Machine Learning Classifiers Evaluation for Automatic Karyogram Generation from G-Banded Metaphase Images
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
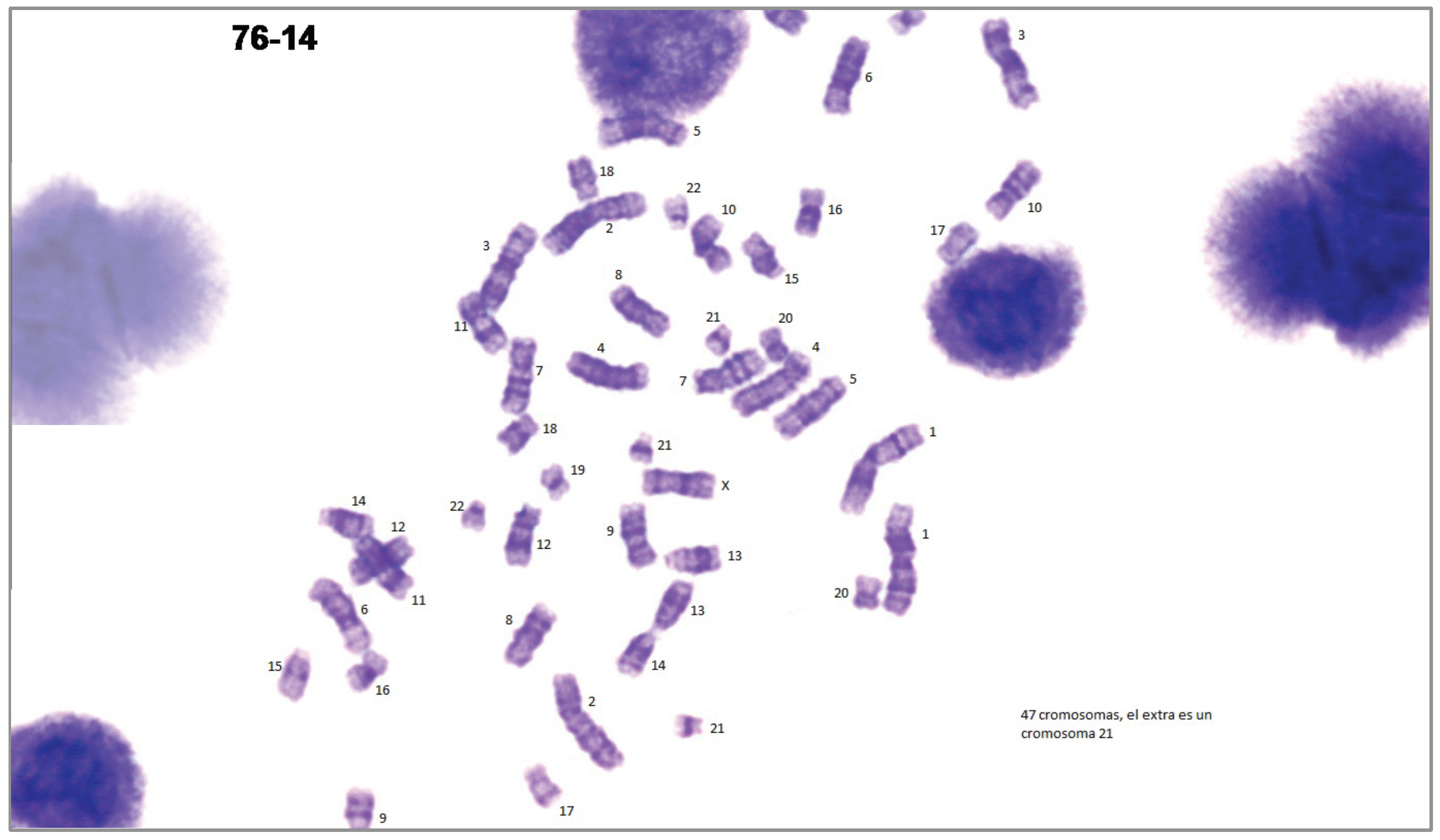
- A dataset obtained in collaboration with cytogeneticists of the CHT. The database consists of 24 G-banded prometaphase images acquired from 24 different patients and their corresponding manual karyograms. This dataset is under construction, and results have not been published elsewhere. In order to use the data of the Laboratory of Cytogenetics of the CHT, the Research and Education Department of the CHT reviewed and approved the use of the karyotypes in this work, judging that no appreciable risks or ethical issues were encountered, since no personal data associated to the karyotypes were issued.
