Effects of an Eco-Friendly Sanitizing Wash on Spinach Leaf Bacterial Community Structure and Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Sanitizers Treatment of Spinach Leaves
2.2. Samples Preparation for Variable V3-V4 Region Sequencing
2.3. Statistical Analysis
2.4. Data Submission
3. Results
3.1. Composition of Spinach Bacterial Community
3.2. Alpha Diversity
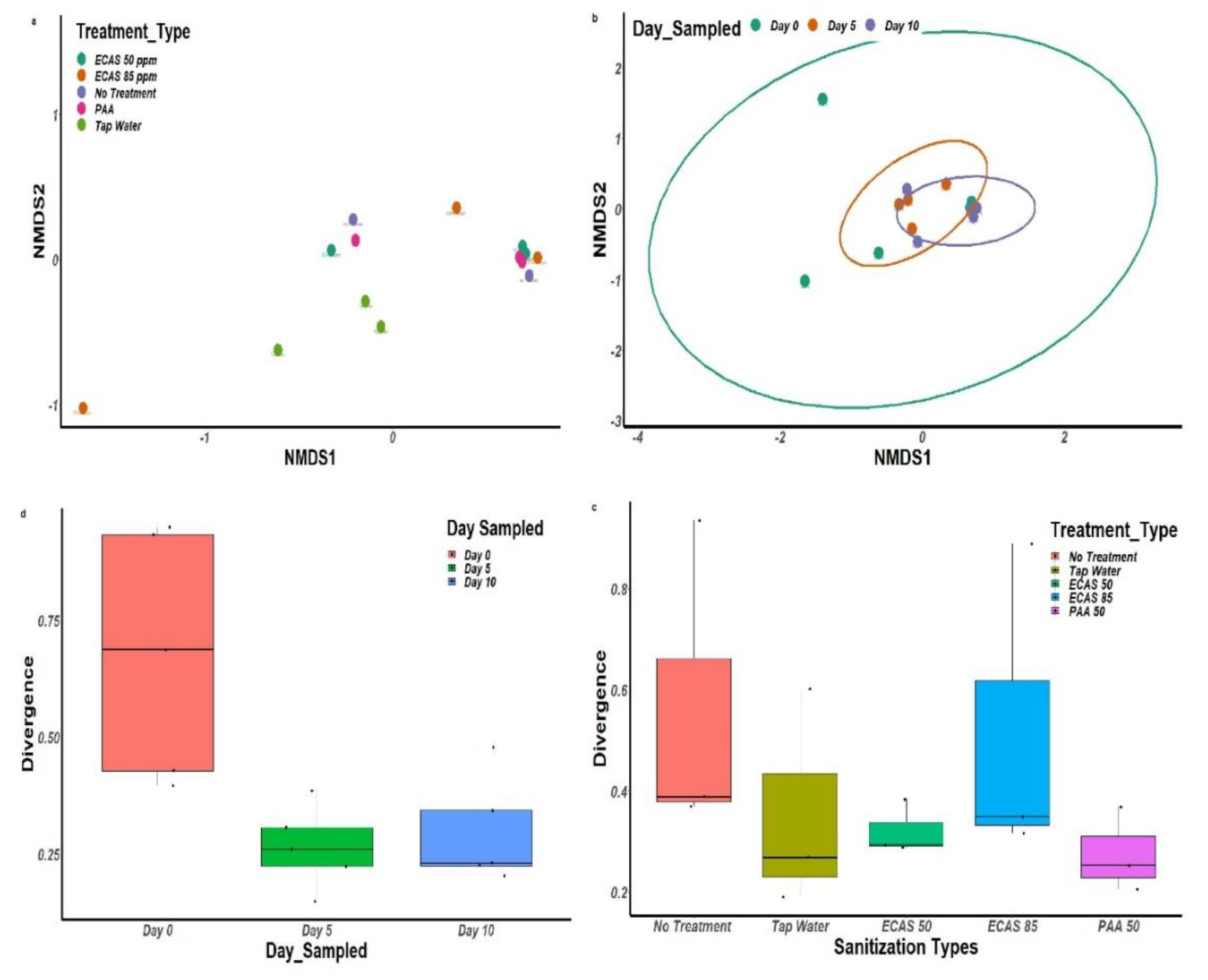
3.3. Bacterial Diversity Associated with Treatment Type and Sampling Day
3.4. Taxa Differences Among the Different Sampling Days
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lindow, S.E.; Brandl, M.T. Microbiology of the Phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, N.; Knief, C.; Chaffron, S.; Innerebner, G.; Roschitzki, B.; Schlapbach, R.; Von Mering, C.; Vorholt, J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 16428–16433. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Velasco, G.; Welbaum, G.; Boyer, R.; Mane, S.; Ponder, M.A. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J. Appl. Microbiol. 2011, 110, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef]
- Whipps, J.; Hand, P.; Pink, D.; Bending, G. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 105, 1744–1755. [Google Scholar] [CrossRef] [PubMed]
- Leff, J.W.; Fierer, N. Bacterial Communities Associated with the Surfaces of Fresh Fruits and Vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef]
- Cordano, A. Occurrence of Listeria monocytogenes in food in Chile. Int. J. Food Microbiol. 2001, 70, 175–178. [Google Scholar] [CrossRef]
- De Oliveira, M.A.; De Souza, V.M.; Bergamini, A.M.M.; De Martinis, E.C.P. Microbiological quality of ready-to-eat minimally processed vegetables consumed in Brazil. Food Control. 2011, 22, 1400–1403. [Google Scholar] [CrossRef]
- Klerks, M.M.; Franz, E.; Van Gent-Pelzer, M.; Zijlstra, C.; Van Bruggen, A.H.C. Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 2007, 1, 620–631. [Google Scholar] [CrossRef]
- Mitra, R.; Cuesta-Alonso, E.; Wayadande, A.; Talley, J.; Gilliland, S.; Fletcher, J. Effect of Route of Introduction and Host Cultivar on the Colonization, Internalization, and Movement of the Human Pathogen Escherichia coli O157:H7 in Spinach. J. Food Prot. 2009, 72, 1521–1530. [Google Scholar] [CrossRef]
- Hackl, E.; Hölzl, C.; Konlechner, C.; Sessitsch, A. Food of plant origin: Production methods and microbiological hazards linked to food-borne disease. Reference: CFT/EFSA/BIOHAZ/2012/01 Lot 1 (Food of plant origin with high water content such as fruits, vegetables, juices and herbs). EFSA Support. Publ. 2013, 10, 402E. [Google Scholar] [CrossRef]
