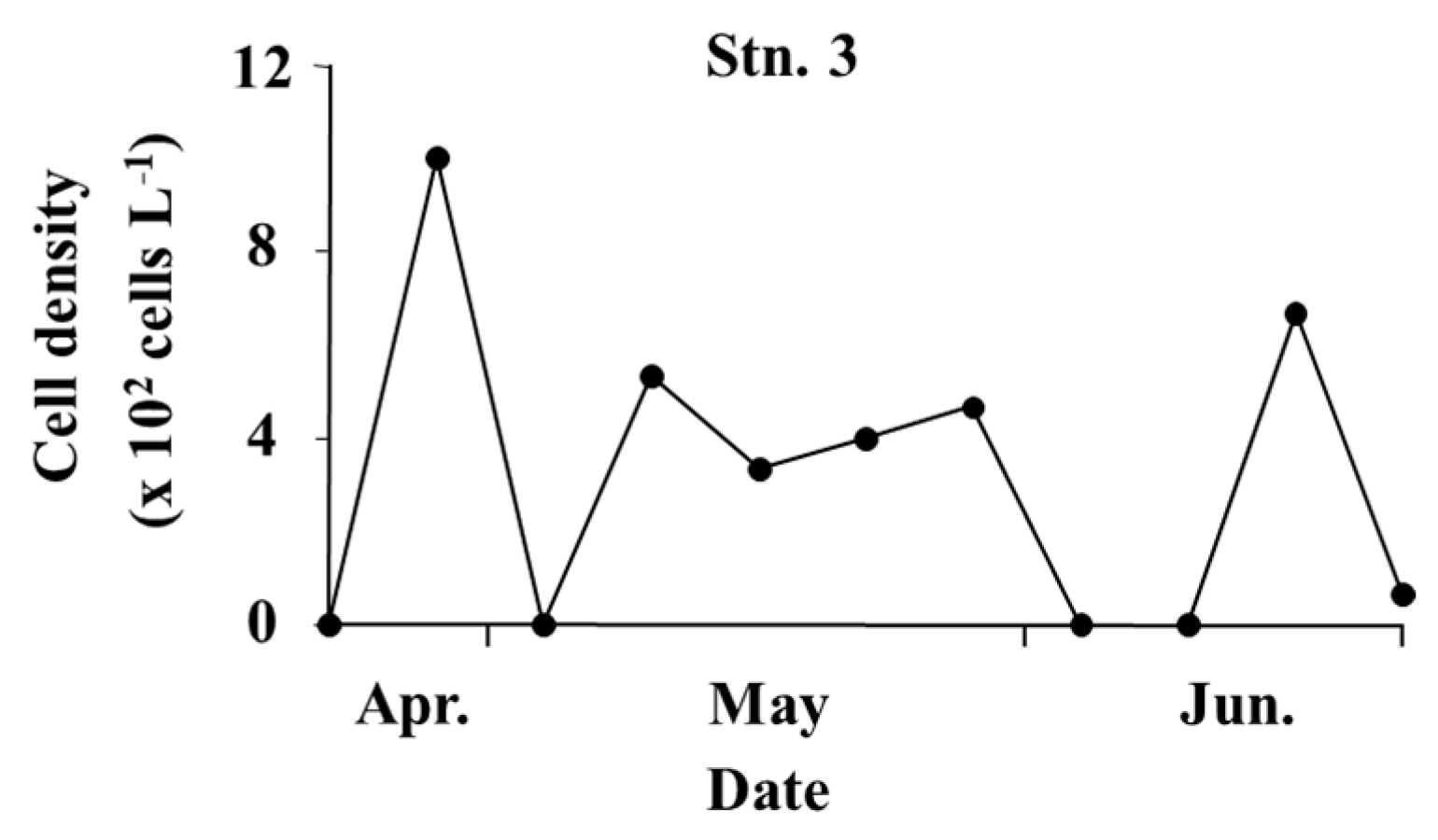
3.2. Fluctuations of Phytoplankton
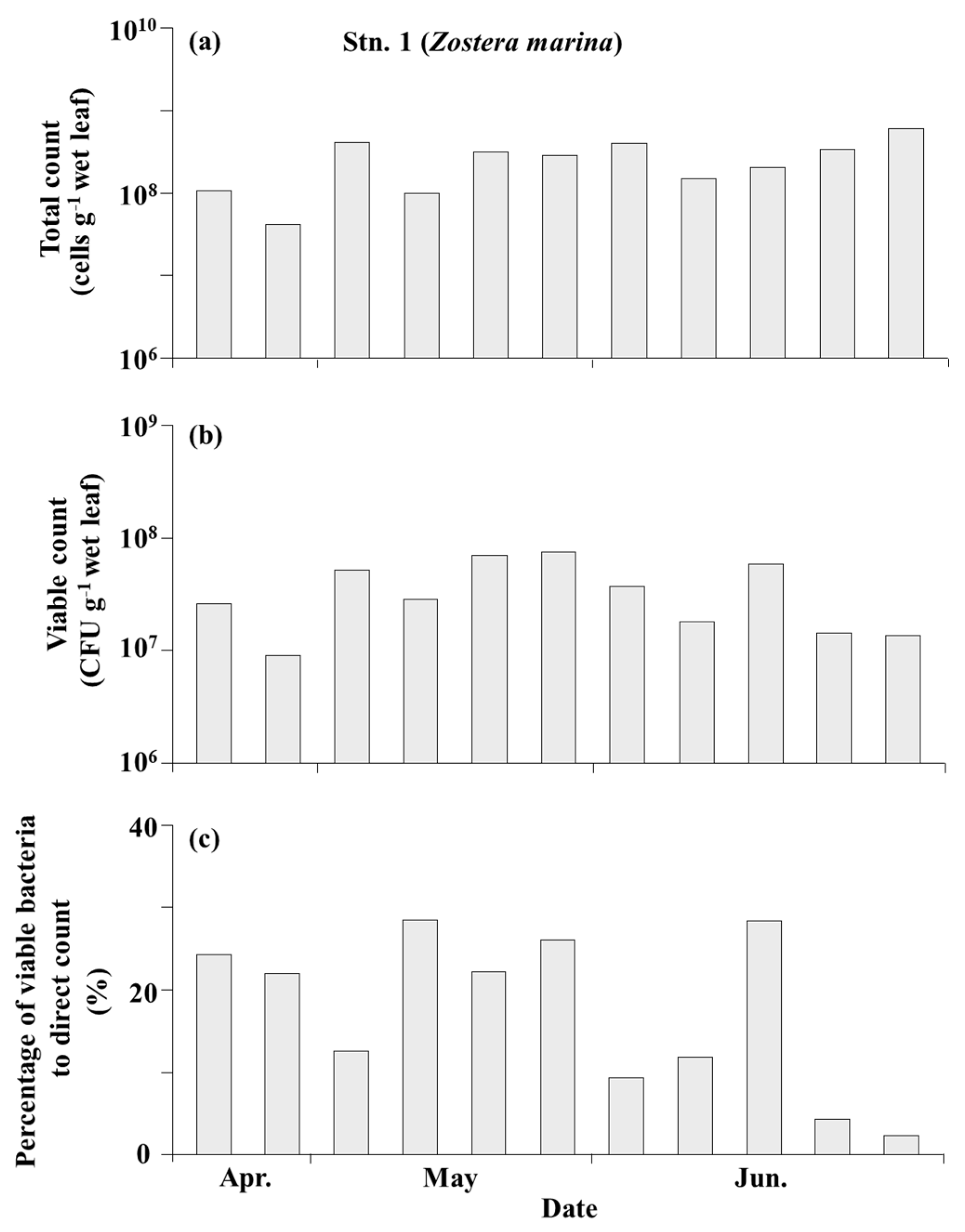
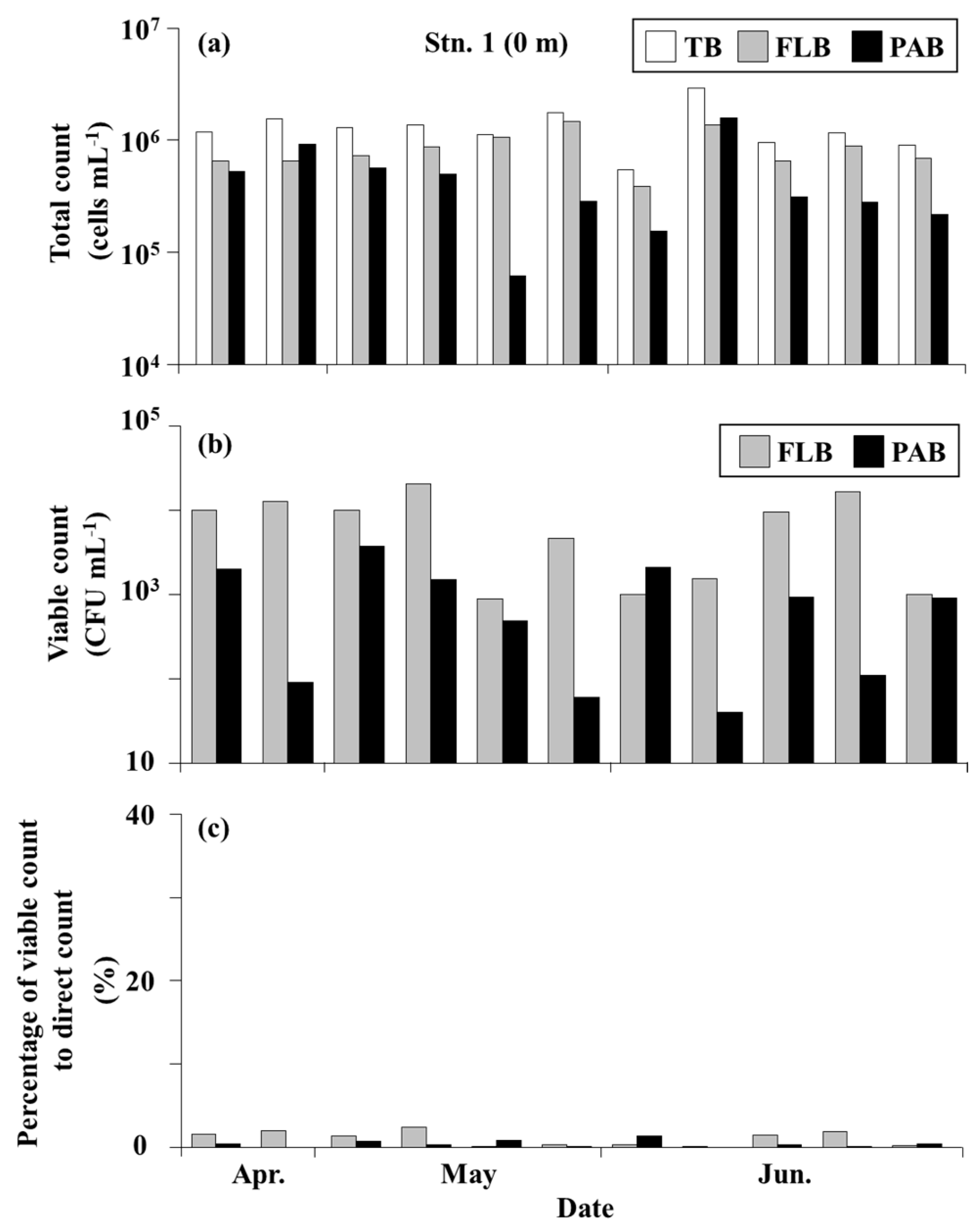
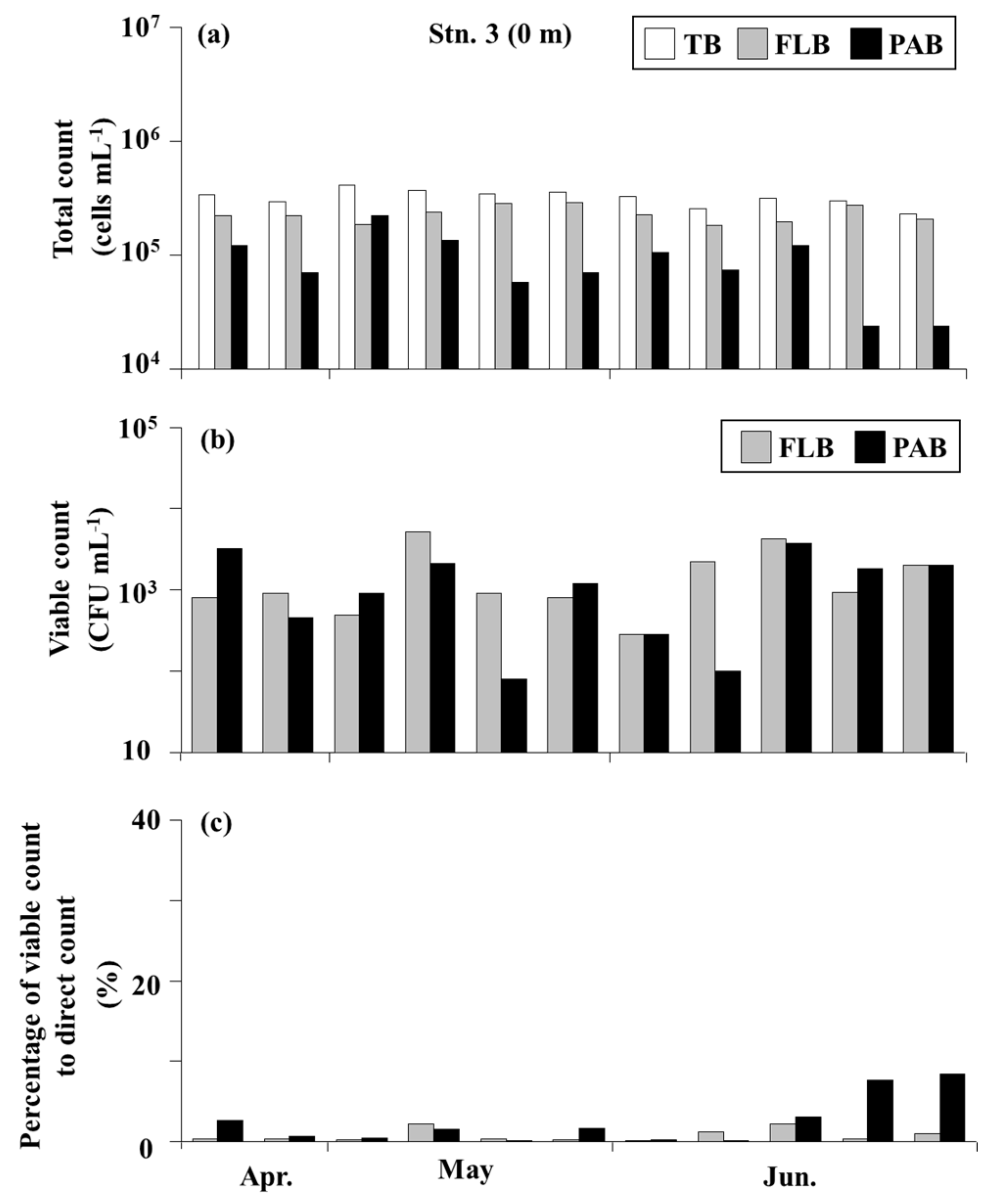
3.3. Bacteria on the Seagrass Leaves
3.4. Bacteria in the Seawater
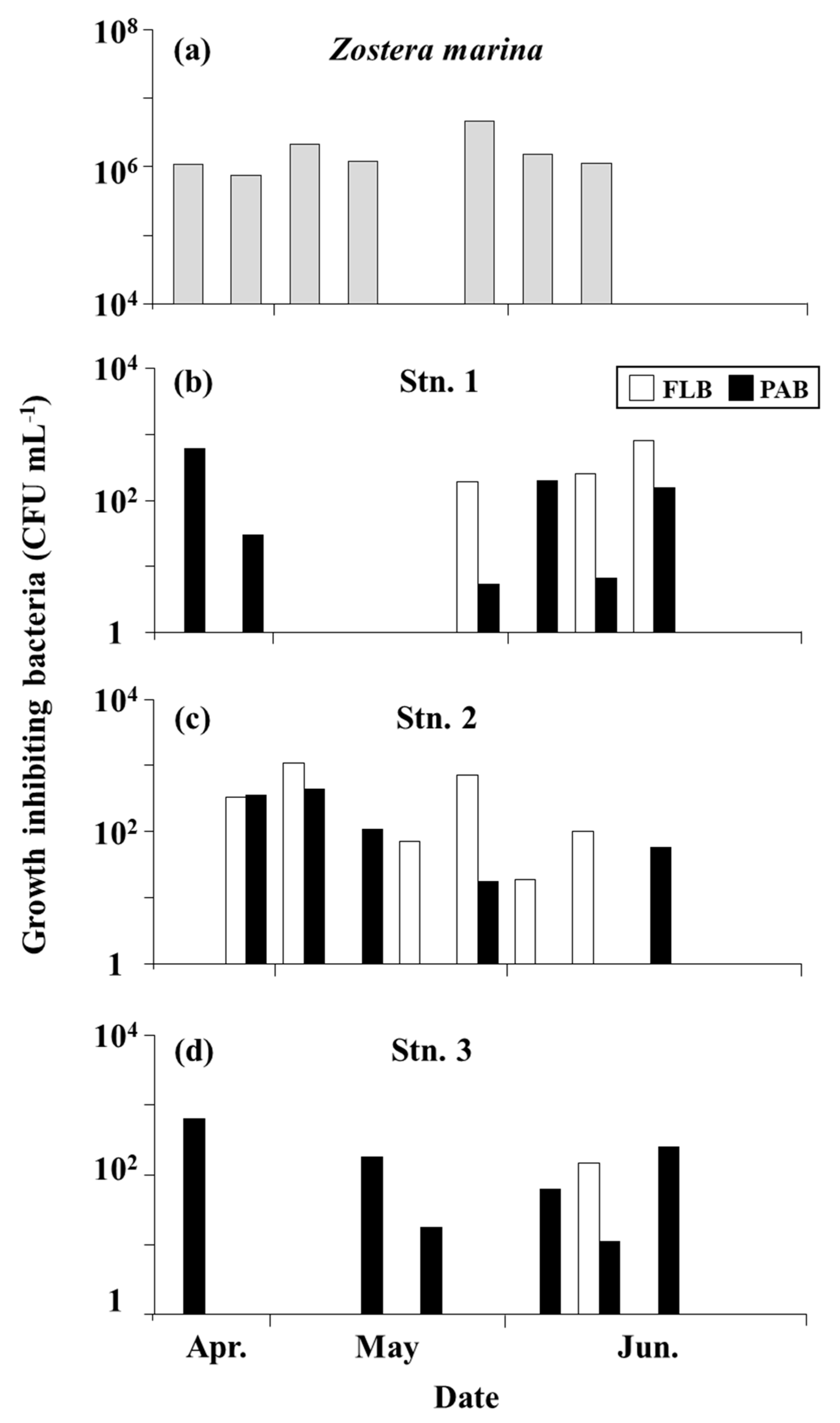
3.5. Bacteria that Inhibit the Growth of A. catenella (Group I)
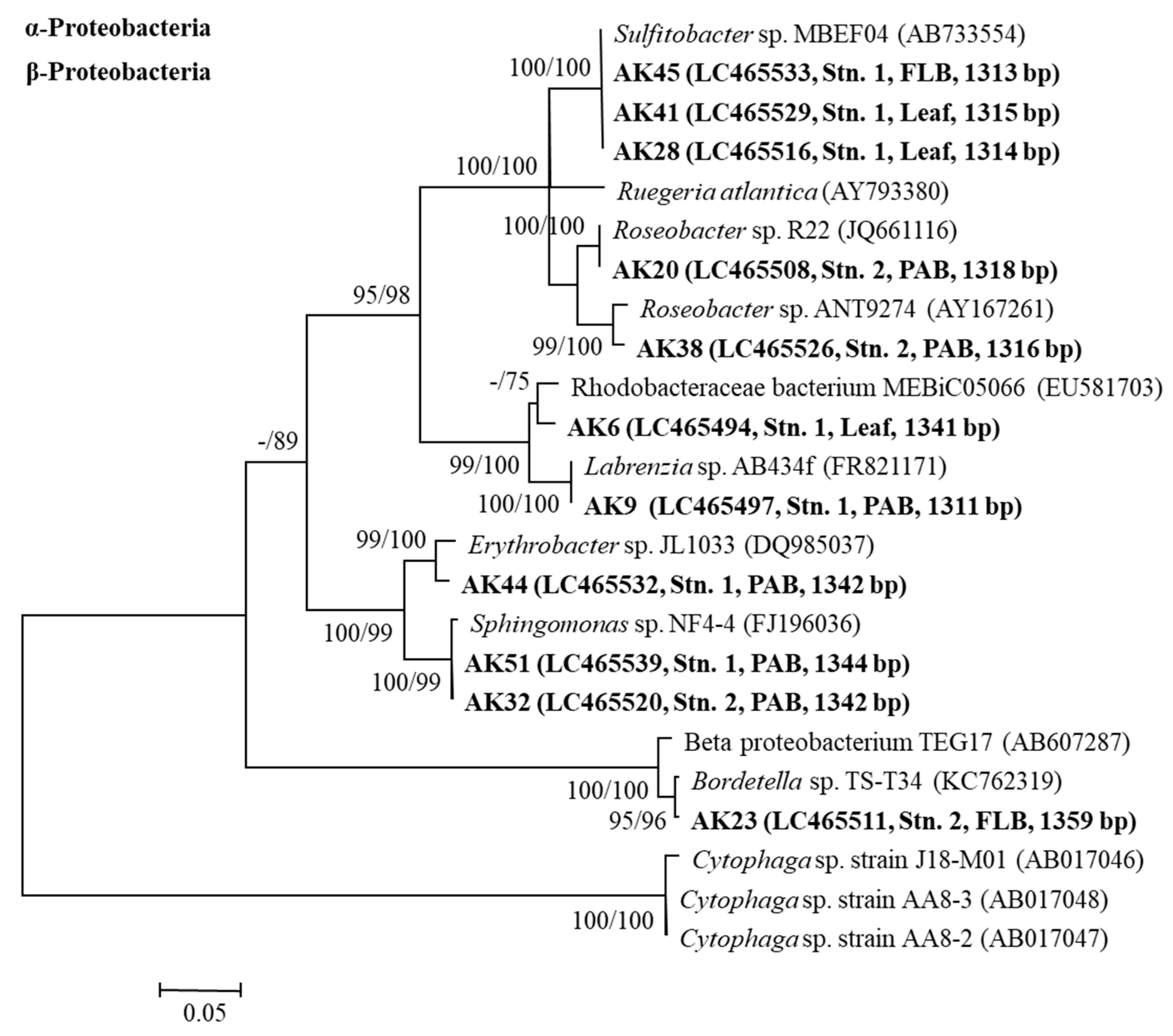
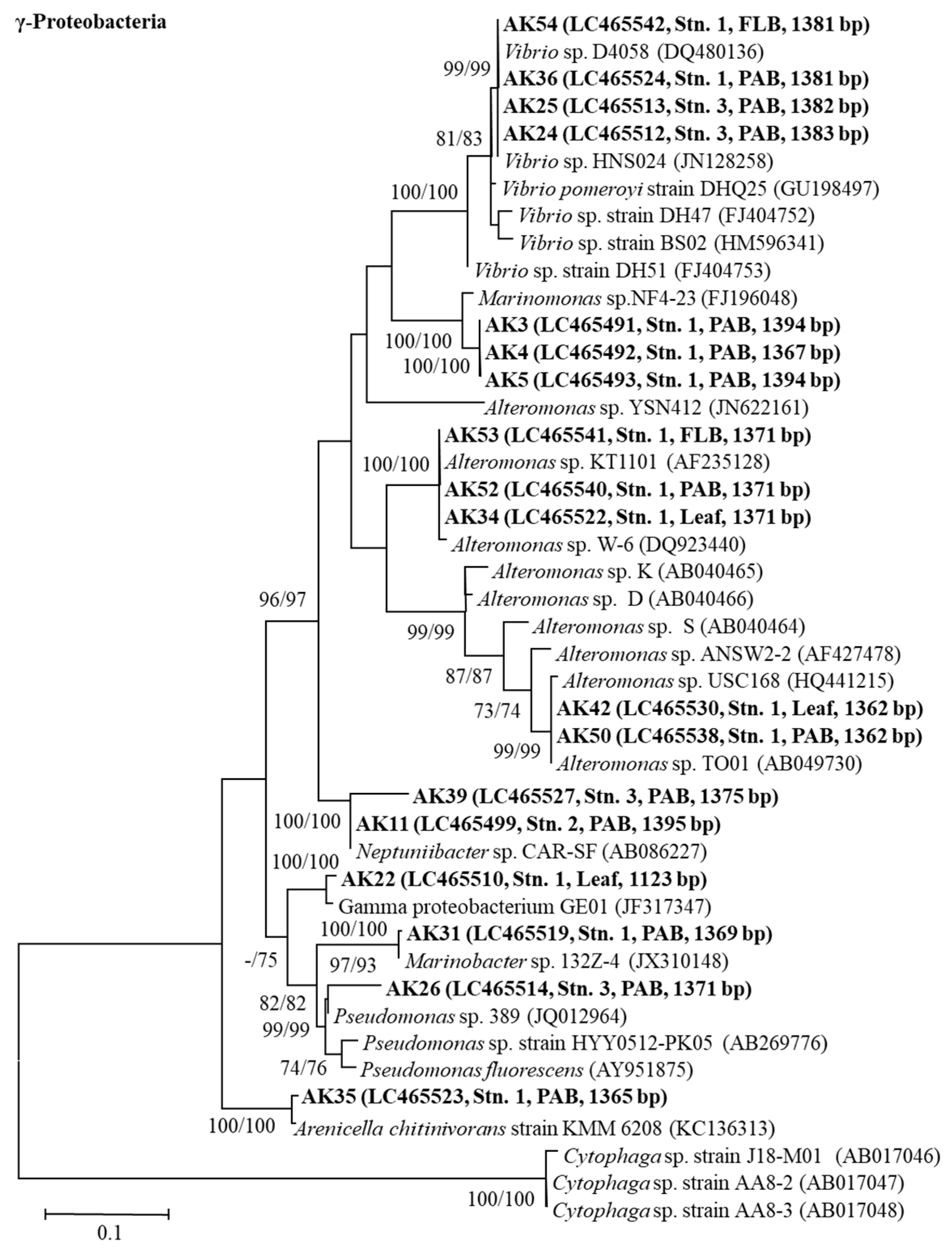
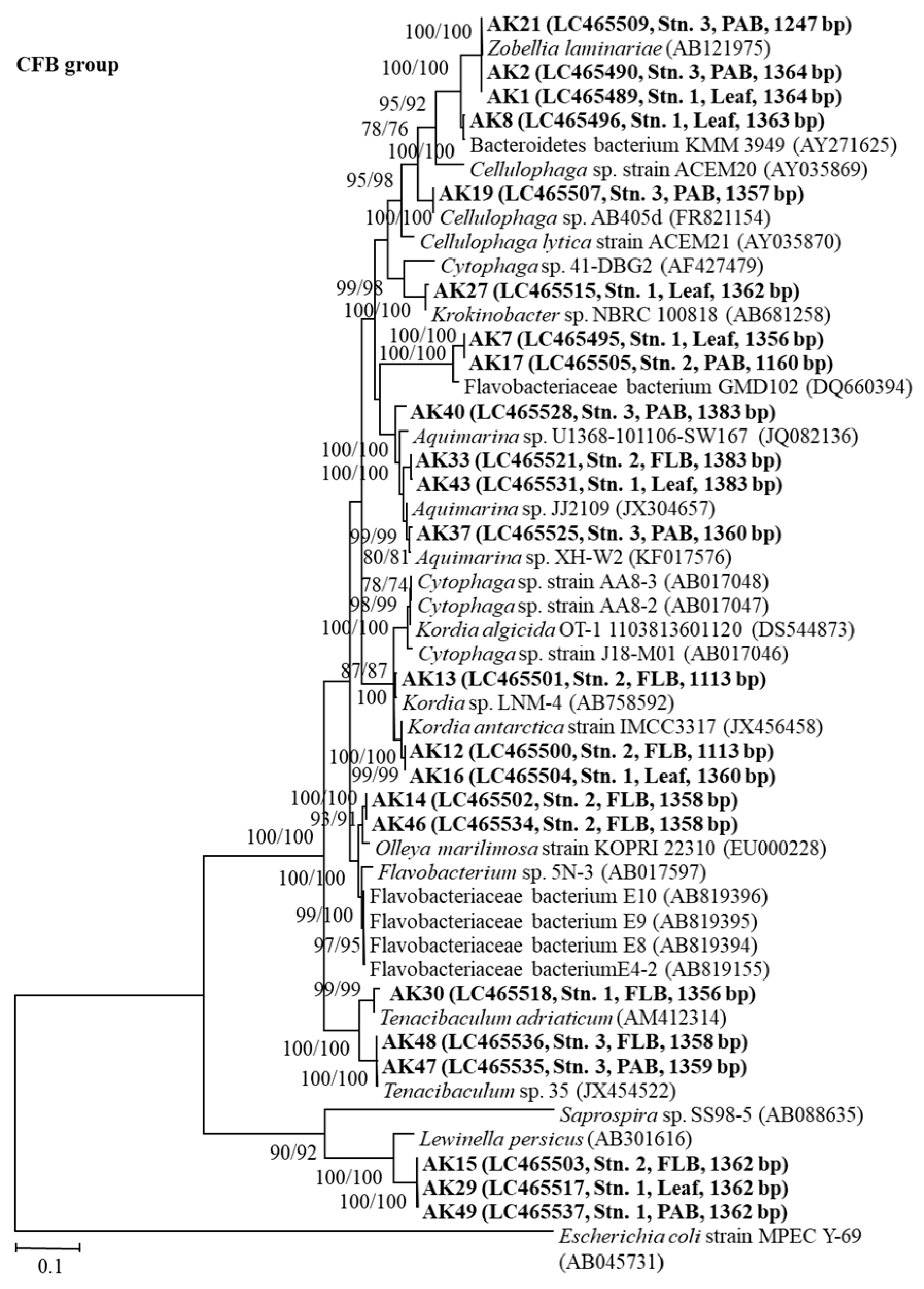
3.6. The 16S rRNA Gene Sequence Analyses of the Bacteria
4. Discussion
4.1. Hydrological Parameters
4.2. Total Bacteria and Culturable Bacteria
4.3. Distribution of Growth-Inhibiting Bacteria against A. catenella (Group I)
4.4. 16S rRNA Gene Sequence Analyses of Growth-Inhibiting Bacteria against A. catenella (Group I)
4.5. The Distribution and Density of Microalgae
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prud’homme van Reine, W. Report of the Nomenclature Committee for Algae: 15. Taxon 2017, 66, 191–192. [Google Scholar] [CrossRef]
- Imai, I.; Fukuyo, Y.; Hiroishi, S. Advanced Researches on Shellfish Poisonings: Current Status and Overview; Kouseisha-Kouseikaku: Tokyo, Japan, 2007. (In Japanese) [Google Scholar]
- Anderson, D.M.; Alpermann, T.J.; Cembella, A.D.; Collos, Y.; Masseret, E.; Montresor, M. The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 2012, 14, 10–35. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H. Biological oceanographic study on method for predicting the occurrence of paralytic shellfish toxin along the Okhotsk Sea coast off Hokkaido. Sci. Rep. Hokkaido Fish Res. Inst. 2018, 93, 1–49. (In Japanese) [Google Scholar]
- Imai, I.; Yamamoto, T.; Ishii, K.I.; Yamamoto, K. Promising prevention strategies for harmful red tides by seagrass beds as enormous sources of algicidal bacteria. In Proceedings of the 5th World Fisheries Congress, Tokyo, Japan, 20−25 October 2008. 6c_0995_133. [Google Scholar]
- Imai, I.; Yamamoto, T.; Ishii, K.I.; Inaba, N.; Yamamoto, K. Prevention of harmful algal blooms by algicidal bacteria associated with eelgrasses. J. Water Waste 2016, 58, 295–300. (In Japanese) [Google Scholar]
- Imai, I.; Inaba, N.; Sakami, T. Environmentally friendly strategies for the prevention of harmful algal blooms using algicidal bacteria associated with seagrass beds. In Marine and Fresh-Water Harmful Algae; Proença, L.A.O., Hallegraeff, G.M., Eds.; Intergovernmental Oceanographic Commission of UNESCO: Florianópolis, Brazil, 2017; pp. 160–165. [Google Scholar]