- A dataset retrieved from Laboratory of Biomedical Imaging (BioImLab) from the University of Padova. The database consists of 119 Q-banded prometaphase images and their corresponding manual karyograms acquired from the same number of cells [38].
- Matlab R2014 has been used for the preprocessing, segmentation and transformation required in order to separate the chromosomes from the input prometaphase image. This software was alsa used to test multiple MLPs with several neurons in the hidden layer in order to find the optimal network configuration that yields the best performance.
- Weka 3.6.7 has been used to train and test classifiers selected for the comparison reported in this work.
- A Carl Zeiss Axioscope A1 microscope.
- An Axiocam ICC1 camera coupled with the microscope, with USB interface.
- A gateway desktop PC with Intel Core-2 Duo processor, 4GB RAM, and Linux 64-bit Mint OS.
2.2. Methods
2.2.1. Outline of the Proposed Automatic Classification
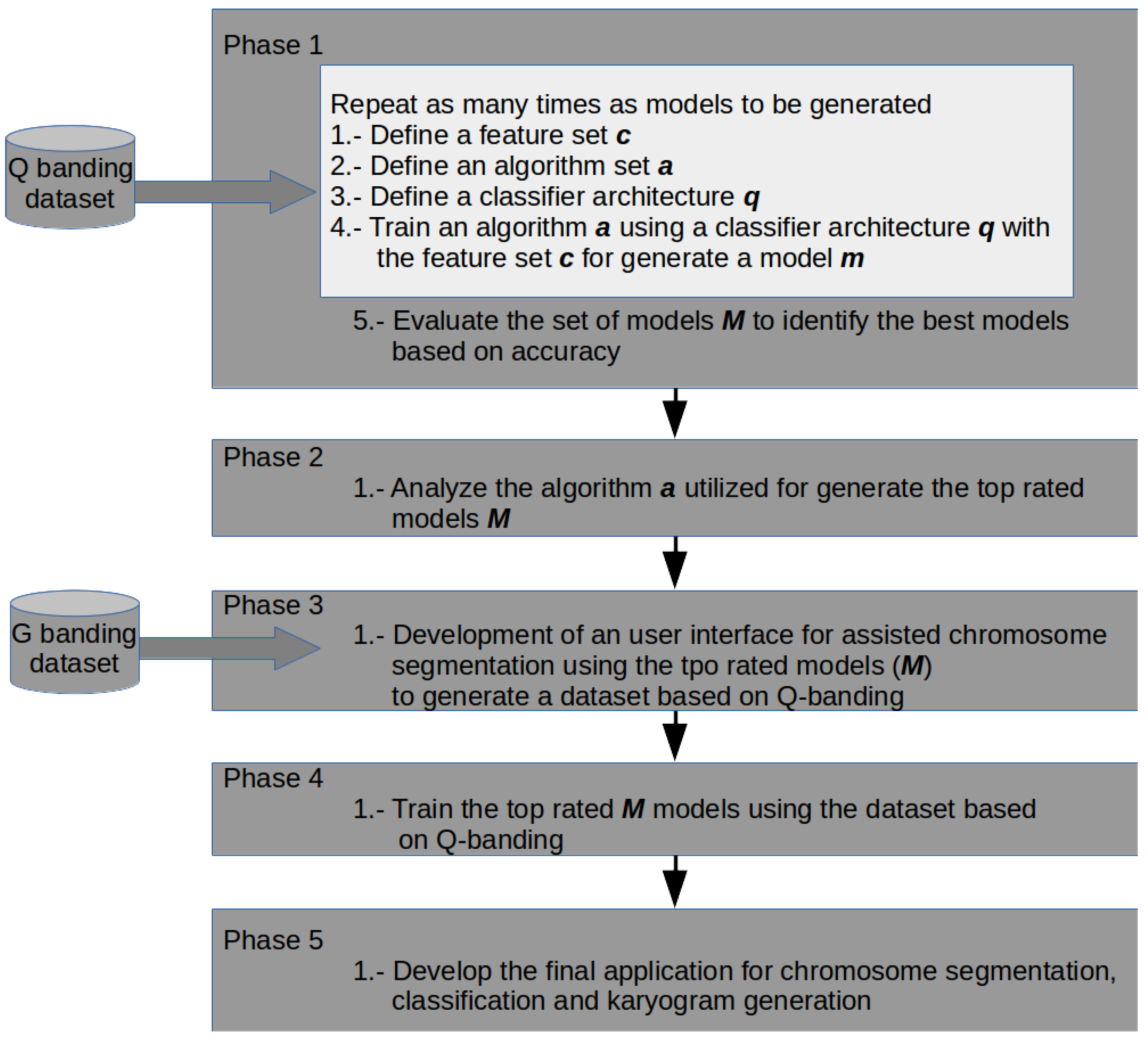
Phase 1. Classifiers Training
- 1.
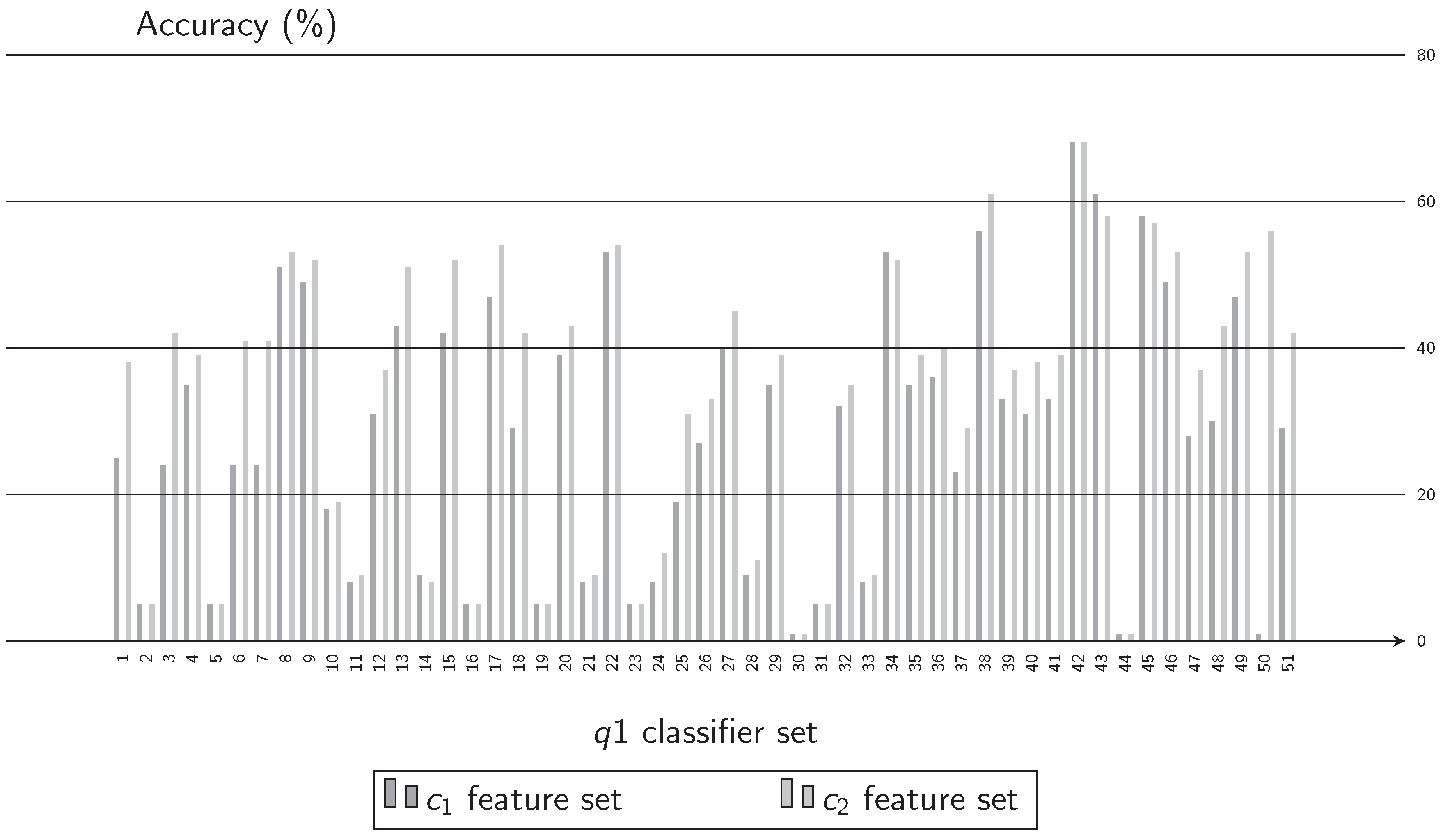
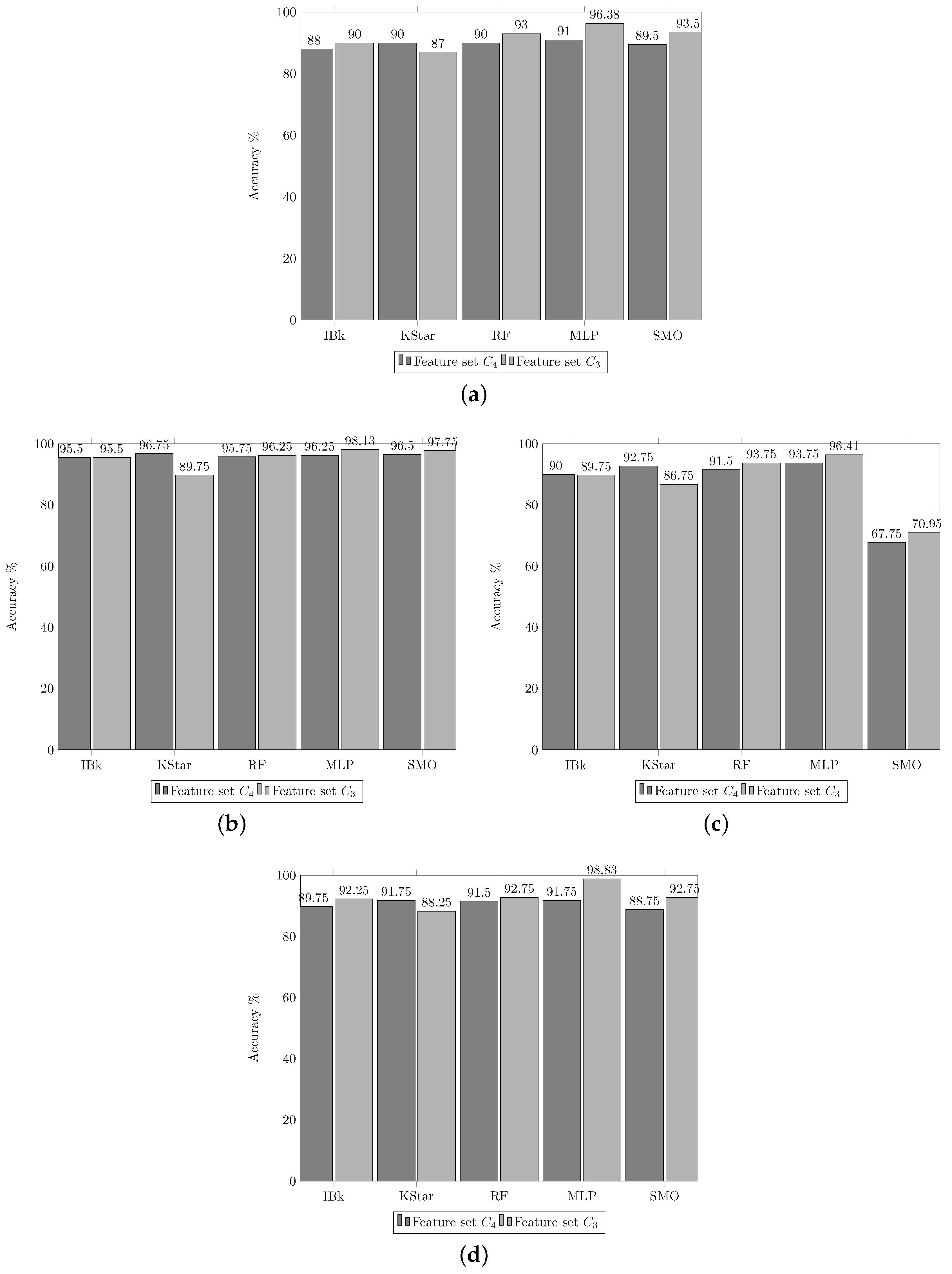
- From the literature, groups of features that can be extracted from Q-band chromosome images where formed. These were listed as the subset of features , , , and and are presented in Table 1. In this activity, one of the feature groups is selected to be extracted from every chromosome in the image database. With these features, two sets are generated in separated datasets to be utilized in training. The first dataset is used to train the classifiers and the second one as test dataset.
- 2.
- In this activity, a group of classification algorithms available in the Weka platform is selected and identified as the set A. These elements conform the subset . According to the experimental results, the elements of the subset were modified to form the subset of algorithms, . The algorithms of subset are enlisted in Table 2, and for subset in Table 3.
- 3.
- This activity defines the architectures that will be used in chromosomes classification. An architecture represents how the chromosomes are going to be classified in one of the 24 output classes. For example, one architecture could be a multiclass classification, where the chromosome would be assigned directly to 1 of the 24 classes. Another option is to divide the group into autosome and sex chromosomes, and identify if it belongs to one of this groups or not (binary classification). Table 4 summarizes the defined architectures for this activity.
- 4.
- Training and testing of a model is performed in this activity. The training and testing dataset, the set of algorithms and the classification architecture are the elements of the current model, . The training and testing accuracies are reported, and they are used as evaluation metric for the next activity.
- 5.
- Once several models were generated (through activities 1 to 4), the best chromosome classification model, , is identified, based on the chromosome classification accuracy. Current could represent, either the output of phase 1, or the final classification model, m.