- Gaglio, R.; Miceli, A.; Sardina, M.T.; Francesca, N.; Moschetti, G.; Settanni, L. Evaluation of microbiological and physico-chemical parameters of retail ready-to-eat mono-varietal salads. J. Food Process. Preserv. 2019, 43, e13955. [Google Scholar] [CrossRef]
- Andrews, J.H.; Harris, R.F. The Ecology and Biogeography of Microorganisms on Plant Surfaces. Annu. Rev. Phytopathol. 2000, 38, 145–180. [Google Scholar] [CrossRef] [PubMed]
- Janisiewicz, W.J.; Korsten, L. Biological Control of Postharvest Diseases of Fruits. Annu. Rev. Phytopathol. 2002, 40, 411–441. [Google Scholar] [CrossRef]
- Barth, M.; Hankinson, T.R.; Hong, Z.; Breidt, F. Microbiological Spoilage of Fruits and Vegetables. In Compendium of the Microbiological Spoilage of Foods and Beverages; Springer: Berlin, Germany, 2009; pp. 135–183. [Google Scholar]
- Schuenzel, K.M.; Harrison, M.A. Microbial Antagonists of Foodborne Pathogens on Fresh, Minimally Processed Vegetables. J. Food Prot. 2002, 65, 1909–1915. [Google Scholar] [CrossRef]
- Scolari, G.; Vescovo, M. Microbial antagonism of Lactobacillus Casei added to fresh vegetables. Ital. J. Food Sci. 2004, 16, 465–475. [Google Scholar]
- Johnston, M.A.; Harrison, M.A.; Morrow, R.A. Microbial Antagonists of Escherichia coli O157:H7 on Fresh-Cut Lettuce and Spinach. J. Food Prot. 2009, 72, 1569–1575. [Google Scholar] [CrossRef]
- Cooley, M.B.; Miller, W.G.; Mandrell, R.E. Colonization of Arabidopsis thaliana with Salmonella enterica and Enterohemorrhagic Escherichia coli O157:H7 and Competition by Enterobacter asburiae. Appl. Environ. Microbiol. 2003, 69, 4915–4926. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Randolph, K.C.; Osborn, S.L.; Tyler, H.L. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 2013, 13, 274. [Google Scholar] [CrossRef] [PubMed]
- Rudi, K.; Flateland, S.L.; Hanssen, J.F.; Bengtsson, G.; Nissen, H. Development and Evaluation of a 16S Ribosomal DNA Array-Based Approach for Describing Complex Microbial Communities in Ready-To-Eat Vegetable Salads Packed in a Modified Atmosphere. Appl. Environ. Microbiol. 2002, 68, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Handschur, M.; Piñar, G.; Gallist, B.; Lubitz, W.; Haslberger, A.G. Culture free DGGE and cloning based monitoring of changes in bacterial communities of salad due to processing. Food Chem. Toxicol. 2005, 43, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Truchado, P.; Gil, M.I.; Suslow, T.; Allende, A. Impact of chlorine dioxide disinfection of irrigation water on the epiphytic bacterial community of baby spinach and underlying soil. PLoS ONE 2018, 13, e0199291. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Velasco, G. Molecular Characterization of Spinach (Spinacia Oleracea) Microbial Community Structure and Its Interaction with Escherichia Coli O157:H7 in Modified Atmosphere Conditions. Ph.D. Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2010. Available online: https://vtechworks.lib.vt.edu/handle/10919/37601 (accessed on 19 September 2019).
