A Transfer Learning Method for Pneumonia Classification and Visualization
Abstract
Featured Application
Abstract
1. Introduction
2. Related Works
2.1. Convolutional Neural Networks
2.2. Transfer Learning and Chest Diseases
3. Materials and Methods
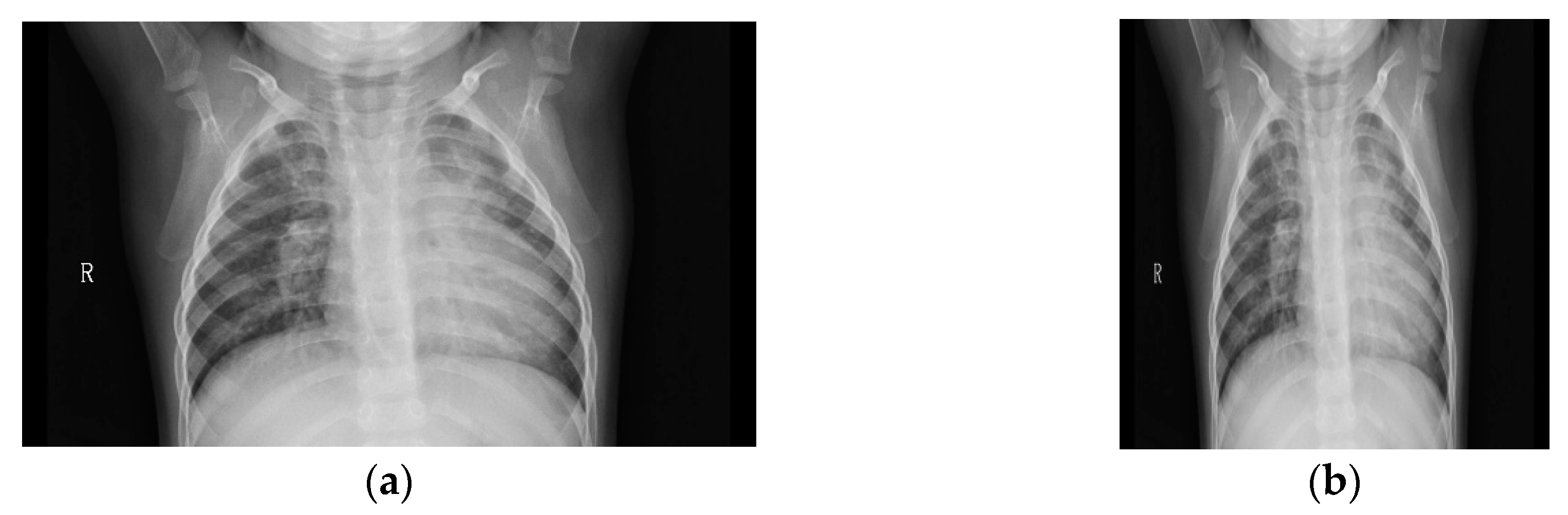
3.1. Pneumonia Dataset
