1. Introduction
Since African swine fever (ASF) was first found in China in August 2018, 28 provinces have experienced outbreaks of the epidemic, with a total of more than 1,160,000 pigs culled, according to statistical data from the Ministry of Agriculture and Rural Affairs, PRC, released on 3 July 2019. The International Office of Epizootics (IOE) reported that there were outbreaks of African swine fever in 37 countries. The Canadian Food Inspection Agency has claimed that the African swine fever virus (ASFV) can be spread by contamination of livestock feed. The risks for ASFV transmission in feed were evaluated by Niederwerder et al. Their findings demonstrated that the ASFV Georgia 2007 strain can easily be transmitted orally when consumed in liquid, but requires a much higher dose when provided in feed [
1]. Recently, Zhao et al. detected the first ASFV, named Pig/HLJ/18, in Heilongjiang, and this virus is highly virulent and transmissible in domestic pigs in China [
2]. Wen et al. adopted gene sequencing analysis and indicated that the ASFVs derived from a pig sample and the SDPP of pig feed, in Jiamusi, Heilongjiang, were identical—both were Pig/HLJ/18 ASFV—which was the same strain found in previous studies. These findings demonstrated that using SDPP as a creep feed supplement could result in a substantial potential hazard for the spread of ASF [
3]. To respond to the extremely grim situation in China, Regulation 64/2018, which was issued by the Ministry of Agriculture and Rural Affairs, mandated that the use of SDPP in creep feed be suspended to prevent and control the spread of ASF. Although SDPP plays a crucial role in creep feed, as it can promote intestinal development in piglets and prevent diarrhea [
4], it was important to discriminate SDPP from other feed additives. Whey protein concentrate (WPC) is the best substitute for SDPP, but WPC is much more expensive, with a cost of nearly twice that of SDPP. Furthermore, studies have shown that WPC is only half as efficient as SDPP [
5]. Later, the new Regulation 91/2018 was issued and implemented, under which ASFV-genome-free porcine blood products are permitted for use again in feed for swine. Hence, the quantitative analysis of the SDPP concentration detection also becomes meaningful. Thus, inspecting the usage of SDPP is an effective measure for verifying declared ingredients used in commercial feed.
The traditional physicochemical detection methods for detecting animal protein sources include feed microscopy, enzyme-linked immunosorbent assays (ELISAs), and polymerase chain reaction [
6]. Feed microscopy is a method used to identify the morphological characteristics of feed particles from animal sources by optical microscopy. ELISA is an immunological kit method that was developed based on the immunological reaction between protein antibodies and antigens to detect specific proteins in feed samples. Polymerase chain reaction, which is the most common method, is a type of molecular biology technique that can detect protein sources in creep feed by amplifying the DNA from a single cell [
7]. The above methods are time-consuming, incur high costs, and involve complex operational processes. Therefore, it is necessary to explore rapid and efficient techniques for qualitative detection and quantitative prediction methods to enforce the legislation of Regulation 64/2018, which limits the use of SDPP in creep feed to prevent further ASF outbreaks.
Electronic nose detection [
8,
9] and near-infrared spectroscopy (NIRS) [
10,
11,
12] are the new and intelligent feed quality detection methods. An electronic nose is able to simulate the biological sense of smell based on an array of gas sensors that have distinct sensitivities to various volatile organic compounds (VOCs) [
13]. Thus, an electronic nose is capable of identifying the differences between test samples based on their volatilities. Electronic nose technology was first applied in the analysis of ruminant meat and bone meal (MBM), which caused the spread of bovine spongiform encephalopathy (BSE) in 2004 [
14]. In response to feed protein shortages and feed safety requirements, researchers have implemented electronic nose testing of feed materials, such as fish meal, soybean, maize, and wheat for animal nutrition research, and to prevent commercial fraud in the last decade [
9,
15,
16,
17]. To the best of our knowledge, no research has been published to date regarding the application of electronic noses for SDPP detection.
NIRS, which is a promising technique for nondestructive analysis, can be utilized to correlate chemical concentrations with the absorption intensities in the NIR region at specific wavelengths, which can be attributed to the molecular vibrations of functional groups, such as C-H, O-H, N-H, H-S, and inorganic compounds [
18]. In recent years, previous studies have investigated the use of NIRS to detect and predict parameters of nutritional interest for different feeds, including MBM, animal protein byproducts (APBPs), animal fat byproducts (AFBPs), multivitamins, and perennial ryegrass [
19,
20,
21,
22]. However, limited studies have investigated the application of NIRS in the qualitative and quantitative detection of SDPP.
Although there were a lot of applications for the electronic nose and NIRS in feed detection, very few researchers have focused on compared the detecting features between electronic noses and NIRS. In this study, it was assumed that there were significantly different chemical compositions between SDPP and WPC. According to the related official limitation of SDPP used in feed, pure and impure samples were used to explore the classification model for qualitative detection. A series of different concentration samples were used to identify the predictive capacity for quantitative detection. Hence, both the mixture discrimination experiment and mixture concentration experiment were designed to verify the feasibility of detecting SDPP in mixture samples. Thus, the chemometric analysis was applied to develop a discriminant analysis model and a quantitative prediction model to monitor the usage of SDPP by separately using electronic nose and NIRS techniques. The main objectives of the present study are as follows:
(1) To explore the feasibility of utilizing electronic noses and NIRS technologies to perform cluster analysis in mixed samples with principal component analysis (PCA) [
23].
(2) To develop a robust classification model for SDPP discrimination based on electronic nose data and NIRS data by adopting the PLS-DA [
24] algorithm with different data preprocessing methods.
(3) To investigate the optimum regression model of the electronic nose and NIRS for the quantitative prediction of SDPP in mixed samples by employing the CR method [
25].
(4) To establish the quantitative predictive models and identify the optimal regression method for SDPP concentrations in mixed samples by utilizing electronic nose and NIRS techniques, respectively.
2. Materials and Methods
2.1. Experimental Materials and Sample Preparation
Feed enterprises often conduct the test of raw protein materials before feed production. Thus, the actual test sample mainly contains one or more different protein raw material, rather than the finished feed product. In this study, the test samples were compounded, which consists of WPC and SDPP at different concentrations to investigate the feasibility of detecting SDPP by both EN and NIRS. WPC was provided by Guangzhou Wheysubime Biotechnology Company Limited (No. 3, Guquan Road, Guangzhou High-Tech Industrial Development Zone, Guangzhou, China) and had a protein content of more than 80%, a fat content under 8%, and water content below 5%. SDPP was provided by Wuhan Yuancheng Gongchuang Technology Company Limited and had a protein content greater than 76% and water content below 10%. All materials were sealed and stored at 4 ± 1 °C and 65% ± 3% RH.