- Onishi, Y.; Mohri, Y.; Tuji, A.; Ohgi, K.; Yamaguchi, A.; Imai, I. The seagrass Zostera marina harbors growth-inhibiting bacteria against the toxic dinoflagellate Alexandrium tamarense. Fish. Sci. 2014, 80, 353–362. [Google Scholar] [CrossRef]
- Imai, I. Interactions between harmful algae and algicidal and growth-inhibiting bacteria associated with seaweeds and seagrasses. In Marine Protists Diversity and Dynamics; Ohtsuka, S., Suzaki, T., Horiguchi, T., Suzuki, N., Not, F., Eds.; Springer: Tokyo, Japan, 2015; pp. 597–619. [Google Scholar]
- Inaba, N.; Trainer, V.L.; Onishi, Y.; Ishii, K.I.; Willie-Echieverria, S.; Imai, I. Algicidal and Growth-inhibiting bacteria associated with seagrass and macroalgae beds in Puget Sound WA, USA. Harmful Algae 2017, 62, 136–147. [Google Scholar] [CrossRef]
- Amaro, A.M.; Fuentes, M.S.; Ogalde, S.R.; Venegas, J.A.; Suàerz-Isla, A.B. Identification and characterization of potentially algal-lytic marine bacteria strongly associated with the toxic dinoflagellate Alexandrium catenella. J. Eukaryotic Microbiol. 2005, 52, 191–200. [Google Scholar] [CrossRef]
- Su, J.Q.; Yang, X.R.; Zheng, T.L.; Tian, Y.; Jiao, N.Z.; Cai, L.Z.; Hong, H.S. Isolation and characterization of a marine algicidal bacterium against the toxic dinoflagellate Alexandrium tamarense. Harmful Algae 2007, 6, 799–810. [Google Scholar] [CrossRef]
- Su, J.; Yang, X.; Zhou, Y.; Zheng, T. Marine bacteria antagonistic to the harmful algal bloom species Alexandrium tamarense (Dinophyceae). Biol. Control 2011, 56, 132–138. [Google Scholar] [CrossRef]
- Wang, B.X.; Zhou, Y.Y.; Bai, S.J.; Su, J.Q.; Tian, Y.; Zheng, T.L.; Yang, X.R. A novel marine bacterium algicidal to the toxic dinoflagellate Alexandrium tamarense. Lett. Appl. Microbiol. 2010, 51, 552–557. [Google Scholar] [CrossRef]
- Bai, S.J.; Huang, L.P.; Su, J.Q.; Tian, Y.; Zheng, T.L. Algicidal effects of a novel marine actinomycete on the toxic Dinoflagellate Alexandrium tamarense. Curr. Microbiol. 2011, 62, 1774–1781. [Google Scholar] [CrossRef] [PubMed]
- Imai, I.; Kawaguchi, M.; Inaba, N.; Yamamoto, K. Biological control of toxic and noxious algal blooms with environment friendly strategies. Kaiyo Mon. 2020, 52, 217–226. (In Japanese) [Google Scholar]
- Fritz, L.; Triemer, R.E. A rapid simple technique utilizing calcofluor white M2R for the visualization of dinoflagellate thecal plates. J. Phycol. 1985, 21, 662–664. [Google Scholar] [CrossRef]
- Ishida, Y.; Eguchi, M.; Kadota, H. Existence of obligatory oligotrophic bacteria as a dominant population in South China Sea and the West Pacific Ocean. Mar. Ecol. Prog. Ser. 1986, 30, 197–203. [Google Scholar] [CrossRef]