3.2. Class Imbalance
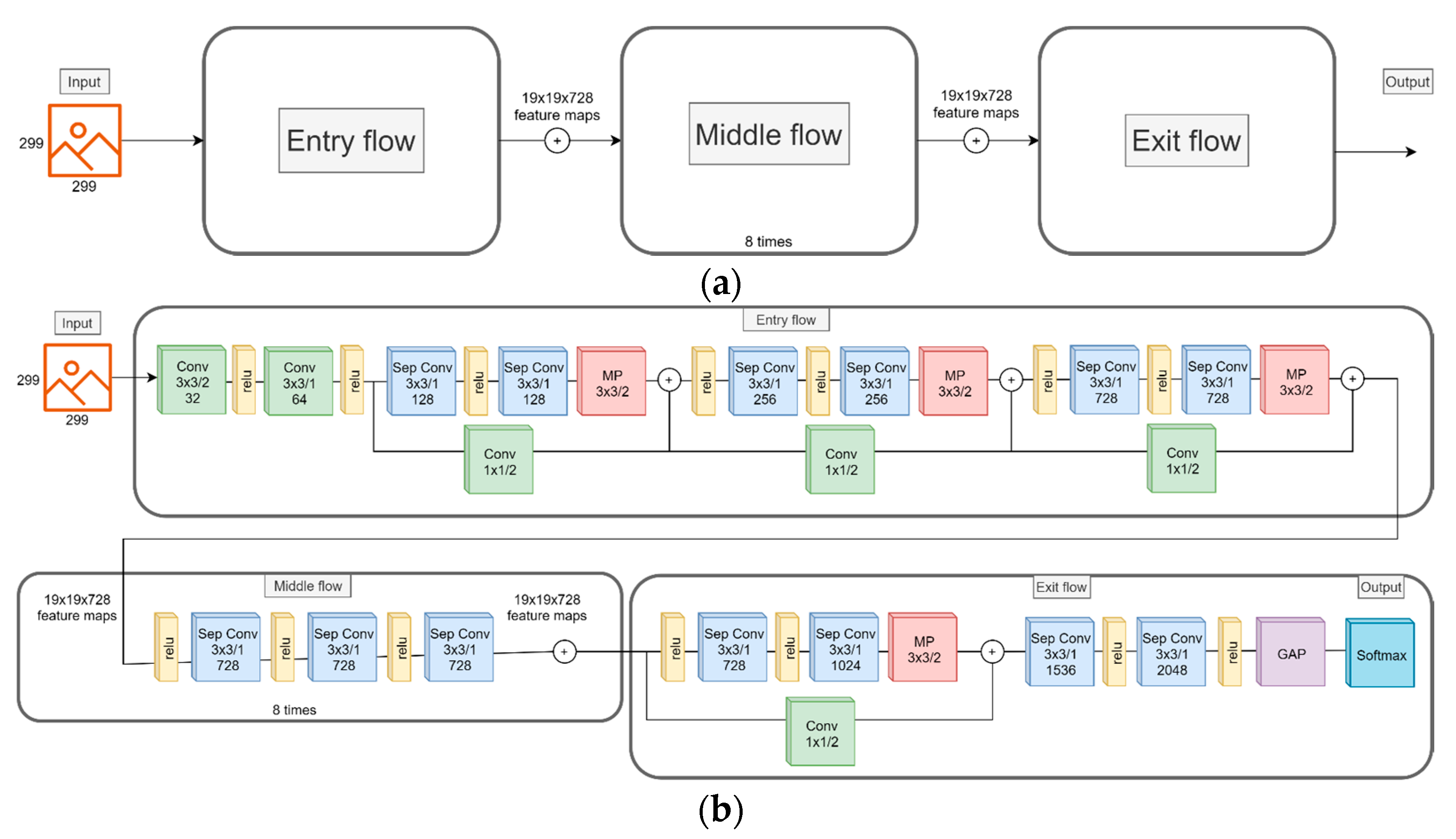
3.3. CNN Model
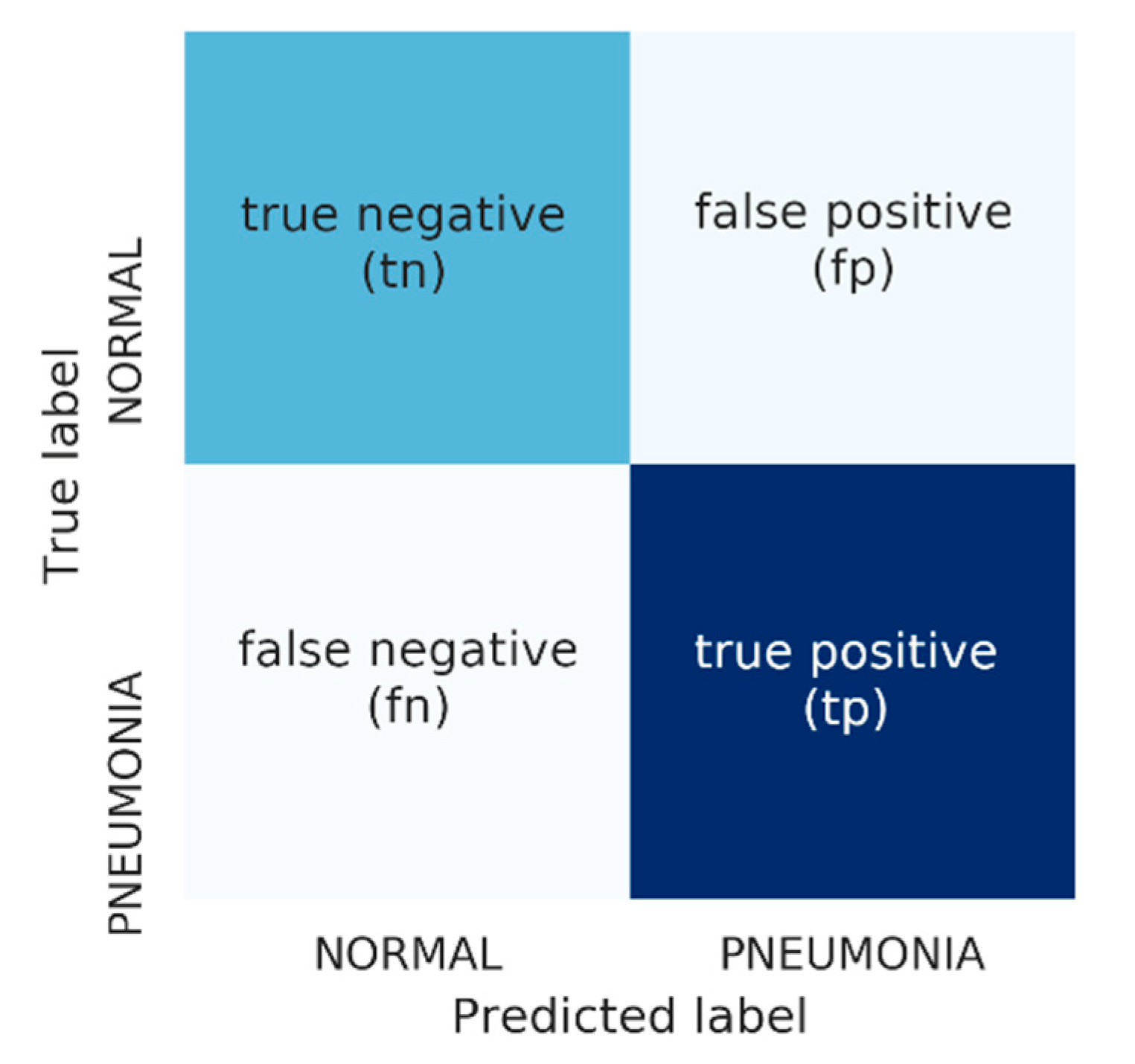
3.4. Performance Measures
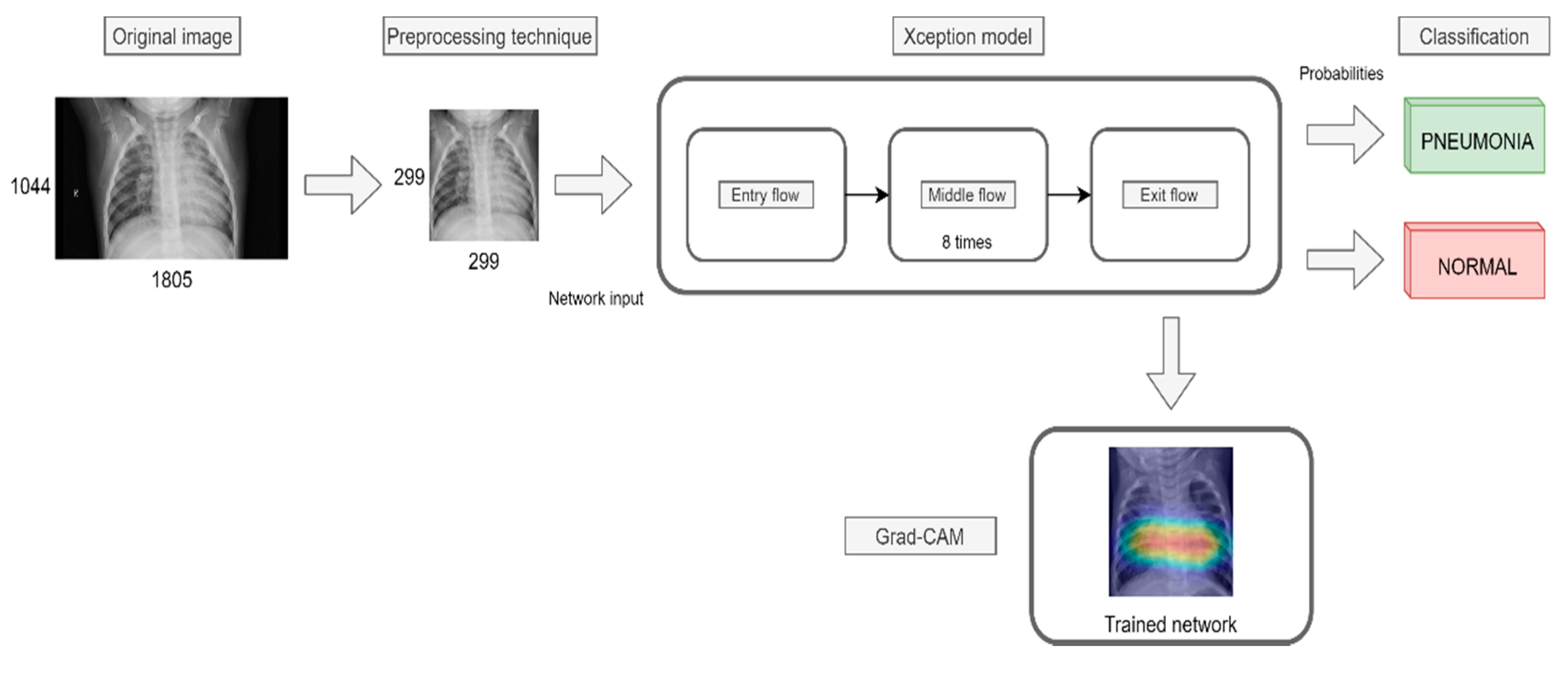
4. Proposal
4.1. Image Preprocessing
- Remove any possible black band from the edges of the image.
- Resize the image to achieve that the smaller edge is (in our case) 299 pixels long.