- Gu, L.; Bai, Z.; Jin, B.; Hu, Q.; Wang, H.; Zhuang, G.; Zhang, H. Assessing the impact of fungicide enostroburin application on bacterial community in wheat phyllosphere. J. Environ. Sci. 2010, 22, 134–141. [Google Scholar] [CrossRef]
- Gu, G.; Ottesen, A.; Bolten, S.; Ramachandran, P.; Reed, E.; Rideout, S.; Luo, Y.; Patel, J.; Brown, E.; Nou, X. Shifts in spinach microbial communities after chlorine washing and storage at compliant and abusive temperatures. Food Microbiol. 2018, 73, 73–84. [Google Scholar] [CrossRef]
- Söderqvist, K.; Osman, O.A.; Wolff, C.; Bertilsson, S.; Vågsholm, I.; Boqvist, S. Emerging microbiota during cold storage and temperature abuse of ready-to-eat salad. Infect. Ecol. Epidemiol. 2017, 7, 1328963. [Google Scholar] [CrossRef]
- Izumi, H.; Kiba, T.; Hashimoto, S. Efficacy of electrolyzed water as a disinfectant for fresh-cut spinach. ACIAR Proc. 2000, 100, 216–221. [Google Scholar]
- Rahman, S.; Ding, T.; Oh, D.-H. Inactivation effect of newly developed low concentration electrolyzed water and other sanitizers against microorganisms on spinach. Food Control. 2010, 21, 1383–1387. [Google Scholar] [CrossRef]
- Gómez-López, V.M.; Marín, A.; Medina-Martínez, M.S.; Gil, M.I.; Allende, A. Generation of trihalomethanes with chlorine-based sanitizers and impact on microbial, nutritional and sensory quality of baby spinach. Postharvest Boil. Technol. 2013, 85, 210–217. [Google Scholar] [CrossRef]
- Guentzel, J.L.; Lam, K.L.; Callan, M.A.; Emmons, S.A.; Dunham, V.L. Reduction of bacteria on spinach, lettuce, and surfaces in food service areas using neutral electrolyzed oxidizing water. Food Microbiol. 2008, 25, 36–41. [Google Scholar] [CrossRef]
- Park, E.-J.; Alexander, E.; Taylor, G.; Costa, R.; Kang, D.-H. Effects of organic matter on acidic electrolysed water for reduction of foodborne pathogens on lettuce and spinach. J. Appl. Microbiol. 2008, 105, 1802–1809. [Google Scholar] [CrossRef]
- Babic, I.; Watada, A.E.; Buta, J.G. Growth of Listeria monocytogenes Restricted by Native Microorganisms and Other Properties of Fresh-Cut Spinach. J. Food Prot. 1997, 60, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Tatsika, S.; Karamanoli, K.; Karayanni, H.; Genitsaris, S. Metagenomic Characterization of Bacterial Communities on Ready-to-Eat Vegetables and Effects of Household Washing on their Diversity and Composition. Pathogens 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Garcia, A.; Santos-Ferro, L.; Ramirez-Orejel, J.C.; Agredano-Moreno, L.T.; Jimenez-Garcia, L.F.; Paez-Esquiliano, D.; Andrade-Esquivel, E.; Cano-Buendia, J.A. The effect of neutral electrolyzed water as a disinfectant of eggshells artificially contaminated with Listeria monocytogenes. Food Sci. Nutr. 2019, 7, 2252–2260. [Google Scholar] [CrossRef] [PubMed]