- Zimmermann, R.; Iturriaga, R.; Becker, B.J. Simultaneous determination of the total number of aquatic bacteria and the number of thereof involved in respiration. Appl. Environ. Microbiol. 1978, 36, 926–935. [Google Scholar] [CrossRef]
- Imai, I. Size distribution, number and biomass of bacteria in intertidal sediments and seawater of Ohmi Bay, Japan. Bull. Jpn. Soc. Microb. Ecol. 1987, 2, 1–11. [Google Scholar] [CrossRef][Green Version]
- Chen, L.C.M.; Edelstein, T.; McLachlan, J. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 1969, 5, 211–220. [Google Scholar] [CrossRef]
- Imai, I.; Itakura, S.; Matsuyama, Y. Selenium requirement for growth of a novel red tide flagellate Chattonella verruculosa (Raphidophyceae) in culture. Fish. Sci. 1996, 62, 834–835. [Google Scholar] [CrossRef]
- Imai, I.; Kido, T.; Yoshinaga, I.; Ohgi, K.; Nagai, S. Isolation of Microcystis-killer bacterium Agrobacterium vitis from the biofilm on the surface of the water plant Egeria densa. In Proceedings of the 14th International Conference on Harmful Algae, Crete, Greece, 1–5 November 2010; pp. 150–152. [Google Scholar]
- Turner, S.; Pryer, K.M.; Miao, V.P.R.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukuryotic Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Shinoda, Y.; Kato, N.; Morita, N. Phylogenetic analysis of a bacterium by sequencing its 16S ribosomal RNA gene. Shimadzu Rev. 2000, 57, 121–132. (In Japanese) [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Steker, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Iizumi, H.; Suzuki, K. Distribution of stable isotope ratios in Akkeshi-ko Estuary ecosystem. Kaiyo Mon. 2002, 34, 412–416. (In Japanese) [Google Scholar]
- Mukai, H.; Iizumi, H.; Kishi, M.J. Steady-state and non-steady-state material inflows of water system in Akkeshi—A case study connecting the forest and the sea. Kaiyo Mon. 2002, 34, 449–457. (In Japanese) [Google Scholar]
- Kasim, M.; Mukai, H. Contribution of benthic and epiphytic diatoms to clam and oyster production in the Akkeshi-ko Estuary. J. Oceanogr. 2006, 62, 267–281. [Google Scholar] [CrossRef]
- Ezura, Y.; Daiku, K.; Tajima, K.; Kimura, T.; Sakai, M. Seasonal differences in bacterial counts and heterotrophic bacterial flora in Akkeshi Bay. In Effect of the Ocean Environment on Microbial Activities; Colwell, R.R., Morita, R.Y., Eds.; University Park Press: Baltimore, MD, USA, 1974; pp. 112–123. [Google Scholar]
- Motoda, S.; Kawamura, T.; Nishizawa, S. Akkeshi Bay. In Productivity of Biocenoses in Coastal Regions of Japan; Hogetsu, K., Hatanaka, M., Hanaoka, T., Kawamura, T., Eds.; University of Tokyo Press: Tokyo, Japan, 1977; pp. 223–265. [Google Scholar]