- Extract the central 299 × 299 region.
4.2. Undersampling
4.3. Cost-Sensitive Learning
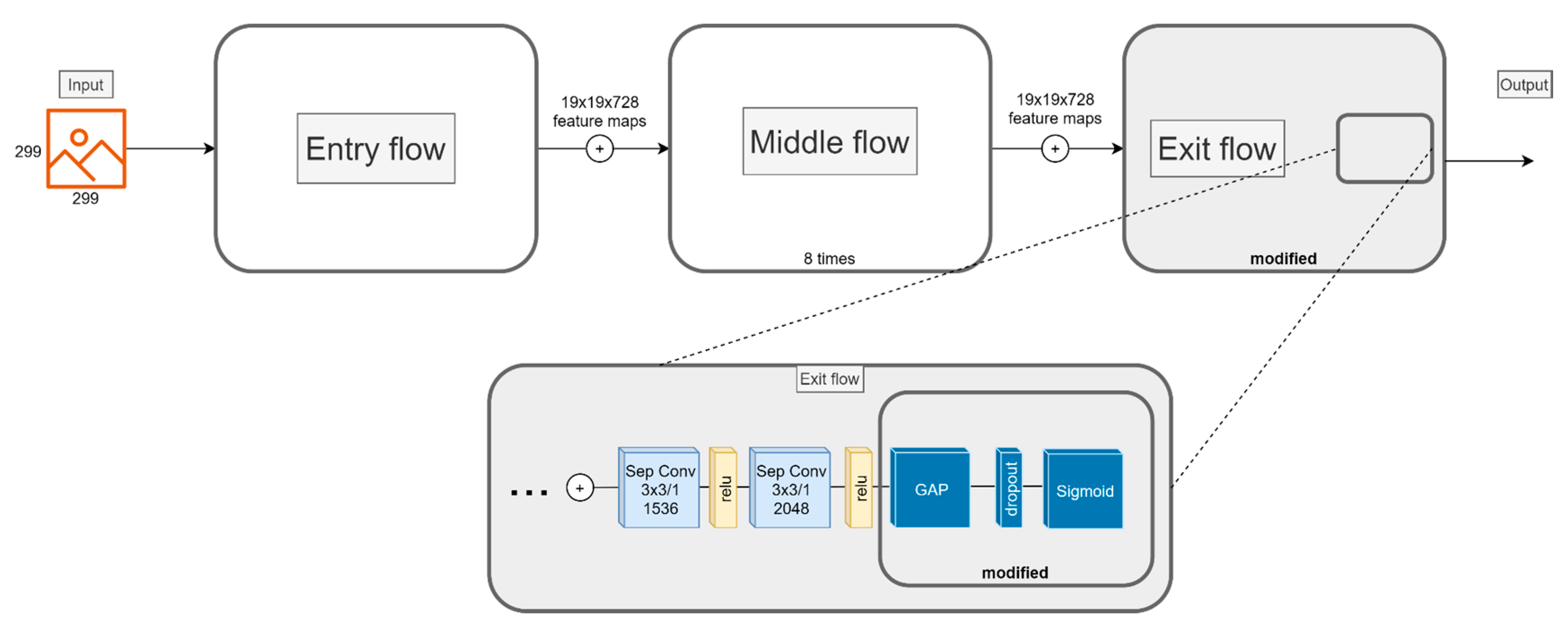
4.4. Transfer Learning
4.5. Data Augmentation and Hyperparameter Tuning
- Horizontal flipping
- Zoom range of ±10%
- Random rotation of ±0.1 degrees.