- Premier, R. Evaluation of Vegetable Washing Chemicals. In Australia: Horticulture Australia Ltd. 2013. Available online: https://www.ausvegvic.com.au/pdf/r%26d_VG09086_vegetable_wash_water_extract.pdf (accessed on 12 September 2019).
- Ogunniyi, A.D.; Tenzin, S.; Khazandi, M.; Ferro, S.; Venter, H.; Pi, H.; Amorico, T.; Deo, P.; Trott, D.J. A pH-neutral electrolyzed oxidizing water significantly reduces microbial contamination of fresh spinach leaves. Food Microbiol. 2020. under review (Accessed on 25/02/2020). [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef]
- Amplicon, P.; Clean-Up, P.; Index, P. 16s Metagenomic Sequencing Library Preparation. 2013. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 17 November 2018).
- Andrews, S. FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 22 January 2019).
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef]
- Core Team, R. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. 2019. Available online: https://www.R-project.org/ (accessed on 14 February 2019).
- Callahan, B.J.; McMurdie, P.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Callahan, B. Silva taxonomic training data formatted for DADA2 (Silva version 132). Zenodo 2018. [Google Scholar] [CrossRef]
- Wright, E.S. Using DECIPHER v2.0 to analyze big biological sequence data in R. R Journal 2016, 8, 352–359. Available online: https://journal.r-project.org/archive/2016-1/wright.pdf (accessed on 3 March 2019). [CrossRef]
- Schliep, K. phangorn: Phylogenetic analysis in R. Bioinformatics 2010, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.; Holmes, S. Shiny-phyloseq: Web application for interactive microbiome analysis with provenance tracking. Bioinformatics 2014, 31, 282–283. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Knight, R. Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 2008, 32, 557–578. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; González, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Genet. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P. Ordination methods, diversity analysis and other functions for community and vegetation ecologists. In Vegan: Community Ecology Package. 2019. Available online: https://www.researchgate.net/publication/319525938_vegan_Community_Ecology_Package_Ordination_methods_diversity_analysis_and_other_functions_for_community_and_vegetation_ecologists_Version_22-4_URL_httpsCRANR-projectorgpackagevegan/link/59b0e308a6fdcc3f888bf5da/download (accessed on 3 March 2019).