- Kondo, R.; Imai, I.; Fukami, K.; Minami, A.; Hiroishi, S. Phylogenetic analysis of algicidal bacteria (Family Flavobacteriaceae) and selective detection by PCR using a specific primer set. Fish. Sci. 1999, 65, 432–435. [Google Scholar] [CrossRef]
- Imai, I. Microbial ecology in coastal systems. Bull. Coast. Oceanogr. 1989, 27, 85–101. (In Japanese) [Google Scholar]
- Kogure, K.; Simidu, U.; Taga, N.; Colwell, R. Correlation of direct viable counts with heterotrophic activity of marine bacteria. Appl. Environ. Microbiol. 1987, 53, 2332–2337. [Google Scholar] [CrossRef]
- Crump, B.C.; Armbrust, E.V.; Baross, J.A. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 1999, 65, 3192–3204. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Riquelme, C.E.; Fukami, K.; Ishida, Y. Annual fluctuations of phytoplankton and bacterial communities in Maizuru Bay and their interrelationship. Bull. Jpn. Soc. Microb. Ecol. 1987, 2, 29–37. [Google Scholar] [CrossRef]
- Zaccone, R.; Caruso, G.; Calì, C. Heterotrophic bacteria in the northern Adriatic Sea: Seasonal changes and ectoenzyme profile. Mar. Environ. Res. 2002, 54, 1–19. [Google Scholar] [CrossRef]
- Nakamura, T.; Yoshikura, T. Total bacteria and viable bacteria in Lake Biwa and Yodo River. Bull. Jpn. Soc. Microbiol. Ecol. 1989, 5, 13–20. (In Japanese) [Google Scholar] [CrossRef][Green Version]
- Kojima, S.; Miyashita, Y.; Hagiwara, T.; Yamaguchi, A.; Imai, I. Possible control strategy of cyanobacterial blooms by bacteria from reed (Phragmites australis) communities in lakes of Ohnuma Quasi-National Park, Hokkaido. Bull. Fish. Sci. Hokkaido Univ. 2016, 66, 19–28. (In Japanese) [Google Scholar]
- Kamagata, Y. How to overcome the limitation of conventional cultivation of organisms. Soil Microorg. 2017, 71, 2–5. (In Japanese) [Google Scholar]
- Park, J.H.; Yoshinaga, I.; Nishikawa, T.; Imai, I. Algicidal bacteria in particle-associated form and in free-living form during a diatom bloom in the Seto Inland Sea, Japan. Aquat. Microb. Ecol. 2010, 60, 151–161. [Google Scholar] [CrossRef][Green Version]
- Inaba, N.; Trainer, V.L.; Nagai, S.; Kojima, S.; Sakami, T.; Takagi, S.; Imai, I. Dynamics of seagrass bed microbial communities in artificial Chattonella blooms: A laboratory microcosm study. Harmful Algae 2019, 84, 139–150. [Google Scholar] [CrossRef]
- Imai, I.; Ishida, Y.; Sakaguchi, K.; Hata, Y. Algicidal marine bacteria isolated from northern Hiroshima Bay, Japan. Fish. Sci. 1995, 61, 628–636. [Google Scholar] [CrossRef]
- Sakami, T.; Sakamoto, S.; Takagi, S.; Inaba, N.; Imai, I. Distribution of three algicidal Alteromonas sp. strains in seagrass beds and surrounding areas in the Seto Inland Sea, Japan. Fish. Sci. 2017, 83, 113–121. [Google Scholar] [CrossRef]