The test samples were compound raw materials used for creep feed, consisting of WPC and SDPP at different concentrations. All test samples were ground 3 times (10 seconds each) by a grinding miller (MFJ-W300) and sieved through a 0.5 mm standard screen hole. An electronic analytical balance (BSA224S-CW) was adopted to ensure that each sample contained 8 g of the compound sample. To construct the qualitative discrimination model, 90 pure samples and 90 impure samples were selected to detect whether the samples contained SDPP. Twenty concentrations from 1%−20% were selected to build the quantitative prediction model to predict the concentrations of SDPP in the samples (
Table 1). The calibration samples and prediction samples were prepared separately. In total, 360 test samples were prepared for the electronic nose and NIRS tests.
2.2. Electronic Nose Detection
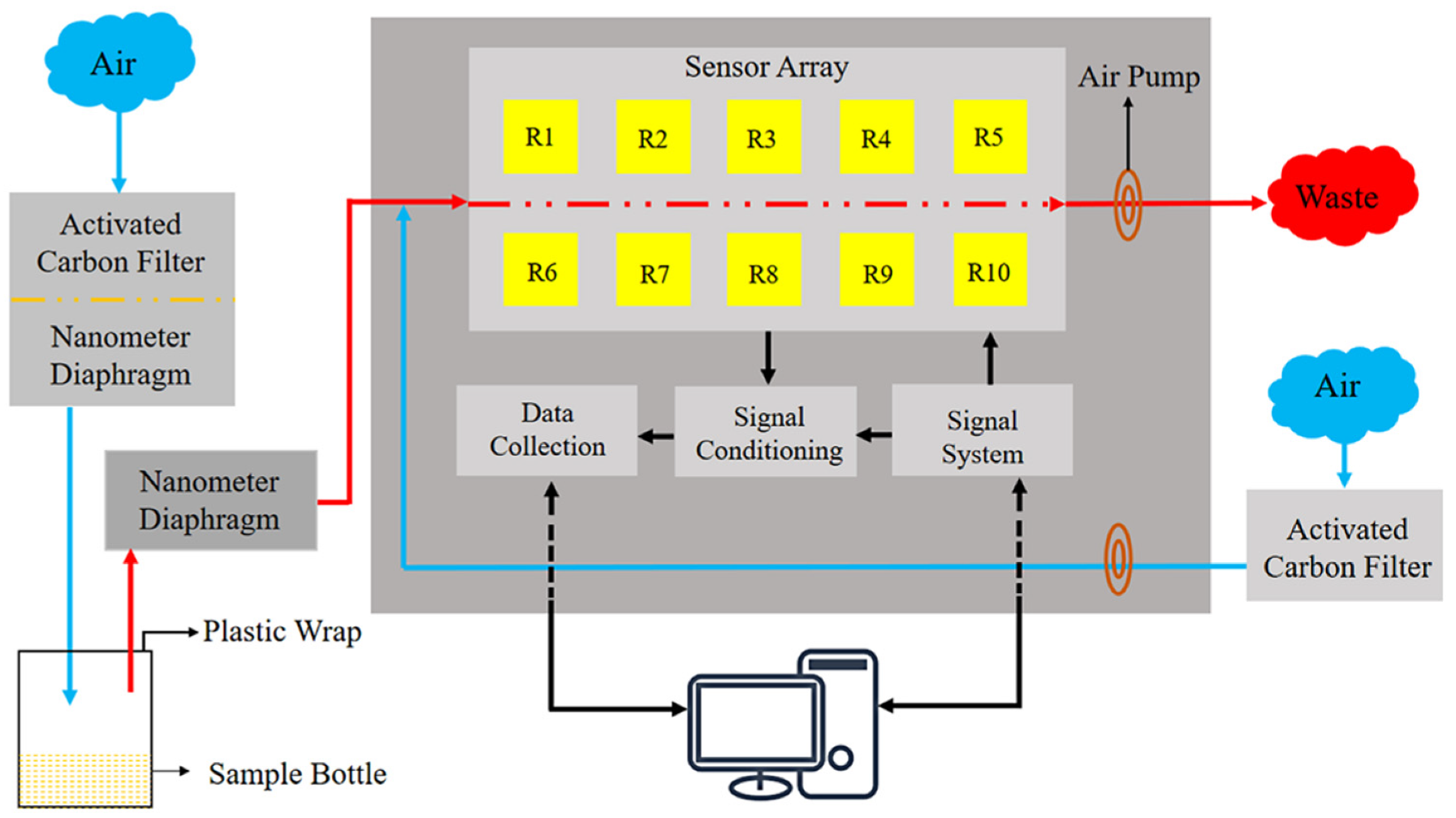
A PEN3 electronic nose (Airsense Analytics GmbH, Schwerin, Germany) was adopted in this research. The electronic nose is mainly composed of a sensor array, sampling and cleaning channel, and a data processing system. The sensor array is composed of 10 metal-oxide sensors, which are the core components of the electronic nose. Each sensor is sensitive to different volatiles (
Figure 1). A series of physical and chemical reactions occur when the active material of the sensor interacts with volatile compounds. The voltage signal produced by the reaction is translated into a data signal recorded via a computer and sent to a signal processing unit for analysis.
The electronic nose was preheated for 30 mins before measurement to ensure that the sensor array worked at an optimal temperature. Zero gas (ambient air in the laboratory room) filtered through standard activated carbon was pumped into the electronic nose to normalize the gas sensors. The parameter settings for the electronic nose are shown in
Table 2 [
26].
In total, 360 electronic nose data points (90 samples at 0% + 15 samples × 6 increments between 0% and 3% + 9 samples × 20 increments between 1% and 20% = 360) were measured by the electronic nose test platform (the environmental temperature was 24 ± 1 °C, and the environmental humidity was 65% ± 3%). All beakers were cleaned by ultrasonic cleaning and natural drying in a room without any unusual smell. Each sample was stored in a 50-ml beaker sealed with double plastic wrap for 30 min. The built-in software WinMuster1.6.2.22 was used to record data. The data at 30 s were selected as the feature values for further analysis. The response features and threshold value of the electronic nose sensor array are illustrated in
Table 3 [
27].