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; pp. 1–15. [Google Scholar]
- Lahti, L.; Shetty, S.; Blake, T.; Salojarvi, J. Microbiome R package. 2012–2019. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Jarvis, K.; Daquigan, N.; White, J.R.; Morin, P.M.; Howard, L.M.; Manetas, J.E.; Ottesen, A.; Ramachandran, P.; Grim, C.J. Microbiomes Associated With Foods From Plant and Animal Sources. Front. Microbiol. 2018, 9, 2540. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.; Van Horn, D.J.; Buelow, H.N.; Okie, J.G.; Gooseff, M.; Barrett, J.E.; Takacs-Vesbach, C. Characterization of growing bacterial populations in McMurdo Dry Valley soils through stable isotope probing with (18) O-water. FEMS Microbiol. Ecol. 2014, 89, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.; Dunlap, C.; Barnett, S.; Franco, C. Decoding Wheat Endosphere-Rhizosphere Microbiomes in Rhizoctonia solani-Infested Soils Challenged by Streptomyces Biocontrol Agents. Front. Plant Sci. 2019, 10, 1038. [Google Scholar] [CrossRef] [PubMed]


| Phylum | Phyla Abundance | Percentage Abundance |
|---|---|---|
| Actinobacteria | 756 | 12.59 |
| Bacteroidetes | 1876 | 31.25 |
| Deinococcus-Thermus | 7 | 0.12 |
| Firmicutes | 396 | 6.60 |
| Fusobacteria | 4 | 0.07 |
| Patescibacteria | 8 | 0.13 |
| Planctomycetes | 7 | 0.12 |
| Proteobacteria | 2949 | 49.13 |
| Treatment Type | Sampling Day | Chao1 | ACE | Shannon | InvSimpson | Fisher |
|---|---|---|---|---|---|---|
| Tap water | 0 | 128.00 | 128.29 | 3.86 | 26.38 | 22.58 |
| 5 | 371.08 | 371.89 | 4.45 | 39.49 | 61.56 | |
| 10 | 337.42 | 338.75 | 4.38 | 41.90 | 51.58 | |
| ECAS 50 mg/L | 0 | 771.06 | 771.86 | 5.59 | 125.72 | 128.92 |
| 5 | 487.38 | 488.94 | 4.47 | 38.84 | 75.21 | |
| 10 | 542.10 | 542.79 | 5.42 | 118.15 | 96.19 | |
| ECAS 85 mg/L | 0 | 60.00 | 60.00 | 3.42 | 17.66 | 13.74 |
| 5 | 445.04 | 446.17 | 5.31 | 134.56 | 77.66 | |
| 10 | 630.76 | 633.55 | 5.42 | 113.40 | 133.90 | |
| PAA 50 mg/L | 0 | 577.32 | 578.44 | 5.28 | 91.59 | 94.20 |
| 5 | 368.25 | 369.16 | 4.69 | 50.61 | 58.87 | |
| 10 | 468.20 | 469.06 | 5.19 | 88.46 | 83.08 | |
| No treatment (control) | 0 | 64.00 | 64.00 | 3.54 | 22.51 | 15.04 |
| 5 | 342.00 | 342.00 | 4.50 | 35.73 | 53.66 | |
| 10 | 415.60 | 416.57 | 5.11 | 80.86 | 73.09 |
| Df | Sum Sq | F.Model | R2 | P | |
|---|---|---|---|---|---|
| Treatment Type | 4 | 0.455 | 1.813 | 0.330 | 0.053 |
| Sampling Day | 2 | 0.421 | 3.359 | 0.305 | 0.006 |
| Residual | 8 | 0.502 | |||
| Total | 14 | 1.378 |
| Sampling Day | p-Value (Observed) | p-Value (Permutated) |
|---|---|---|
| Day 0–Day 5 | 0.022 | 0.029 |
| Day 0–Day 10 | 0.031 | 0.027 |
| Day 5–Day 10 | 0.856 | 0.854 |
| ASVs * | Relative Abundance (RA) | Reject Hypothesis | |||
|---|---|---|---|---|---|
| Taxon a | W | Day 0 | Day 5 | Day 10 | |