- Imai, I. Algicidal ranges in killer bacteria of direct attack type for marine phytoplankton. Bull. Plankton Soc. Jpn. 1997, 44, 3–9. [Google Scholar]
- Meyer, N.; Bigalke, A.; Kaulfuß, A.; Pohnert, G. Strategies and ecological roles of algicidal bacteria. FEMS Microbiol. Rev. 2017, 41, 880–899. [Google Scholar] [CrossRef]
- Lee, S.O.; Kato, J.; Takiguchi, N.; Kuroda, A.; Ikeda, T.; Mitsutani, A.; Ohtake, H. Involvement of an extracellular protease in algicidal activity of the marine bacterium Pseudoalteromonas sp. strain A28. Appl. Environ. Microbiol. 2000, 66, 4334–4339. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Miyazaki, Y.; Matsuyama, Y.; Muraoka, W.; Yamaguchi, K.; Oda, T. Producing mechanism of an algicidal compound against red tide phytoplankton in a marine bacterium γ-proteobacterium. Appl. Microb. Biotechnol. 2006, 73, 684–690. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Yoshikawa, T.; Nishitarumizu, S. Algicidal activity and identification of an algicidal substance produced by marine Pseudomonas sp. C55a-2. Fish. Sci. 2011, 77, 397–402. [Google Scholar] [CrossRef]
- Umetsu, S.; Kanda, M.; Imai, I.; Sakai, R.; Fujita, M.J. Questiomycins, algicidal compounds produced by the marine bacterium Alteromonas sp. D and their production cue. Molecules 2019, 24, 4522. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Miyazono, A. Horizontal distribution of toxic Alexandrium spp. (Dinophyceae) resting cysts around Hokkaido, Japan. Plankton Biol. Ecol. 2005, 52, 76–84. [Google Scholar]
- Lee, S.Y.; Lee, I.W.; Choi, C.I. Seasonal changes of community structure of phytoplankton in three Korean seagrass beds. Ocean Polar Res. 2006, 28, 95–105. [Google Scholar] [CrossRef][Green Version]
- Wit, R.; Troussellier, M.; Courties, C.; Buffan-Dubau, E.; Lemaire, E. Short-term interactions between phytoplankton and intertidal seagrass vegetation in a coastal lagoon (Bassin d’Arcachon, SW France). Hydrobiologia 2012, 699, 55–68. [Google Scholar] [CrossRef]
- Cummins, S.P.; Roberts, D.E.; Ajani, P.; Underwood, A.J. Comparisons of assemblages of phytoplankton between open water and seagrass habitats in a shallow coastal lagoon. Mar. Freshwater Res. 2004, 55, 447–456. [Google Scholar] [CrossRef]
- Jacobs-Palmer, E.; Gallego, R.; Ramón-Laca, A.; Kunselman, E.; Cribari, K.; Horwith, M.; Kelly, R.P. A halo of reduced dinoflagellate abundances in and around eelgrass beds. PeerJ 2020, 8, e8869. [Google Scholar] [CrossRef]
- Iizumi, H.; Taguchi, S.; Minami, T.; Mukai, H.; Maekawa, S. Distribution and variability of nutrients, chlorophyll a, particulate organic matters, and their carbon and nitrogen contents, in Akkeshi-Ko, an estuariy in northern Japan. Bull. Hokkaido Natl. Fish. Res. Inst. 1995, 59, 43–67. [Google Scholar]