2.3. FT-NIR Spectral Analysis
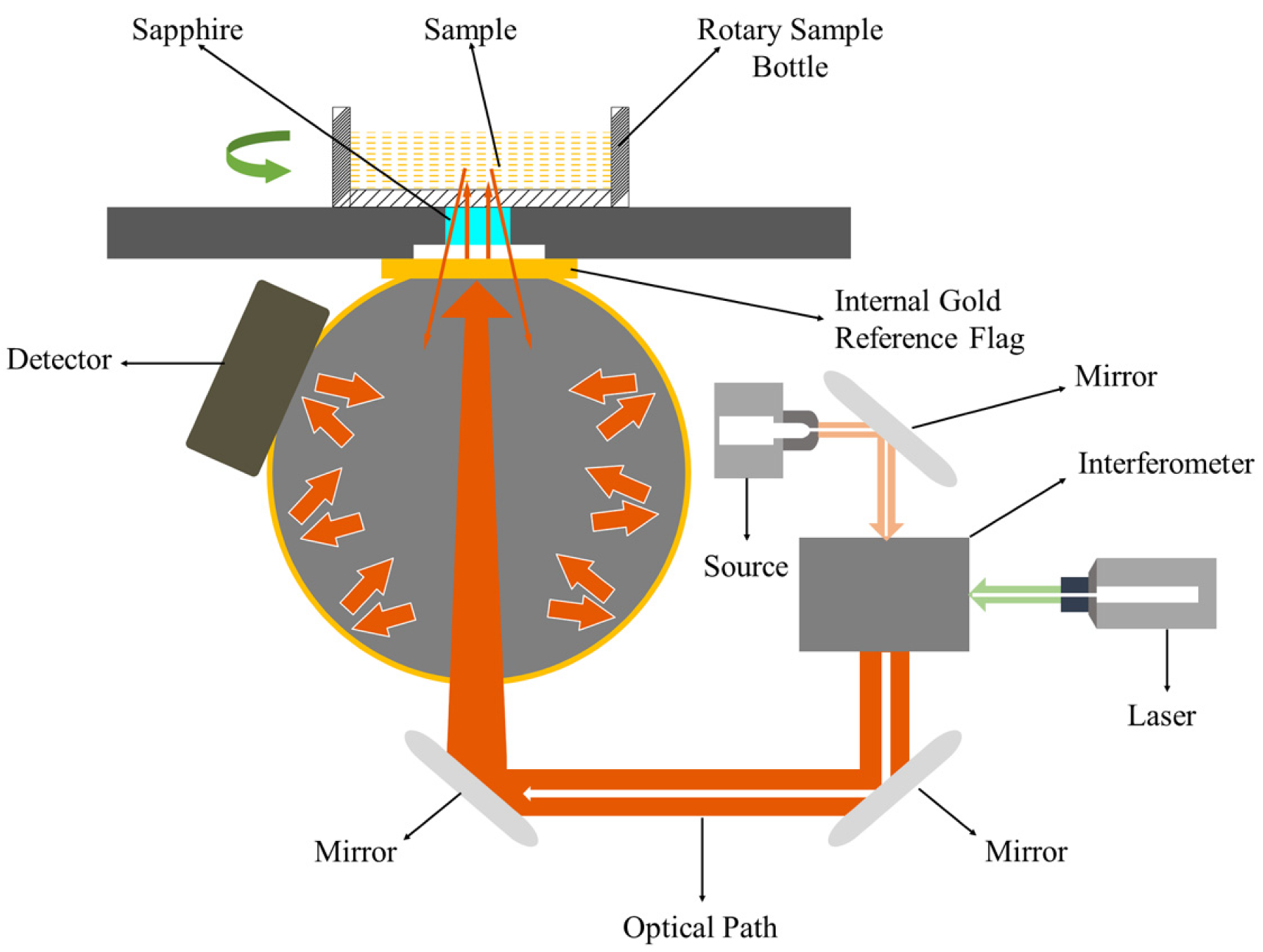
NIR diffuse reflectance spectrometry was applied to acquire NIR spectra for each sample. A schematic diagram of the Fourier transform NIR (FT–NIR) spectroscopy system is displayed in
Figure 2. The energy beam produced by a halogen light entered the interferometer, where the spectrum was encoded (an internal reference laser was used to provide internal wavelength calibration). The encoded spectrum was transmitted to the surface of the sample, where specific frequencies of energy were absorbed. The signal generated by the beam passed to the detector was measured and digitized in the computer for Fourier transformation to form a final NIR spectrum. An integrating sphere was utilized to improve the signal-to-noise ratio.
In total, 360 samples were scanned using an Antaris II FT-NIR Analyzer (Thermo Scientific Co., Waltham, MA, US). Ground samples were scanned to measure the NIR reflectance spectra from 1000 to 2500 nm (wavenumber range from 10,000 to 4000 cm
−1) at 0.96-nm intervals using a 5-cm sample cup spinner accessory. The resolution was 8 cm
−1. Absorbance values are expressed as log(1/R), where R is the sample reflectance. The average spectrum of each sample was obtained from 64 successive scan data recorded by the built-in software RESULT Integration [
28].
2.4. Data Preprocessing, Sample Partition, and Cross-Validation
Multiple scattering correction (MSC) is probably the most widely used preprocessing method in NIRS; it was originally proposed by Martens et al. [
29]. The idea of MSC is to remove those defects or flaws caused by scattering effects from the original data matrix before developing models. The theoretical basis for MSC stems from the fact that the wavelength dependence of scattered light is different from that of light based on compound absorption. Thus, data from multiple wavelengths can be used to distinguish between the absorption of light and the scattering of light.
The derivative preprocessing has the ability to eliminate additivity and multiplicative effects in the spectra and has been used in the spectral analysis for over a decade. The first-order derivative removes only the baseline, while the second-order derivative removes both the baseline and the linear trend. The derivative method proposed by Savitzky and Golay [
30] is probably one of the most popular NIR data preprocessing methods, using smoothing that does not overly reduce the signal-to-noise ratio of the corrected spectra. They popularized a method of numerical derivation of a vector by fitting a polynomial to a symmetrical window of the raw data in order to find the derivative value at the center point. When calculating the parameters of this polynomial, it is easy to find the derivative value of any order of the function by analysis and then use that value as the estimated value of the derivative at that central point, repeat this step, and apply it to all other points in the spectra. The number of points (window size) used to calculate the polynomial and the number of orders to fit the polynomial are specified in advance.
Mean centering subtracts the mean value of the response at each wavelength point in all samples by the spectral response value. Since the response value at each wavelength point is subtracted from the mean value of that column, the data in each row after averaging represents the difference between that sample and the mean sample of the original data.
This study uses two sample partition methods: the Kennard–Stone [
31] method and iterative random partition of samples. The former one selects a subset of samples from the original dataset, which provides uniform coverage of the original dataset and includes samples on the dataset boundaries. The method initially uses geometric distance to find the two samples furthest apart in the original data set. To add another sample to the selected subset, the method selects from the remaining samples the sample that is furthest from the selected sample in the geometric distance. The distance separating a candidate sample from a selected subset refers to the geometric distance between the candidate sample and its closest selected subset. Then, add that candidate sample and repeat the process until the specified required number of samples is added to the selected subset. In fact, this will generate a very evenly distributed space of selected sample points on the dataset. On the contrary, the iterative random partition method is relatively more complex because it randomly generates training sets and test sets for each iteration, and then builds training models and predictions from it, stopping when a specified number of iterations is reached. As the number of iterations increases, the difference between the training set and the test set of the model gradually decreases, and the overall predictive performance of the model becomes more and more stable, thus better reflecting the true predictive ability of the model. However, the disadvantage is the high computational complexity.