5. Results
5.1. Experimental Framework
- RUS is performed over the original training data of the Pneumonia dataset.
- Hold-out 80-20 was performed in order to obtain the training and validation sets.
- Pre-processing technique was applied to training, validation and original test sets.
- Training and validation sets were fed into the network.
- The test set was fed into the trained network to obtain the classification results.
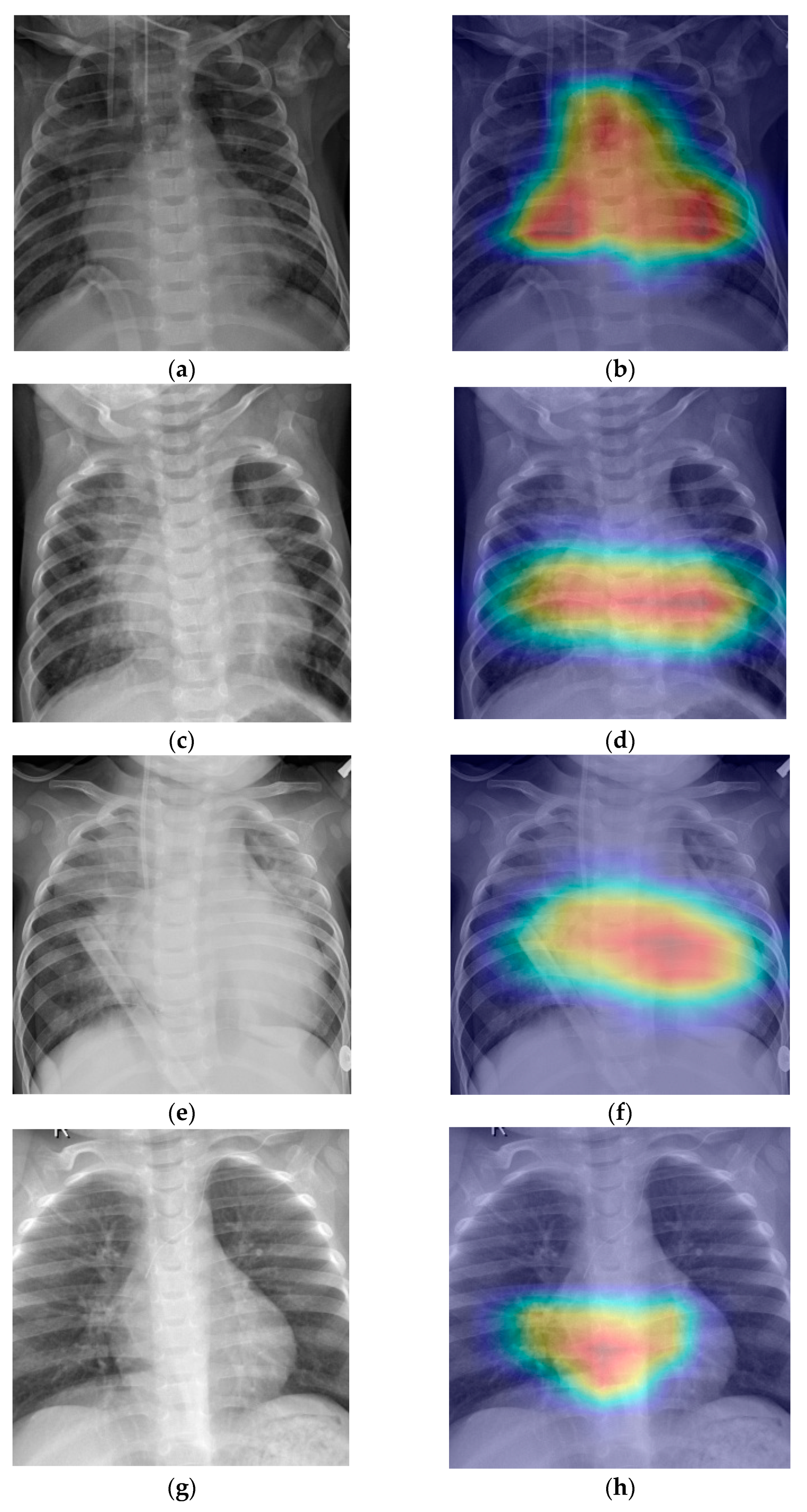
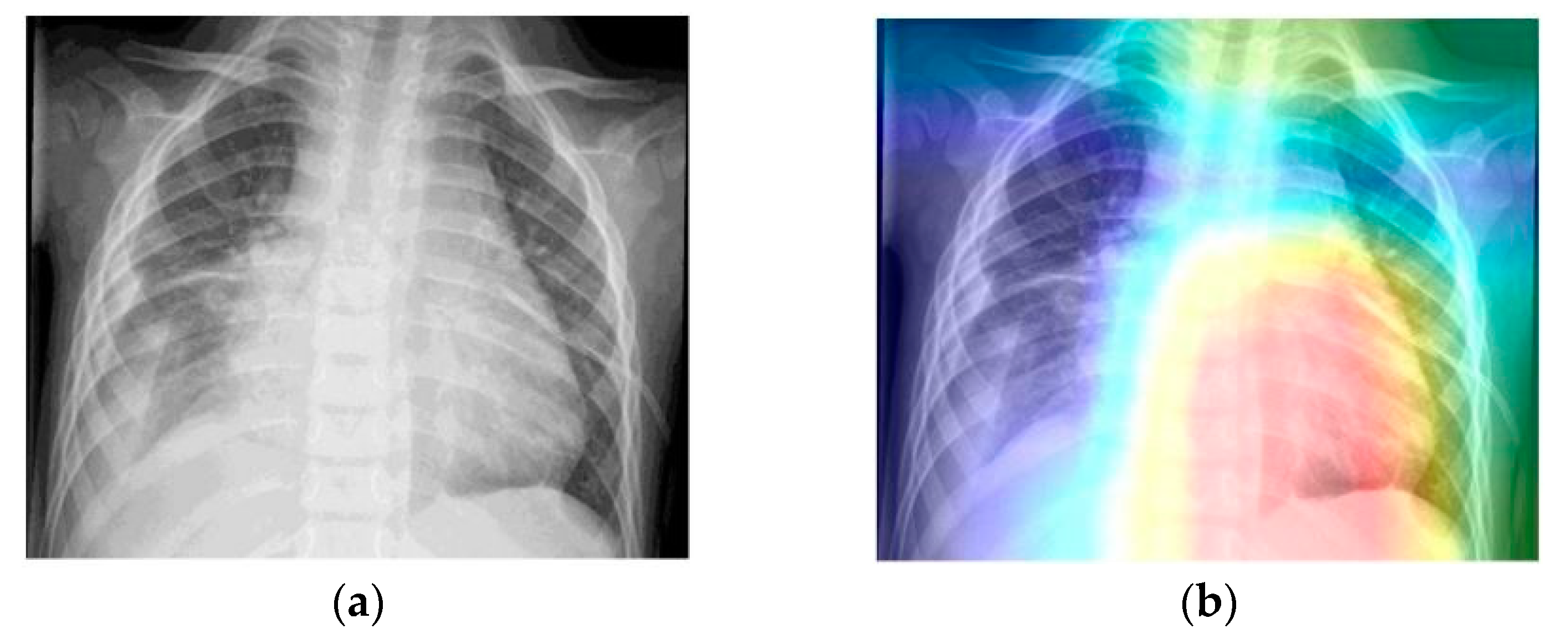
- Grad-CAM was used in order to generate a heatmap of the possible localization of pneumonia manifestations on test images.
5.2. Validation Results
5.3. Test Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mahomed, N.; van Ginneken, B.; Philipsen, R.H.; Melendez, J.; Moore, D.P.; Moodley, H.; Sewchuran, T.; Mathew, D.; Madhi, S.A. Computer-aided diagnosis for World Health Organization-defined chest radiograph primary-endpoint pneumonia in children. Pediatr. Radiol. 2020, 50, 482–491. [Google Scholar] [CrossRef]
- Doi, K. Computer-Aided Diagnosis in Medical Imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Suetens, P. Fundamentals of Medical Imaging, 2nd ed.; Cambridge University Press: New York, NY, USA, 2009; pp. 14–32. [Google Scholar]
- Aliyu, G.; El-Kamary, S.S.; Abimiku, A.L.; Hungerford, L.; Obasanya, J.; Blattner, W. Cost-effectiveness of point-of-care digital chest-x-ray in HIV patients with pulmonary mycobacterial infections in Nigeria. BMC Infect. Dis. 2014, 14, 675. [Google Scholar] [CrossRef] [PubMed]
- Sutton, D. Textbook of Radiology and Imaging, 7th ed.; Churchill Livingstone Elsevier: London, UK, 2003; pp. 131–135. [Google Scholar]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [PubMed]
- Talo, M.; Baloglu, U.B.; Yıldırım, Ö.; Rajendra Acharya, U. Application of deep transfer learning for automated brain abnormality classification using MR images. Cogn. Syst. Res. 2019, 54, 176–188. [Google Scholar] [CrossRef]