- Anderson, D.M.; Glibert, P.M.; Burkholder, J. Harmful algal blooms and eutrophication: Nutrient sources, composition, and consequences. Estuaries 2002, 25, 704–726. [Google Scholar] [CrossRef]
- Imai, I.; Yamaguchi, M.; Hori, Y. Eutrophication and occurrences of harmful algal blooms in the Seto Inland Sea, Japan. Plankton Benthos Res. 2006, 1, 71–84. [Google Scholar] [CrossRef]
- Hallegraeff, G.M.; Bolch, C.J. Transport of toxic dinoflagellate cysts via ships’ ballast water. Mar. Pollut. Bull. 1991, 22, 27–30. [Google Scholar] [CrossRef]
- Matsuyama, Y. Impacts of the harmful dinoflagellate Heterocapsa curcularisquama bloom on shellfish aquaculture in Japan and some experimental studies on invertebrates. Harmful Algae 2012, 14, 144–155. [Google Scholar] [CrossRef]
- Hallegraeff, G.M. Ocean climate change, phytoplankton community responses, and harmful algal blooms: A formidable predictive challenge. J. Phycol. 2010, 46, 220–235. [Google Scholar] [CrossRef]
- Abdennadher, M.; Hamza, A.; Fekih, W.; Hannachi, I.; Zouari-Belaaj, A.; Bradai, N.; Aleya, L. Factors determining the dynamics of toxic blooms of Alexandrium minutum during a 10-year study along the shallow southwestern Mediterranean coasts. Estuar. Coast. Shelf Sci. 2012, 106, 102–111. [Google Scholar] [CrossRef]
- Mukai, H. Seagrass bed ecosystem in a total system of land– and coastal marine ecosystems. Kaiyo Mon. 2005, 34, 449–457. (In Japanese) [Google Scholar]
- Duffy, J.E. Biodiversity and the functioning of seagrass ecosystems. Mar. Ecol. Prog. Ser. 2006, 311, 233–250. [Google Scholar] [CrossRef]
- Orth, R.J.; Carruthers, T.J.B.; Dennison, W.C.; Duarte, C.M.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A global crisis for seagrass ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef]
- Yanagi, T. “Sato-Umi”—A new concept for sustainable fisheries. In Fisheries for Global Welfare and Environment; Tsukamoto, K., Kawamura, T., Takeuchi, T., Beard, T.D., Jr., Kaiser, M.J., Eds.; TERRAPUB: Tokyo, Japan, 2008; pp. 351–358. [Google Scholar]



| Strain | Origin | Closely Related Strain | Classification | Homology | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | DDBJ Accession Number | Date | Area | Station | FLB/PAB/Leaf | Name | DDBJ Accession Number | ||||
| AK1 | LC465489 | April | 21 | 2011 | Akkeshi Bay | Stn. 3 | PAB | Zobellia laminariae | AB121975 | Flabobacteria | 100 |
| AK2 | LC465490 | April | 21 | 2011 | Akkeshi Bay | Stn. 3 | PAB | Zobellia laminariae | AB121975 | Flabobacteria | 100 |