Cross-validation is a model validation method used to evaluate how the results of statistical analysis will be generalized to independent test sets. In chemometrics, it is a very useful tool to evaluate both the complexity of the model, such as the number of latent variables in a partial least squares discriminant analysis model and the performance of the model when applied to an unknown sample. In this study, a Venetian blinds method belonging to k-fold cross-validation was used. In this approach, the raw data set is randomly equated into k-folds. In the k-fold, one subsample was retained for the test data used to validate the model, and the remaining k-1 subsample was used as training data. The cross-validation process was then repeated k times, with each of the k subsamples being used as a test sample only once. These k results are then used to calculate the mean of the root mean square error (RMSEC) for the training set and the root mean square error (RMSECV) for the cross-validation set to assess the pros and cons of the model fitting data and the ability of the model to predict unknown samples.
2.5. Principal Component Analysis
Modern analytical instruments often generate data with too many variables, like spectra data from hundreds to thousands. When there are more variables (features) than observations, the correlated model will be under the risk of being massively over-fitted. This kind of model would generally result in terrible out-of-sample performance. By reducing the dimension of feature space, there will be fewer relationships between variables to consider, and the model is less likely to be over-fitted. In fact, dimensionality reduction represents a tradeoff between a more robust model and lower interpretability of new variables. PCA [
32] is a common method used for linear pattern recognition that adopts the dimensionality reduction method to change the variance distribution of new variables. It is defined as an orthogonal linear transformation that transforms the data to a new coordinate system such that the greatest variance by some scalar projection of the data comes to lie on the first coordinate (the first principal component), the second greatest variance on the second coordinate, and so on. In this research, one or several components accounting for the main variance were selected to replace the original variables of the electronic nose and NIRS [
33,
34].
2.6. Partial Least Squares Discriminant Analysis
Partial least squares discriminant analysis (PLS–DA) is an improved discrimination algorithm based on PLSR that assigns a reference value (dummy variable) to each classification sample. The key to this method is the regression modeling of multiple dependent variables and multiple independent variables. Introducing a residual matrix E, the equation relating the response variable Y and the spectral matrix X is as follows [
35]:
The main step to acquire the least-squares solution B is to decompose both matrix X and matrix Y, and eventually, the predictive value of samples can be calculated.
Because using redundant LVs in the PLS model can lead to model overfitting, the number of LVs is chosen to correspond to that at which the cumulative variance in Y first reached 95%. A threshold is introduced to determine classification. The model performance is evaluated by the accuracy rate (%) as follows:
where TP is the number of true positive samples, TN is the number of true negative samples, FP is the number of false-positive samples, and FN is the number of false-negative samples [
36]. The F
1-Score is the harmonic mean of precision and recall, calculated as follows:
Cohen’s kappa is an alternative measure that can be used to determine the accuracy rate for classification problems. It can be calculated as follows [
37]:
where N is the number of test samples,
is the number of TPs for each class in the main diagonal,
is the sum of counts in the ith column, and
is the sum of counts in the ith row. The calculated value of Cohen’s kappa ranges from −1 (total disagreement) through 0 (random classification) to 1 (complete agreement). The closer the kappa value is to 1, the better the performance of the classification algorithm.
2.7. Continuum Regression
Principal component regression (PCR), PLSR, and multiple linear regression (MLR) can all be unified under the single technique of CR. Lorber et al. [
38] demonstrated the relation of PLS to PCR in an unnamed regression-type method (LWK method), and later, Stone and Brooks introduced the descriptive name “continuum regression” in their paper [
25]. CR is a continuous technique that adopts a cross-validation method to find the optimal regression model within the CR parameter space in which the LVs and the power value corresponding to the minimum predictive residual error sum of squares (PRESS) value. In this study, the continuum power regression method [
39], which significantly simplifies the LWK method, was adopted. In the CR, two parameters, the number of LVs and the power γ (from 0 to ∝), were selected using cross-validation, and MLR (γ = 0), PLS (γ = 1), and PCR (γ = ∝) were used as special cases. Searching the three-dimensional PRESS surface yielded the optimal model for which PRESS was the minimum.
2.8. Analysis of Multivariate Regression Models
All steps of the analysis for the various data preprocessing methods and the classification regression model calculations were carried out in MATLAB (MathWorks, Natick, MA, US) with PLS Toolbox v.8.2.1 (Eigenvector Research Inc., Wenatchee, WA, USA). First, Hotelling’s T2 ellipse was adopted to eliminate abnormal samples. Second, various data preprocessing methods, such as multiplicative scatter correction (MSC), mean-center, and the Savitzky–Golay first derivative with mean-center were implemented to investigate how the predictive accuracies of the regression models were affected by different preprocessing methods for both the electronic nose and NIR datasets. For the Savitzky–Golay first derivative preprocessing method, the number of points in the filter (width = 15), the order of the polynomial to fit the points (order = 2), and the order of the derivative (order = 1) were set.
MLR uses a multiterm linear polynomial to describe a mathematical relationship using several “characteristic” data points measured by ordinary least squares (OLS) [
40]. It refers to a statistical technique that is used to predict the outcome of a dependent variable based on the value of two or more independent variables (predictors). It is sometimes known simply as multiple regression, and it is an extension of linear regression. Once each of the independent factors has been determined to predict the dependent variable, the information on the multiple variables can be used to create an accurate prediction on the level of effect they have on the outcome variable. The model creates a relationship in the form of a straight line (linear) that best approximates all the individual data points. The estimated regression coefficients by the least-squares depend on the predictors in the model, and they can be quite variable when the predictors are correlated. PCR [
41] is an effective method for solving multiple collinearity problems by reducing the dimensionality of a dataset through searching for orthogonal directions in the ordinary data space along which the variance of the data set is maximized. It performs PCA on the observed data matrix for the predictors to obtain the principal components which are used as regressors. Then it will regress the observed vector of outcomes on the selected principal components as covariates, using ordinary least squares regression (same as MLR) to get a vector of estimated regression coefficients (with dimension equal to the number of selected principal components). Finally, it will transform this vector back to the scale of the actual covariates, using the selected PCA loadings (the eigenvectors corresponding to the selected principal components) to get the final PCR estimator (with dimension equal to the total number of covariates) for estimating the regression coefficients characterizing the original model. Due to the PCA process, PCR successfully avoids the collinearity problem, which is very common in NIR data. PLSR is a technique that constructs new predictor variables, known as components, as linear combinations of the original predictor variables. PLSR constructs these components while considering the observed response values, leading to a parsimonious model with reliable predictive power [
27]. PLSR and PCR are both methods to model a response variable when there are a large number of predictors, especially when they are highly correlated or even collinear. PLSR is based on the linear transition from a large number of original descriptors to a new variable space based on a small number of orthogonal factors (latent variables). Unlike PCR, latent variables of PLSR are chosen in such a way as to provide maximum correlation with the dependent variable; thus, the PLS model contains the smallest necessary number of factors. With an increasing number of factors, the PLS model converges to the ordinary MLR model.