Phase 2. Classifier Analysis
Phase 3. Application Development
Phase 4. Classifiers Training
Phase 5. Application Integration
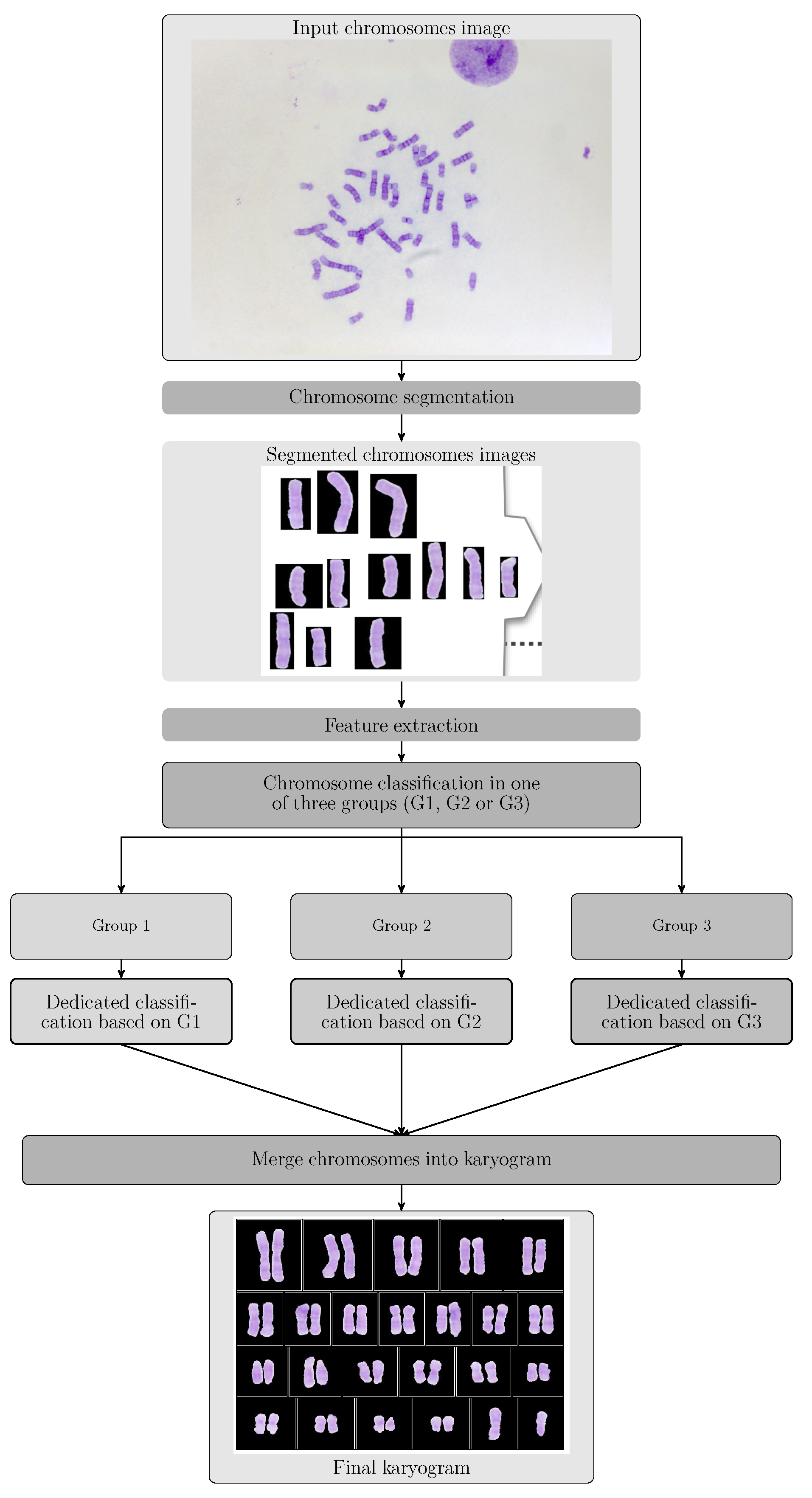
- Module A:
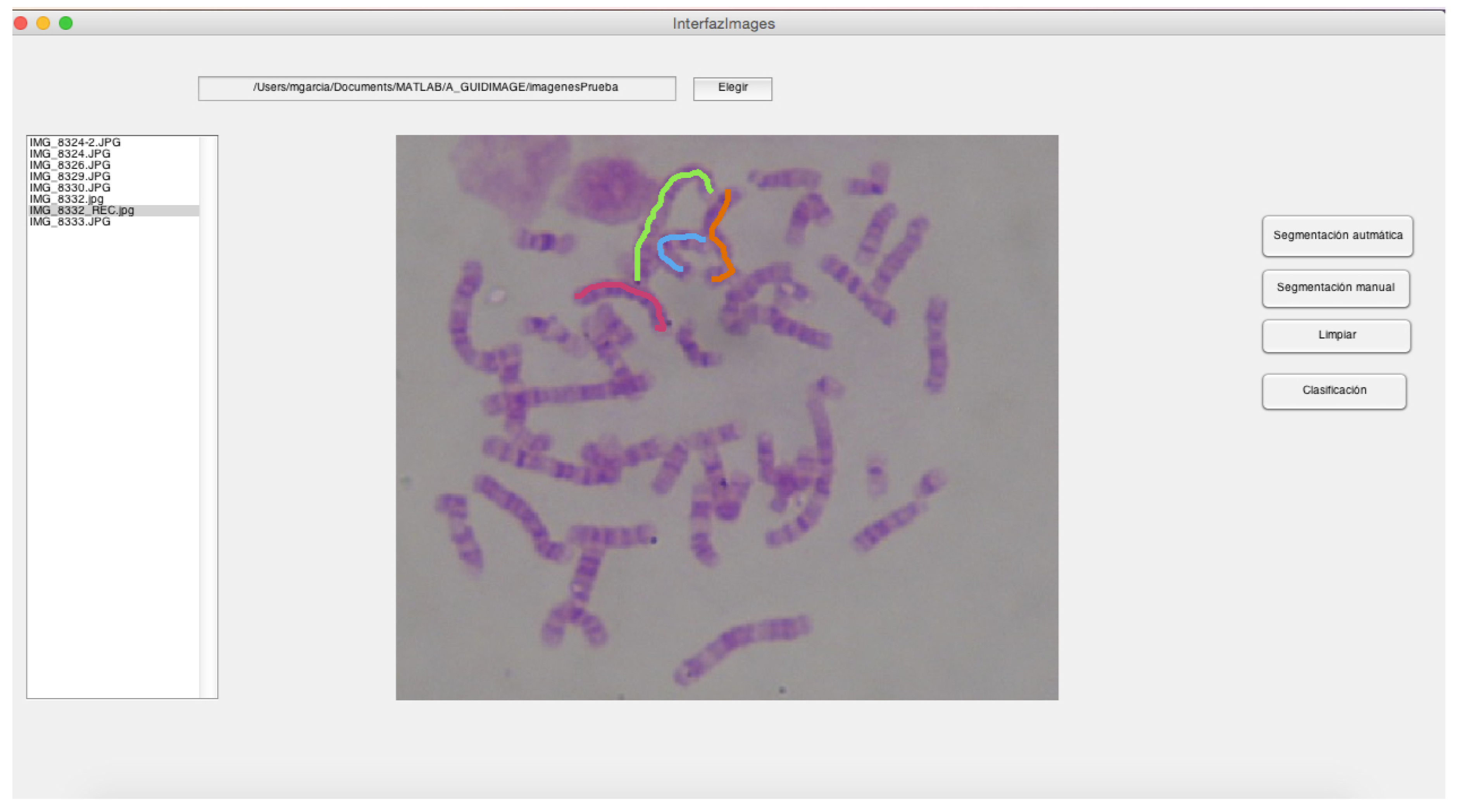
- This module includes a GUI and the segmentation related operations. It allows the user to generate the segmented chromosomes and arrange them in directories.
- Module B:
- It comprises the automatic classification operations, including the development of a GUI that allows the user to: (i) Enter the segmented chromosomes obtained by the module A; (ii) Use the classification model m, obtained in phase 4. Its output is composed of the classified chromosomes.