| AK8 | LC465496 | April | 27 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Bacteroidetes bacterium KMM 3949 | AY271625 | Flabobacteria | 98.92 |
| AK9 | LC465497 | April | 27 | 2011 | Akkeshi-ko Estuary | Stn. 1 | PAB | Labrenzia sp. AB434f | FR821171 | α-Proteobacteria | 100 |
| AK10 | LC465498 | April | 27 | 2011 | Akkeshi-ko Estuary | Stn. 1 | PAB | Leifsonia sp. SS12.6 | KC160899 | Actinobacteria | 99.42 |
| AK11 | LC465499 | April | 27 | 2011 | Akkeshi Bay | Stn. 2 | PAB | Neptuniibacter sp. CAR-SF | AB086227 | γ-Proteobacteria | 99.86 |
| AK12 | LC465500 | April | 27 | 2011 | Akkeshi Bay | Stn. 2 | FLB | Kordia antarctica strain IMCC3317 | JX456458 | Flabobacteria | 99.64 |
| AK13 | LC465501 | April | 27 | 2011 | Akkeshi Bay | Stn. 2 | FLB | Kordia sp. LNM-4 | AB758592 | Flabobacteria | 99.47 |
| AK16 | LC465504 | May | 6 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Kordia antarctica strain IMCC3317 | JX456458 | Flabobacteria | 99.25 |
| AK18 | LC465506 | May | 6 | 2011 | Akkeshi Bay | Stn. 2 | PAB | Bacillus baekryungensis strain YD13 | JN210568 | Bacilli | 99.79 |
| AK19 | LC465507 | May | 12 | 2011 | Akkeshi Bay | Stn. 3 | PAB | Cellulophaga sp. AB405d | FR821154 | Flabobacteria | 99.93 |
| AK20 | LC465508 | May | 12 | 2011 | Akkeshi Bay | Stn. 2 | PAB | Roseobacter sp. r22 | JQ661116 | α-Proteobacteria | 99.93 |
| AK21 | LC465509 | May | 12 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Zobellia laminariae | AB121975 | Flabobacteria | 99.92 |
| AK24 | LC465512 | May | 18 | 2011 | Akkeshi Bay | Stn. 3 | PAB | Vibrio sp. D4058 | DQ480136 | γ-Proteobacteria | 99.66 |
| AK25 | LC465513 | May | 18 | 2011 | Akkeshi Bay | Stn. 3 | PAB | Vibrio sp. D4058 | DQ480136 | γ-Proteobacteria | 99.51 |
| AK27 | LC465515 | May | 25 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Krokinobacter sp. NBRC 100818 | AB681258 | Flabobacteria | 99.71 |
| AK28 | LC465516 | May | 25 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Sulfitobacter sp. MBEF04 | AB733554 | α-Proteobacteria | 100 |
| AK31 | LC465519 | May | 25 | 2011 | Akkeshi-ko Estuary | Stn. 1 | PAB | Marinobacter sp. 132Z-4 | JX310148 | γ-Proteobacteria | 99.58 |
| AK32 | LC465520 | May | 25 | 2011 | Akkeshi Bay | Stn. 2 | PAB | Sphingomonas sp. NF4-4 | FJ196036 | α-Proteobacteria | 99.49 |
| AK34 | LC465522 | June | 2 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Alteromonas sp. KT1101 | AF235128 | γ-Proteobacteria | 99.79 |
| AK36 | LC465524 | June | 2 | 2011 | Akkeshi-ko Estuary | Stn. 1 | PAB | Vibrio sp. HNS024 | JN128258 | γ-Proteobacteria | 99.86 |