Several statistical metrics were adopted to evaluate the performance of the regression models combined with different pretreatment methods. The selection of the optimal models depended mainly on the coefficient of determination for the calibration set (
) and prediction set (
), as well as the root-mean-square error of calibration (RMSEC). The residual predictive deviation (RPD; the ratio of the standard error of cross-validation to the standard deviation), and RMSEP were employed to evaluate the performance of the regression model in predicting the concentrations of SDPP in mixed samples [
42]. The computational formulas for R
2, RMSEC, RMSEP, and RPD can be described by the following equations:
where
is the value predicted by the calibration model,
is the value predicted by the cross-validation model,
is the reference value,
is the mean of the reference values,
is the mean of the predicted values, n is the number of samples in the calibration or validation steps, m is the number of predicted samples, and SD is the standard deviation [
43].
3. Results and Discussion
3.1. Interpretation of the Raw Data of the Electronic Nose and Near-Infrared Spectroscopy
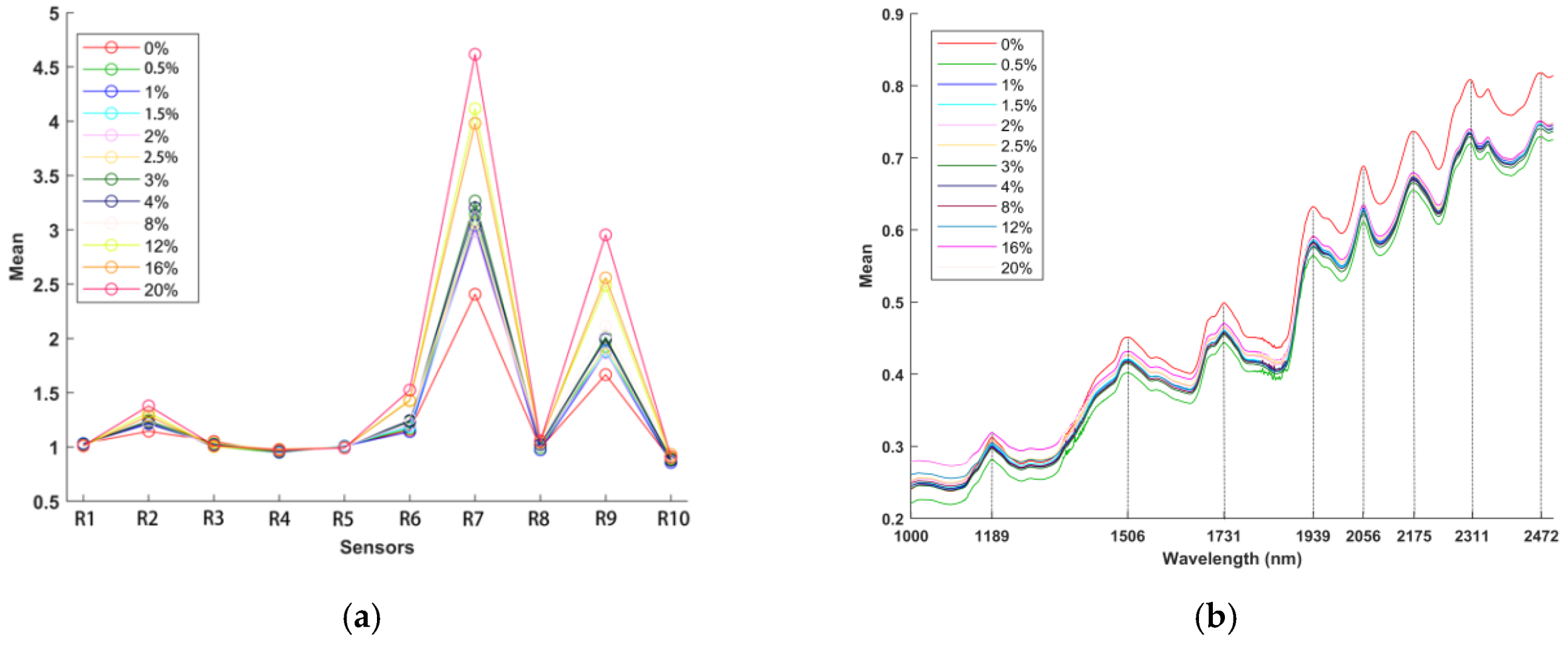
In total, 360 samples were measured by the electronic nose, and the mean data for several samples selected as feature values are plotted in
Figure 3a to show the variation trends in the data from different sensors (as there were too many concentrations; therefore, only a few concentrations are shown). Additionally, 360 samples were scanned by the NIRS analyzer, and the mean spectra are plotted in
Figure 3b to show the variation trends in the data for different wavelengths (the selected concentrations are the same as those used in the electronic nose test).
The average values of 15 samples at different concentrations are shown as the response values for the electronic nose in
Figure 3a (0%, 0.5%, 1%, 1.5%, 2%, 2.5%, 3%, 4%, 8%, 12%, 16%, and 20%). The response values of R1, R3, R4, R5, R8, and R10 showed slight changes, and the response values of R2, R6, R7, and R9 increased gradually; however, the response values of R7 and R9 showed the greatest changes.
The target substances of the R7 and R9 sensors are aromatics and sulfur- and chlorine-containing organics, as shown in
Table 3. Thus, SDPP is rich in sulfur-containing organics, such as sulfur-containing amino acids, which are essential aroma components. For instance, immunoglobulin (IgG) in SDPP contains dimethyl disulfide, which originates from L-cysteine and is an odorous compound. Moreover, the aroma can be a critical ingredient for improving the palatability of creep feed [
44]. Thus, the results proved that utilizing an electronic nose for SDPP detection is feasible.
The mean spectral data are plotted in
Figure 3b to show the variation tendencies of different wavelengths (the selected concentrations are the same as those used in the electronic nose test). The major absorbance differences between the samples of different concentrations were surveyed in the second overtone region (1100−1300 nm), the first overtone region (1600−1950 nm), and the combination vibrational band region (2050−2500 nm). The single absorption centered at approximately 1189 nm was the second overtone of the -CH3, -CH2, and -CH functional group vibration bands. The absorption bands at 1506, 1730, and 1939 nm were assigned to the vibrations of the first overtones of symmetric and asymmetric -NH, -CH, -CH2, -CH3, -SH and -OH functional group stretching. Furthermore, the absorption peaks at approximately 2056, 2175, and 2311 nm were attributed to the characteristic bands of the combined vibrational absorption of -OH and -NH2 groups and -CH3, -CH2, and -CH functional groups, respectively [
40,
42].