- Li, Z.; Wang, C.; Han, M.; Xue, Y.; Wei, W.; Li, L.J.; Fei-Fei, L. Thoracic Disease Identification and Localization with Limited Supervision. In Deep Learning and Convolutional Neural Networks for Medical Imaging and Clinical Informatics. Advances in Computer Vision and Pattern Recognition; Springer: Cham, Swizterland, 2019; pp. 139–161. [Google Scholar]
- Fang, W.; Zhong, B.; Zhao, N.; Love, P.E.; Luo, H.; Xue, J.; Xu, S. A deep learning-based approach for mitigating falls from height with computer vision: Convolutional neural network. Adv. Eng. Inform. 2019, 39, 170–177. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Zuluaga, M.A.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S.; et al. Interactive Medical Image Segmentation Using Deep Learning With Image-Specific Fine Tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef]
- Li, H.; Li, A.; Wang, M. A novel end-to-end brain tumor segmentation method using improved fully convolutional networks. Comput. Biol. Med. 2019, 108, 150–160. [Google Scholar] [CrossRef]
- Chen, S.; Ding, C.; Liu, M. Dual-force convolutional neural networks for accurate brain tumor segmentation. Pattern Recognit. 2019, 88, 90–100. [Google Scholar] [CrossRef]
- Geng, L.; Zhang, S.; Tong, J.; Xiao, Z. Lung segmentation method with dilated convolution based on VGG-16 network. Comput. Assist. Surg. 2019, 24, 27–33. [Google Scholar] [CrossRef]
- Jung, M.; Chi, S. Human activity classification based on sound recognition and residual convolutional neural network. Autom. Constr. 2020, 114, 103177. [Google Scholar] [CrossRef]
- Yao, Z.; Li, J.; Guan, Z.; Ye, Y.; Chen, Y. Liver disease screening based on densely connected deep neural networks. Neural Netw. 2020, 123, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Keras: The Python Deep Learning library. Available online: https://keras.io/ (accessed on 2 November 2019).