| AK37 | LC465525 | June | 2 | 2011 | Akkeshi Bay | Stn. 3 | PAB | Aquimarina sp. XH-W2 | KF017576 | Flabobacteria | 99.57 |
| AK41 | LC465529 | June | 8 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Sulfitobacter sp. MBEF05 | AB733554 | α-Proteobacteria | 99.56 |
| AK42 | LC465530 | June | 8 | 2011 | Akkeshi-ko Estuary | Stn. 1 | Leaf | Alteromonas sp. TO01 | AB049730 | γ-Proteobacteria | 99.86 |
| AK45 | LC465533 | June | 8 | 2011 | Akkeshi-ko Estuary | Stn. 1 | FLB | Sulfitobacter sp. MBEF04 | AB733554 | α-Proteobacteria | 100 |
| AK47 | LC465535 | June | 8 | 2011 | Akkeshi Bay | Stn. 3 | PAB | Tenacibaculum sp. 35 | JX454522 | Flabobacteria | 99.85 |
| AK48 | LC465536 | June | 8 | 2011 | Akkeshi Bay | Stn. 3 | FLB | Tenacibaculum sp. 35 | JX454522 | Flabobacteria | 99.85 |
| AK50 | LC465538 | June | 15 | 2011 | Akkeshi-ko Estuary | Stn. 1 | PAB | Alteromonas sp. USC168 | HQ441215 | γ-Proteobacteria | 99.78 |
| AK51 | LC465539 | June | 15 | 2011 | Akkeshi-ko Estuary | Stn. 1 | PAB | Sphingomonas sp. NF4-4 | FJ196036 | α-Proteobacteria | 98.75 |
| AK52 | LC465540 | June | 15 | 2011 | Akkeshi-ko Estuary | Stn. 1 | PAB | Alteromonas sp. KT1101 | AF235128 | γ-Proteobacteria | 99.79 |
| AK53 | LC465541 | June | 15 | 2011 | Akkeshi-ko Estuary | Stn. 1 | FLB | Alteromonas sp. W-6 | DQ923440 | γ-Proteobacteria | 99.59 |
| AK54 | LC465542 | June | 15 | 2011 | Akkeshi-ko Estuary | Stn. 1 | FLB | Vibrio sp. HNS024 | JN128258 | γ-Proteobacteria | 99.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onishi, Y.; Tuji, A.; Yamaguchi, A.; Imai, I. Distribution of Growth-Inhibiting Bacteria against the Toxic Dinoflagellate Alexandrium catenella (Group I) in Akkeshi-Ko Estuary and Akkeshi Bay, Hokkaido, Japan. Appl. Sci. 2021, 11, 172. https://doi.org/10.3390/app11010172
Onishi Y, Tuji A, Yamaguchi A, Imai I. Distribution of Growth-Inhibiting Bacteria against the Toxic Dinoflagellate Alexandrium catenella (Group I) in Akkeshi-Ko Estuary and Akkeshi Bay, Hokkaido, Japan. Applied Sciences. 2021; 11(1):172. https://doi.org/10.3390/app11010172
Chicago/Turabian StyleOnishi, Yuka, Akihiro Tuji, Atsushi Yamaguchi, and Ichiro Imai. 2021. "Distribution of Growth-Inhibiting Bacteria against the Toxic Dinoflagellate Alexandrium catenella (Group I) in Akkeshi-Ko Estuary and Akkeshi Bay, Hokkaido, Japan" Applied Sciences 11, no. 1: 172. https://doi.org/10.3390/app11010172
APA StyleOnishi, Y., Tuji, A., Yamaguchi, A., & Imai, I. (2021). Distribution of Growth-Inhibiting Bacteria against the Toxic Dinoflagellate Alexandrium catenella (Group I) in Akkeshi-Ko Estuary and Akkeshi Bay, Hokkaido, Japan. Applied Sciences, 11(1), 172. https://doi.org/10.3390/app11010172