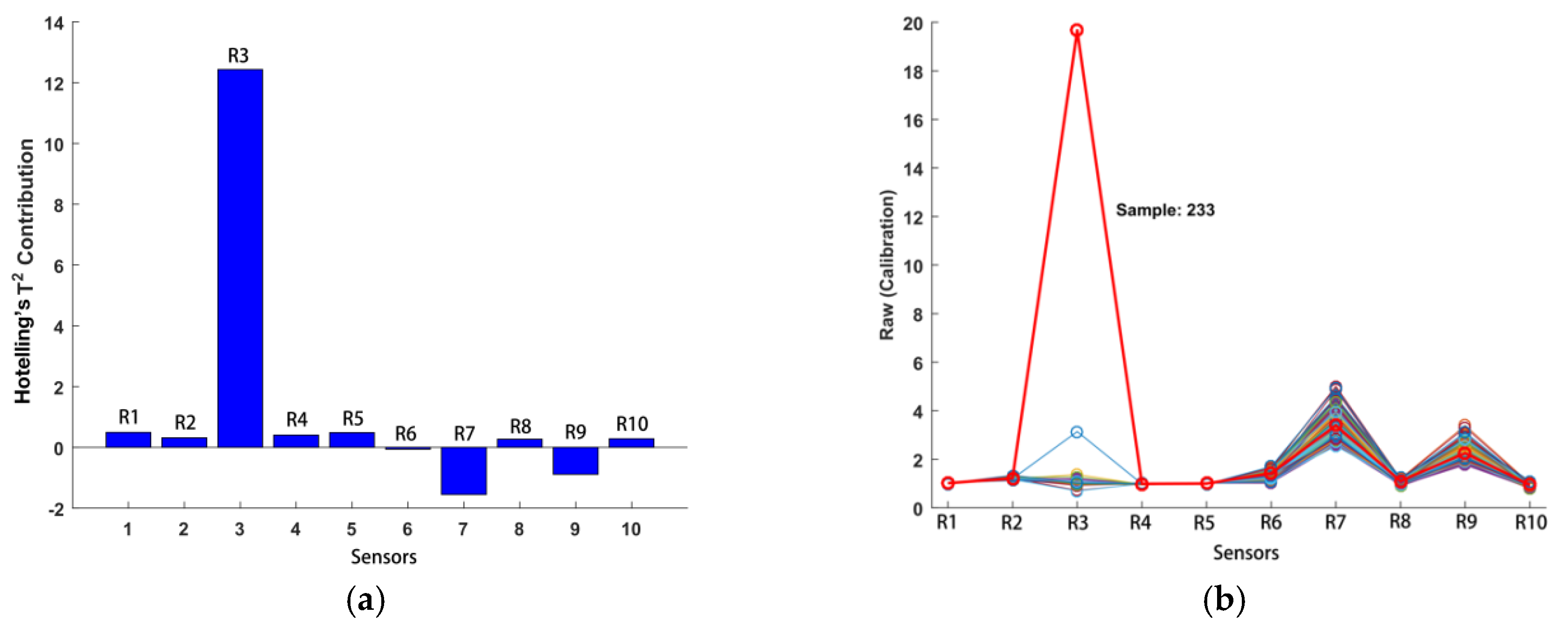
Hotelling’s T
2 test (with a confidence level of 95%) was employed as the measure of the variation among samples in the electronic nose and NIR datasets [
16]. The results indicated that electronic nose sample 233 was indeed unusual, with a large T
2 value of 159 (95% limit = 8.08) and a small Q residual of 0.000179 (95% limit = 0.0402). The T
2 contributions of the electronic nose data are described in
Figure 4a, and the raw data for sample 233 are shown in
Figure 4b. Therefore, electronic nose sample 233 was eliminated as an abnormal sample. The iterative random sampling method was adopted to divide the calibration set and prediction set for both detection models and regression models, and the number of iterations was set to 100. Thus, two-thirds of the samples were selected as the calibration set, and one-third of the samples were selected as the prediction set for each detection model and regression model.
3.2. Principal Component Analysis
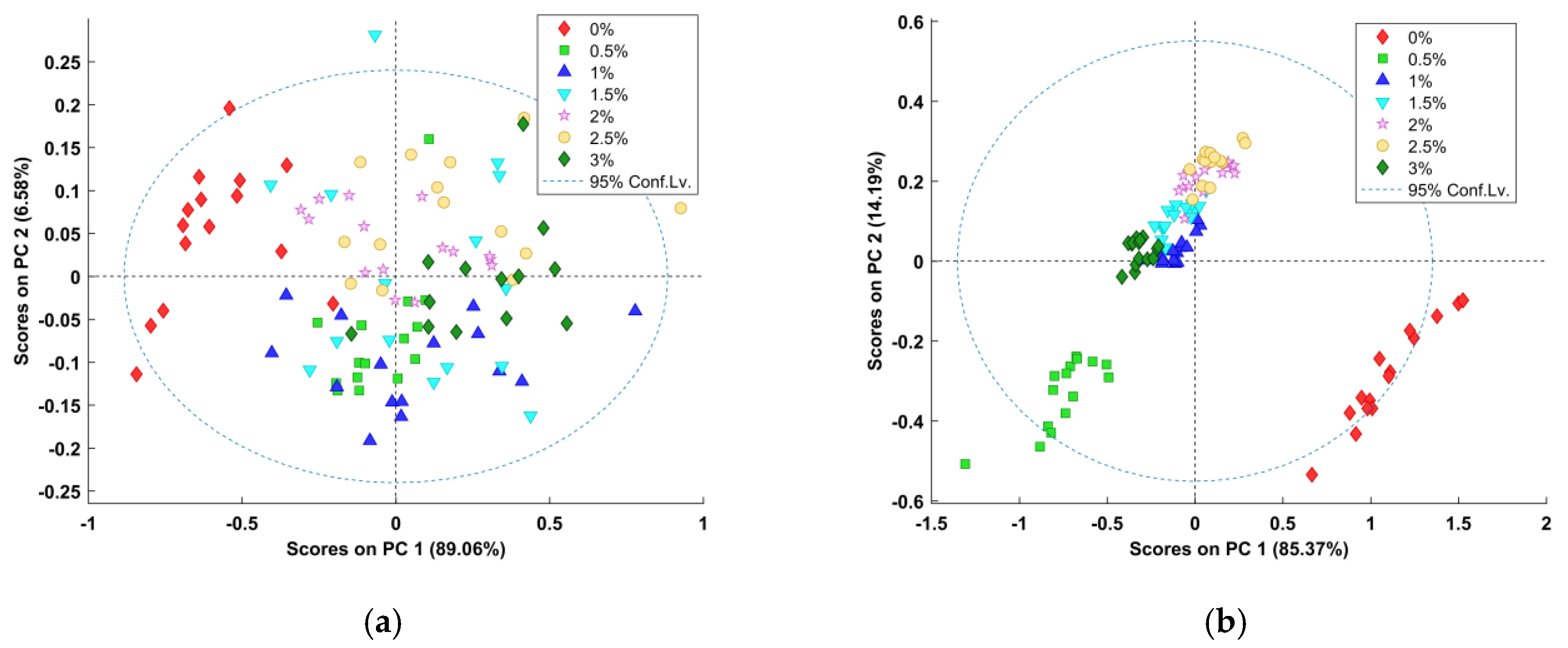
PCA was carried out to discriminate against the presence of SDPP in the test samples. The results of the PCA application to the electronic nose data and all full-spectral data are shown in
Figure 4. The first two principal components (PC) accounted for 95.64% of the variance in the electronic nose data, with PC1 and PC2 explaining 89.06% and 6.58% of the variance, respectively (
Figure 5a). The majority of the variance in the NIRS data was captured by the first two PCs, accounting for 99.56%, with PC1 accounting for 85.37% of the variance and PC2 accounting for 14.19% of the variance (
Figure 5b). The rest of the components did not provide further useful information for determining the presence of SDPP [
45]. According to
Figure 5a, the separation between samples with different concentrations was not substantial. There was an overlap between the “0%” samples and the other samples. As seen in
Figure 5b, although the separation of different samples was not very clear, there was no overlap between the “0%” samples and the other samples. In addition, the clustering quality of the samples was better than that from the electronic nose test. The results indicated that the volatile components of SDPP and WPC might be similar, although the chemical compositions of the materials are different.