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems (NIPS’12), Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 29th IEEE Conference on Computer Vision and Pattern Recognition CVPR 2016, Las Vegas, NV, USA, 26 June–1 July 2016; IEEE: Danvers, MA, USA, 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 29th IEEE Conference on Computer Vision and Pattern Recognition CVPR 2016, Las Vegas, NV, USA, 26 June–1 July 2016; IEEE: Danvers, MA, USA, 2016; pp. 770–778. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; IEEE: Danvers, MA, USA, 2017; pp. 1800–1807. [Google Scholar]
- Bakator, M.; Radosav, D. Deep Learning and Medical Diagnosis: A Review of Literature. Multimodal Technol. Interact. 2018, 2, 47. [Google Scholar] [CrossRef]
- Tsiakmaki, M.; Kostopoulos, G.; Kotsiantis, S.; Ragos, O. Transfer Learning from Deep Neural Networks for Predicting Student Performance. Appl. Sci. 2020, 10, 2145. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning with Python, 1st ed.; Manning Publications Co.: Shelter Island, NY, USA, 2018; pp. 287–295. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; IEEE: Danvers, MA, USA, 2017; pp. 3462–3471. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef]
- Blumenfeld, A.; Greenspan, H.; Konen, E. Pneumothorax detection in chest radiographs using convolutional neural networks. In Proceedings of the Medical Imaging 2018: Computer-Aided Diagnosis, Houston, TX, USA, 27 February 2018. [Google Scholar]
- Que, Q.; Tang, Z.; Wang, R.; Zeng, Z.; Wang, J.; Chua, M.; Gee, T.S.; Yang, X.; Veeravalli, B. CardioXNet: Automated Detection for Cardiomegaly Based on Deep Learning. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–22 July 2018; IEEE: Danvers, MA, USA, 2018; pp. 612–615. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, January 27–1 February 2019; AAAI Press: Palo Alto, CA, USA, 2019; pp. 590–597. [Google Scholar]
- Allaouzi, I.; Ben Ahmed, M. A Novel Approach for Multi-Label Chest X-Ray Classification of Common Thorax Diseases. IEEE Access 2019, 7, 64279–64288. [Google Scholar] [CrossRef]
- Baltruschat, I.M.; Nickisch, H.; Grass, M.; Knopp, T.; Saalbach, A. Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification. Sci. Rep. 2019, 9, 6381. [Google Scholar] [CrossRef]
- Chassagnon, G.; Vakalopolou, M.; Paragios, N.; Revel, M.P. Deep learning: Definition and perspectives for thoracic imaging. Eur. Radiol. 2020, 30, 2021–2030. [Google Scholar] [CrossRef]
- Stephen, O.; Sain, M.; Maduh, U.J.; Jeong, D.-U. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J. Healthc. Eng. 2019, 2019, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing medical imaging data for machine learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef]
- Liang, G.; Zheng, L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Methods Programs Biomed. 2019, 187, 104964. [Google Scholar] [CrossRef] [PubMed]
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, D.; Tiwari, P.; Moreira, C.; Damaševičius, R.; de Albuquerque, V.H.C. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl. Sci. 2020, 10, 559. [Google Scholar] [CrossRef]
- Pasa, F.; Golkov, V.; Pfeiffer, F.; Cremers, D.; Pfeiffer, D. Efficient Deep Network Architectures for Fast Chest X-Ray Tuberculosis Screening and Visualization. Sci. Rep. 2019, 9, 6268. [Google Scholar] [CrossRef]
- Vajda, S.; Karargyris, A.; Jaeger, S.; Santosh, K.C.; Candemir, S.; Xue, Z.; Antani, S.; Thoma, G. Feature Selection for Automatic Tuberculosis Screening in Frontal Chest Radiographs. J. Med. Syst. 2018, 42, 146. [Google Scholar] [CrossRef]
- Hwang, S.; Kim, H.-E.; Jeong, J.; Kim, H.-J. A novel approach for tuberculosis screening based on deep convolutional neural networks. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis, San Diego, CA, USA, 28 February–2 March 2016; Tourassi, G.D., Armato, S.G., III, Eds.; SPIE: Bellingham, WA, USA, 2016; pp. 750–757. [Google Scholar]
- Chauhan, A.; Chauhan, D.; Rout, C. Role of gist and PHOG features in computer-aided diagnosis of tuberculosis without segmentation. PLoS ONE 2014, 9, e112980. [Google Scholar] [CrossRef]
- Kermany, D.; Zhang, K.; Goldbaum, M. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Available online: https://data.mendeley.com/datasets/rscbjbr9sj/2 (accessed on 7 October 2019).