| f Pseudomonadaceae; g unclassified (ASV1) | 37 | 0.0010 | 0.0087 | 0.0032 | TRUE ** |
| g Alkanindiges; s illinoisensis (ASV2) | 14 | 0.0030 | 0.0079 | 0.0017 | TRUE ** |
| f Flavobacteriaceae; g unclassified (ASV3) | 13 | 0.0016 | 0.0000 | 0.0019 | TRUE ** |
| f Flavobacteriaceae; g unclassified (ASV4) | 12 | 0.0002 | 0.0000 | 0.0009 | TRUE ** |
| g Herminiimonas; s aquatilis (ASV5) | 10 | 0.0011 | 0.0000 | 0.0012 | TRUE ** |
| f Micrococcaceae; g Arthrobacter (ASV6) | 10 | 0.0107 | 0.0076 | 0.0162 | TRUE** |
| f Oxalobacteraceae; g unclassified (ASV7) | 10 | 0.0142 | 0.0099 | 0.0187 | TRUE ** |
| f Flavobacteriaceae; g Persicivirga(ASV8) | 10 | 0.0028 | 0.0000 | 0.0001 | TRUE ** |
| o Bacillales; f unclassified (ASV9) | 10 | 0.0010 | 0.0017 | 0.0000 | TRUE ** |
| g Achromobacter; s xylosoxidans (ASV10) | 7 | 0.0009 | 0.0000 | 0.0002 | TRUE ** |
| g Flavobacterium; s frigidarium (ASV11) | 7 | 0.0010 | 0.0000 | 0.0001 | TRUE ** |
| g Alkanindiges; s illinoisensis (ASV12) | 6 | 0.0060 | 0.0388 | 0.0066 | TRUE ** |
| f Flavobacteriaceae; g unclassified (ASV13) | 6 | 0.0013 | 0.0000 | 0.0002 | TRUE ** |
| f Pseudomonadaceae; g unclassified (ASV14) | 6 | 0.0022 | 0.0005 | 0.0000 | TRUE ** |
| g Flavobacterium; s frigidarium (ASV15) | 5 | 0.0013 | 0.0002 | 0.0000 | TRUE ** |
| f Micrococcaceae; g Arthrobacter (ASV16) | 5 | 0.1405 | 0.0667 | 0.1766 | TRUE ** |
| f Flavobacteriaceae; g unclassified (ASV17) | 5 | 0.0010 | 0.0002 | 0.0007 | TRUE ** |
| Family | |||||
| Taxon | W | ||||
| o Sphingobacteriales; f Sphingobacteriaceae | 6 | 0.5810 | 0.1827 | 0.2363 | TRUE ** |
| o Pseudomonadales; f Pseudomonadaceae | 5 | 0.2025 | 0.5894 | 0.2082 | TRUE ** |
| o Xanthomonadales; f Xanthomonadaceae | 2 | 0.4953 | 0.1604 | 0.3443 | TRUE ** |
| o Flavobacteriales; f Flavobacteriaceae | 2 | 0.4343 | 0.1861 | 0.3796 | TRUE ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tenzin, S.; Ogunniyi, A.D.; Ferro, S.; Deo, P.; Trott, D.J. Effects of an Eco-Friendly Sanitizing Wash on Spinach Leaf Bacterial Community Structure and Diversity. Appl. Sci. 2020, 10, 2986. https://doi.org/10.3390/app10082986
Tenzin S, Ogunniyi AD, Ferro S, Deo P, Trott DJ. Effects of an Eco-Friendly Sanitizing Wash on Spinach Leaf Bacterial Community Structure and Diversity. Applied Sciences. 2020; 10(8):2986. https://doi.org/10.3390/app10082986
Chicago/Turabian StyleTenzin, Sangay, Abiodun D. Ogunniyi, Sergio Ferro, Permal Deo, and Darren J. Trott. 2020. "Effects of an Eco-Friendly Sanitizing Wash on Spinach Leaf Bacterial Community Structure and Diversity" Applied Sciences 10, no. 8: 2986. https://doi.org/10.3390/app10082986
APA StyleTenzin, S., Ogunniyi, A. D., Ferro, S., Deo, P., & Trott, D. J. (2020). Effects of an Eco-Friendly Sanitizing Wash on Spinach Leaf Bacterial Community Structure and Diversity. Applied Sciences, 10(8), 2986. https://doi.org/10.3390/app10082986