3.3. PLS-DA Model
To distinguish pure samples (0%) from impure samples, the PLS–DA model was adopted as a discrimination model by combining all mixture samples (0.5%−3%) into one category, the impure category. Before performing PLS–DA calculations, various preprocessing methods were implemented to preprocess both the raw electronic nose data and the raw NIR spectral data. The discrimination results of the PLS–DA models based on different preprocessing methods are summarized in
Table 4. The accuracy of all PLS-DA models is greater than 90%, which indicates that there was a distinct difference between the pure and impure samples. According to
Table 4, in general, the PLS–DA algorithm performed better on the NIR data than it did on the electronic data for all of the three classification metrics. The high classification rate indicated that both the electronic nose and NIR spectral techniques could be adopted to detect the usage of SDPP. The two types of preprocessing methods containing mean centering performed better than the other methods did, with all metrics reaching absolutely 100%. Although the two techniques had good performances in SDPP detection, compared with the electronic nose technique, the NIRS technique established a more accurate discrimination model with a much smaller confidence interval range.
The results of the PLS–DA models based on the electronic nose data and NIRS data are described in
Figure 6a,b, respectively. The sample number was considered to have X varieties, while the Y variable was associated with sample varieties (a dummy variable was introduced to judge whether a sample belonged to the pure or impure sample group). A threshold value was introduced as the class limit [
46]. Furthermore, the appropriate thresholds for the electronic nose data and NIRS data were obtained from the PLS–DA algorithms, as shown in
Figure 6c,d, respectively, which demonstrated good sensitivity without losing too much specificity.
Figure 6c shows that the threshold value for the electronic nose dataset was Y = 0.53, and
Figure 6d shows that the threshold value for the NIR spectral dataset was Y = 0.6. Variable importance in projection (VIP) plots was adopted to inspect the contribution of each variable by describing the relationship between the spectral matrix (X) and the response variable (Y). Two evident peaks from the electronic nose data and five evident peaks from the NIR spectral data (with a score greater than 1) were observed in the VIP score plots in
Figure 6e,f. It was confirmed that R7 and R9 were the most sensitive sensors for detecting the usage of SDPP. It was found that 1731, 1939, 2056, 2173, and 2307 nm were the feature wavelengths for detecting the usage of SDPP. Hence, the VIP score can also provide information for analyzing changes in the chemical properties of samples.
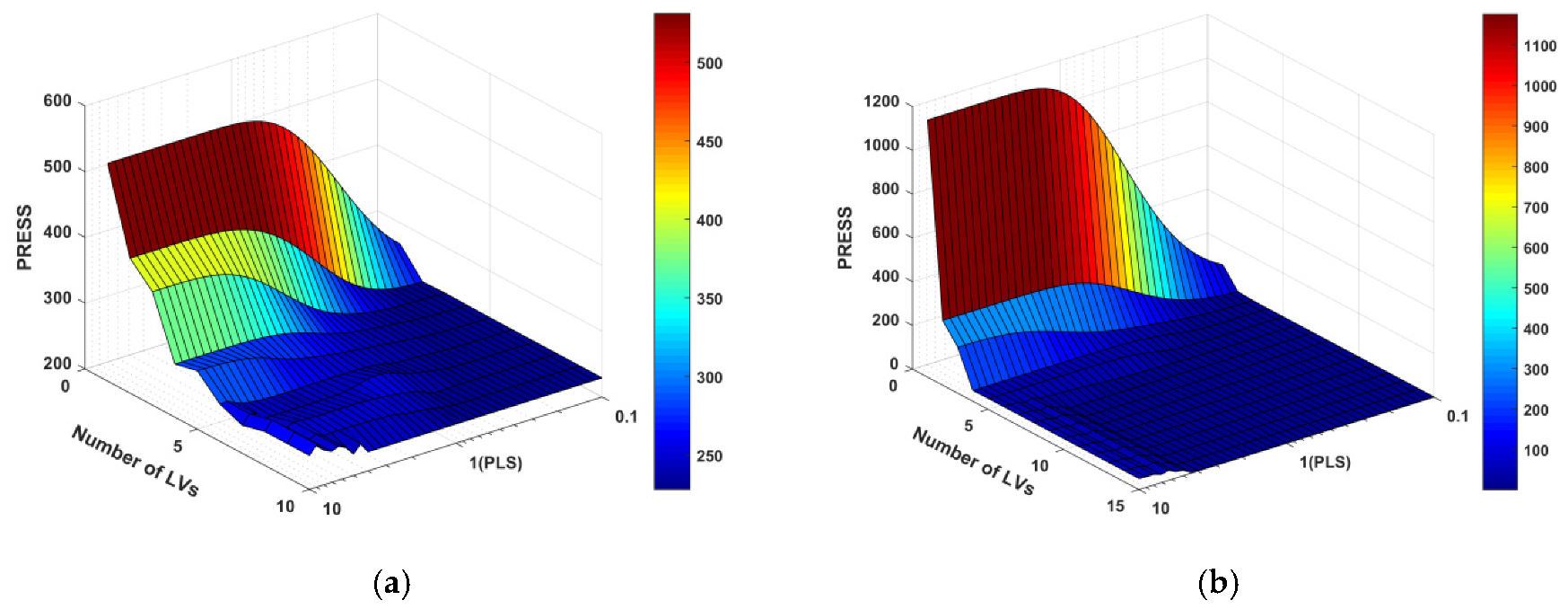
3.4. Continuum Regression Model
The powers used in the investigated CR model ranged from 0.1 to 10 in logarithmically spaced intervals. The maximum numbers of LVs were 10 and 15 for the electronic nose and NIR datasets, respectively. The resulting CR prediction error surfaces are shown in
Figure 7. A given number of LVs and a given value of γ corresponded to a specific height of the PRESS surface.
Considering that there was only one MLR model on the right near the edge, the constant PRESS was identified as the number of LVs varied. In the “MLR flat” region where γ was small and the number of LVs was large, as γ decreased, the CR gave more importance to correlation (as opposed to the structure variance in predictor variables). That is, the data would converge to the MLR results much more quickly as the number of LVs increased. In contrast, the “PCR mountain” region where the number of LVs was few and γ was largely impaired the CR prediction ability greatly. In this context, the cross-validation procedure was used to find the valley located between these two extremes and containing a relatively low PRESS.