- López, V.; Fernández, A.; García, S.; Palade, V.; Herrera, F. An insight into classification with imbalanced data: Empirical results and current trends on using data intrinsic characteristics. Inf. Sci. (N.Y.). 2013, 250, 113–141. [Google Scholar] [CrossRef]
- Szegedy, C.; Wei, L.; Yangqing, J.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 28th IEEE Conference on Computer Vision and Pattern Recognition CVPR 2015, Boston, MA, USA, 7–12 June 2015; IEEE: Danvers, MA, USA, 2015; pp. 1–9. [Google Scholar]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Batista, G.E.A.P.A.; Prati, R.C.; Monard, M.C. A study of the behavior of several methods for balancing machine learning training data. ACM Sigkdd Explor. Newsl. 2004, 6, 20. [Google Scholar] [CrossRef]
- López, V.; Fernández, A.; Moreno-Torres, J.G.; Herrera, F. Analysis of preprocessing vs. cost-sensitive learning for imbalanced classification. Open problems on intrinsic data characteristics. Expert Syst. Appl. 2012, 39, 6585–6608. [Google Scholar]
- OpenCV. Available online: https://opencv.org/ (accessed on 2 November 2019).
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]




| Class | Weight |
|---|---|
| NORMAL | 5.0 |
| PNEUMONIA | 0.5 |
| Cost Function | Learning Rate (Lr) | Optimizer | No. Epochs | Batch Size | Lr Decay |
|---|---|---|---|---|---|
| Binary cross entropy | Adam | 100 | 32 | 10 times after a plateau |
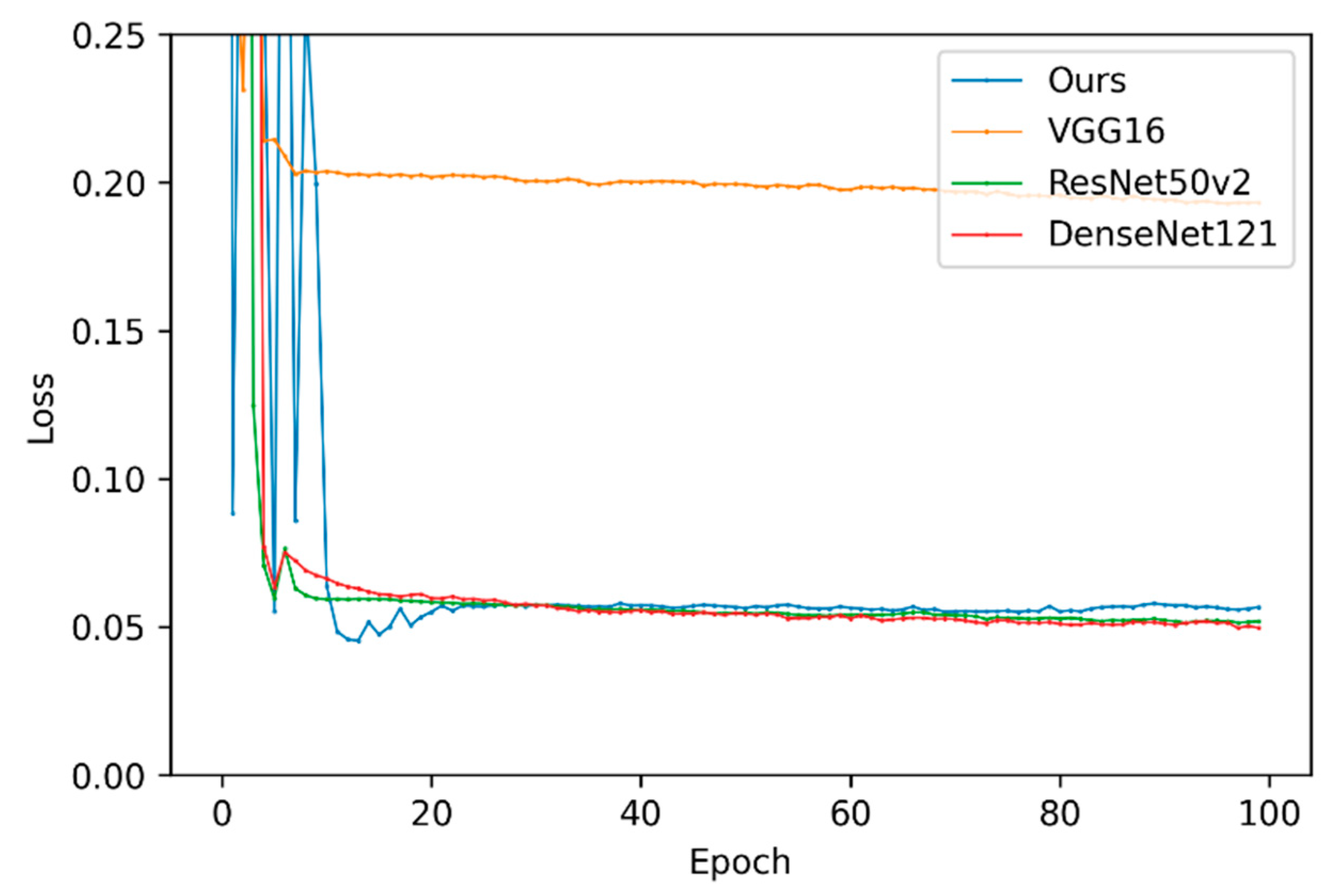
| Model | Best Epoch | Validation Loss | Average Training Time (Epoch) | Average Training Time (Example) | Convergence Time (Best Model) |
|---|---|---|---|---|---|
| VGG-16 | 97 | 0.1928 | 44 s | 0.0202 s | 4268 s |
| ResNet50-v2 | 93 | 0.0514 | 43 s | 0.0198 s | 3999 s |
| DenseNet121 | 98 | 0.0496 | 51 s | 0.0234 s | 4998 s |
| Our model | 14 | 0.0453 | 53 s | 0.0244 s | 742 s |
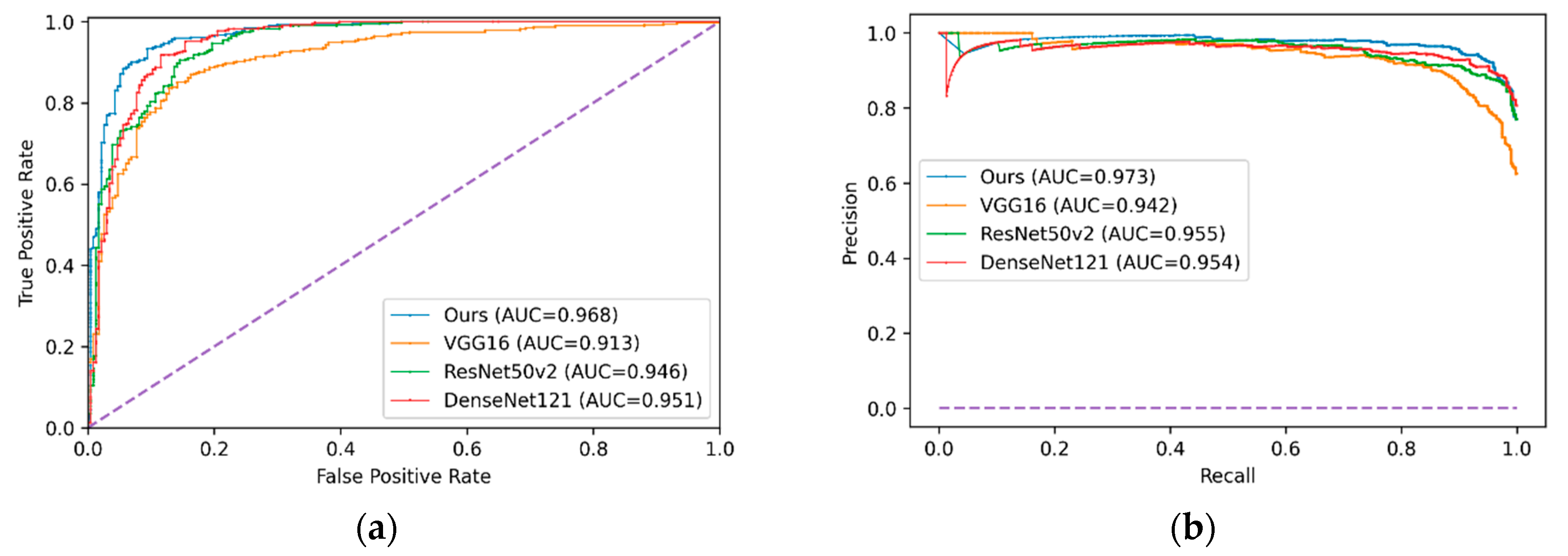
| Model | Confusion Matrix | Precision | Recall | F1-Score | ROC Curve AUC | Precision–Recall AUC | |
|---|---|---|---|---|---|---|---|
| VGG-16 | 184 | 50 | 0.874 | 0.892 | 0.883 | 0.913 | 0.942 |
| 42 | 348 | ||||||
| ResNet50-v2 | 156 | 78 | 0.832 | 0.990 | 0.904 | 0.946 | 0.955 |
| 4 | 386 | ||||||
| DenseNet121 | 159 | 75 | 0.838 | 0.992 | 0.908 | 0.951 | 0.954 |
| 3 | 387 | ||||||
| LZNet2019 | 188 | 46 | 0.891 | 0.967 | 0.927 | 0.953 | - |
| 13 | 377 | ||||||
| Cho2020 | 207 * | 31 * | 0.932 | 0.996 | 0.959 * | 0.993 | - |
| 4 * | 386 * | ||||||
| Our model | 162 | 72 | 0.843 | 0.992 | 0.912 | 0.968 | 0.973 |
| 3 | 387 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luján-García, J.E.; Yáñez-Márquez, C.; Villuendas-Rey, Y.; Camacho-Nieto, O. A Transfer Learning Method for Pneumonia Classification and Visualization. Appl. Sci. 2020, 10, 2908. https://doi.org/10.3390/app10082908
Luján-García JE, Yáñez-Márquez C, Villuendas-Rey Y, Camacho-Nieto O. A Transfer Learning Method for Pneumonia Classification and Visualization. Applied Sciences. 2020; 10(8):2908. https://doi.org/10.3390/app10082908
Chicago/Turabian StyleLuján-García, Juan Eduardo, Cornelio Yáñez-Márquez, Yenny Villuendas-Rey, and Oscar Camacho-Nieto. 2020. "A Transfer Learning Method for Pneumonia Classification and Visualization" Applied Sciences 10, no. 8: 2908. https://doi.org/10.3390/app10082908
APA StyleLuján-García, J. E., Yáñez-Márquez, C., Villuendas-Rey, Y., & Camacho-Nieto, O. (2020). A Transfer Learning Method for Pneumonia Classification and Visualization. Applied Sciences, 10(8), 2908. https://doi.org/10.3390/app10082908