From the CR PRESS surfaces of the electric nose data in the test set, the optimal model parameters were located in the region between MLR and PLSR. Specifically, the minimum PRESS was at γ = 0.63 and 8 LVs. This optimal CR model had an RMSEP of 1.6779, compared to 1.6692 for the best MLR model and 1.6861 for the 5-LV best PLS model. However, the minimum PRESS of the NIR data in the test set was found in the region between PCR and PLSR, which was at γ = 2.82 and 14 LVs. This optimal CR model had an RMSEP of 0.1285, compared to 0.4439 for the 4-PC best PCR model and 0.1706 for the 5-LV best PLS model.
As the γ-value decreased to zero, at which point the CR model eventually converged to MLR, the algorithm prioritized the correlation between the predictors and the predicted variables while minimizing the importance of explaining the variance structure of predictor variables. In addition, no matter what value γ may take, only if the LVs were equal to the number of predictor variables would the algorithm converge to MLR. In terms of the electronic nose dataset, with only 10 variables representing 10 independent sensors, the minimum PRESS surface was located in the region between MLR and PLS, which might suggest that MLR was a good model because the γ value was small (0.63) and the LVs (8) were large. Therefore, it was not a coincidence that the best MLR model had a slightly better predictive performance than that of PLS. In contrast, the NIR dataset with many collinear variables preferred a large γ value, for which the algorithm focused more on explaining the variance structure of the predictor variables. Although the optimal CR model had a slightly better predictive performance than that of PLS, with 15 LVs, its model complexity was much higher than that of the 5-LV best PLS.
3.5. Prediction Model Based on Electric Nose Data
The fitting correlation coefficient (R
2) and the root-mean-square error (RMSE) are the two most important indexes for judging the correlation effects of different regression models. According to previous research results, the prediction value has a high correlation with the actual value when R
2 is greater than 0.8. If R
2 is closer to or further from 1, the correlation is higher or lower, respectively. However, if R
2 is lower than 0.8, the prediction value has a low correlation with the actual value, and the prediction is unfeasible. In addition, the closer the RSME value is to 0, the better the prediction effect [
27]. The RPD is used as a statistical indicator. The RPD should generally be greater than 3. Furthermore, R
2 and RPD cannot be very high when the variance in the reference data is low [
47].
The prediction performances of the MLR and PLSR methods based on the electric nose data are shown in
Table 5. The
values of the electronic nose ranged from 0.7419 to 0.8936, and the RMSEC values ranged from 1.9716 to 3.0709. Because the R
2 value was larger than 0.8, the application of the electronic nose for predicting the concentration of SDPP mixed with WPC is feasible. In addition, the optimal algorithm for the electronic nose was the MLR algorithm with the mean center preprocessing method, which confirmed the accuracy of the CR analysis.
3.6. Prediction Model Based on NIRS Data
The prediction performances of the PCR and PLSR methods based on the NIRS data are shown in
Table 6. The
values of the electronic nose ranged from 0.9797 to 0.9989, and the RMSEC values ranged from 0.2003 to 0.8585. Because the R
2 value was larger than 0.8, the application of NIRS for predicting the concentration of SDPP mixed with WPC is feasible. In addition, the optimal algorithm for NIRS was the PLSR algorithm with the Savitzky–Golay first derivative and mean-center preprocessing method, which again confirmed the accuracy of the CR analysis.
3.7. Contrastive Analysis of Prediction Models Based on Different Techniques
The prediction results of the MLR model based on the electronic nose data preprocessed by the mean-center method are described in
Figure 8a, with an Rc
2 value of 0.8936, an RMSEC value of 1.9716, and an Rp
2 value of 0.9247. The prediction results of the PLSR model based on the NIRS data preprocessed by the Savitzky–Golay first derivative and mean-center method are described in
Figure 8b, with an Rc
2 value of 0.9989, an RMSEC value of 0.2003, and an Rp
2 value of 0.999. These results demonstrate that the regression model based on the NIRS data is superior to that based on electronic nose data.
4. Conclusions
In this research, both the electronic nose and NIRS techniques were first applied for SDPP detection in mixed samples. The results demonstrated that both the electronic nose and NIRS were mature enough to provide rapid and accurate techniques for the detection and quantification of the usage of SDPP. The results of the electronic nose experiment indicated that the vital volatile substances of SDPP that enabled the detection of SDPP in mixed samples by electronic nose technology were the sulfur-containing organics, which were odorous compound and sensitive to sensors R7 and R9. The results of the NIRS experiment implied that there exist significantly different components between SDPP and WPC, which may explain the spectral absorption variations. Compared with traditional detection methods, these two electronic technologies could provide faster and more efficient detection methods for SDPP. The details are as follows:
- (1)
The results of the PCA application indicated that the cluster performance based on electronic nose data was poor. Comparatively, the cluster performance based on NIRS data was better. It revealed that for the electronic nose, the volatile components of SDPP and WPC were similar in large part, but the absorption features of these two types of materials in the near-infrared region existed many differences.
- (2)
The classification result of the PLS–DA model showed that the accuracy of the NIRS discrimination model (the classification accuracy was up to 100%) was higher than that of the electronic nose model (the classification accuracy is 95%). Furthermore, the confidence interval range of the NIRS discrimination model was much smaller, which demonstrate that the discrimination capability of NIRS was superior to that of the electronic nose for SDPP detection.
- (3)
The CR analysis revealed that the optimal regression model based on electronic nose data was MLR, and the optimal regression model based on NIRS data was PLSR.
- (4)
According to the result of multivariate regression analysis, the MLR prediction model with the mean-center preprocessing method was the optimal prediction model for the electronic nose, with Rp2 and RMSEP values of 0.9247 and 1.7441, respectively. The PLSR prediction model with the Savitzky–Golay first derivative and mean-center preprocessing method was the optimal prediction model for NIRS, with Rp2 and RMSEP values of 0.999 and 0.1905, respectively. Both results confirmed the accuracy of the CR analysis. Furthermore, compared with the electronic nose, NIRS showed an excellent capacity for qualitative and quantitative analysis, in which both the analytical precision and prediction accuracy were higher than obtained those using the electronic nose. According to the results, when taking detection accuracy and average time consumption into consideration, NIRS, which has superior accuracy and is less time-consuming relative to the electronic nose, should be adopted. When considering the detection cost, an electronic nose that has less cost should be considered. Furthermore, considering that the PEN3 is portable, while the Antaris II must stay within the laboratory, it is much more convenient to use an electronic nose in many outdoor scenes and its analytical result can also be trusted.
However, further research is needed to consider more protein raw materials, such as yeast hydrolysate (YH), yeast extract (YE), and other animal protein byproducts, which could be used as substitutes for SDPP to enlarge the sample groups and experimental database. Thus, the robustness and accuracy of the qualitative and quantitative analysis model could be improved. Physical and chemical tests could be performed to reveal the critical components for identifying SDPP in other substitutes. Furthermore, more new advanced techniques, such as the electronic tongue, Raman spectroscopy, and hyperspectral imaging techniques, could be applied.