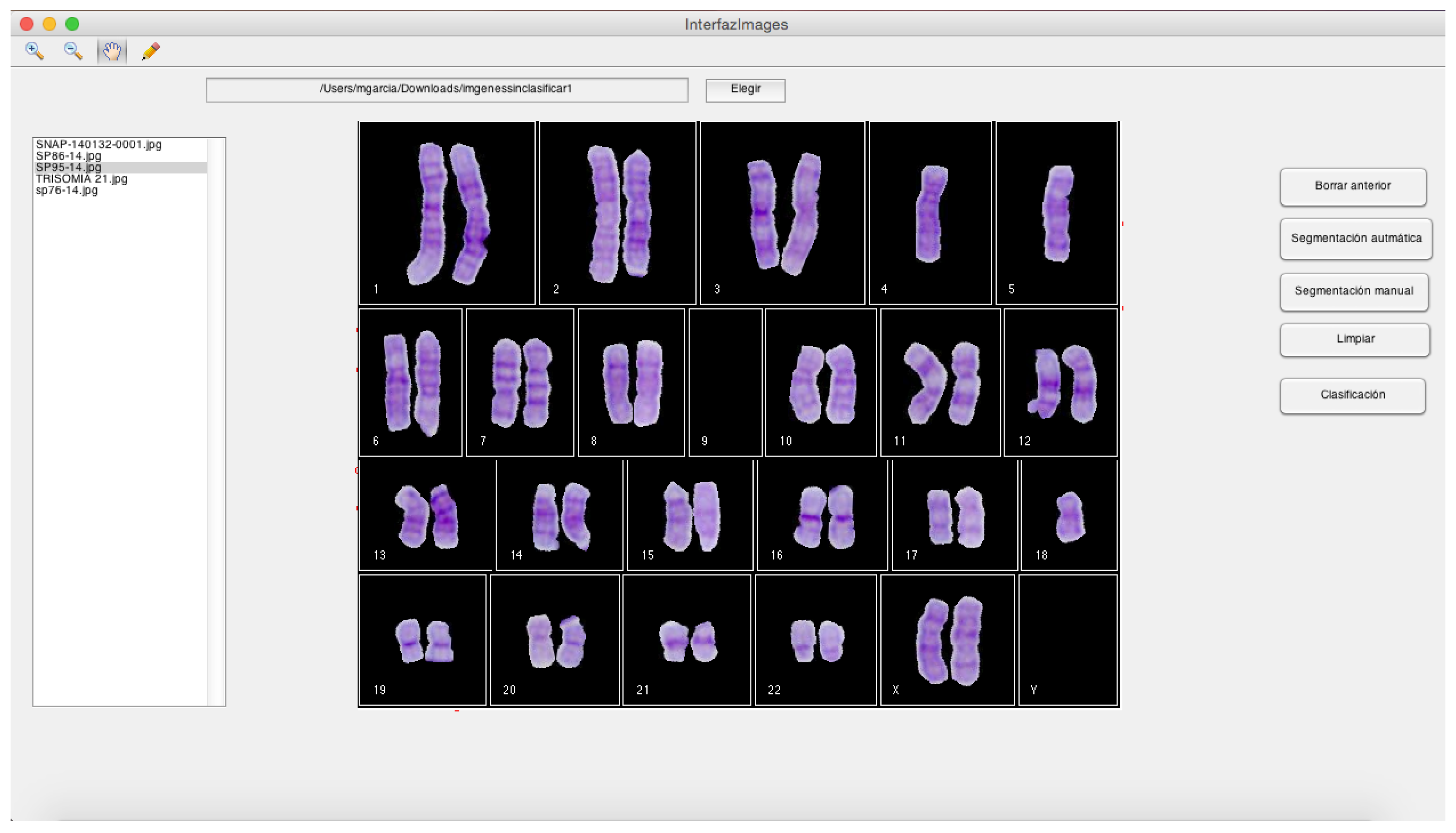
- Module C:
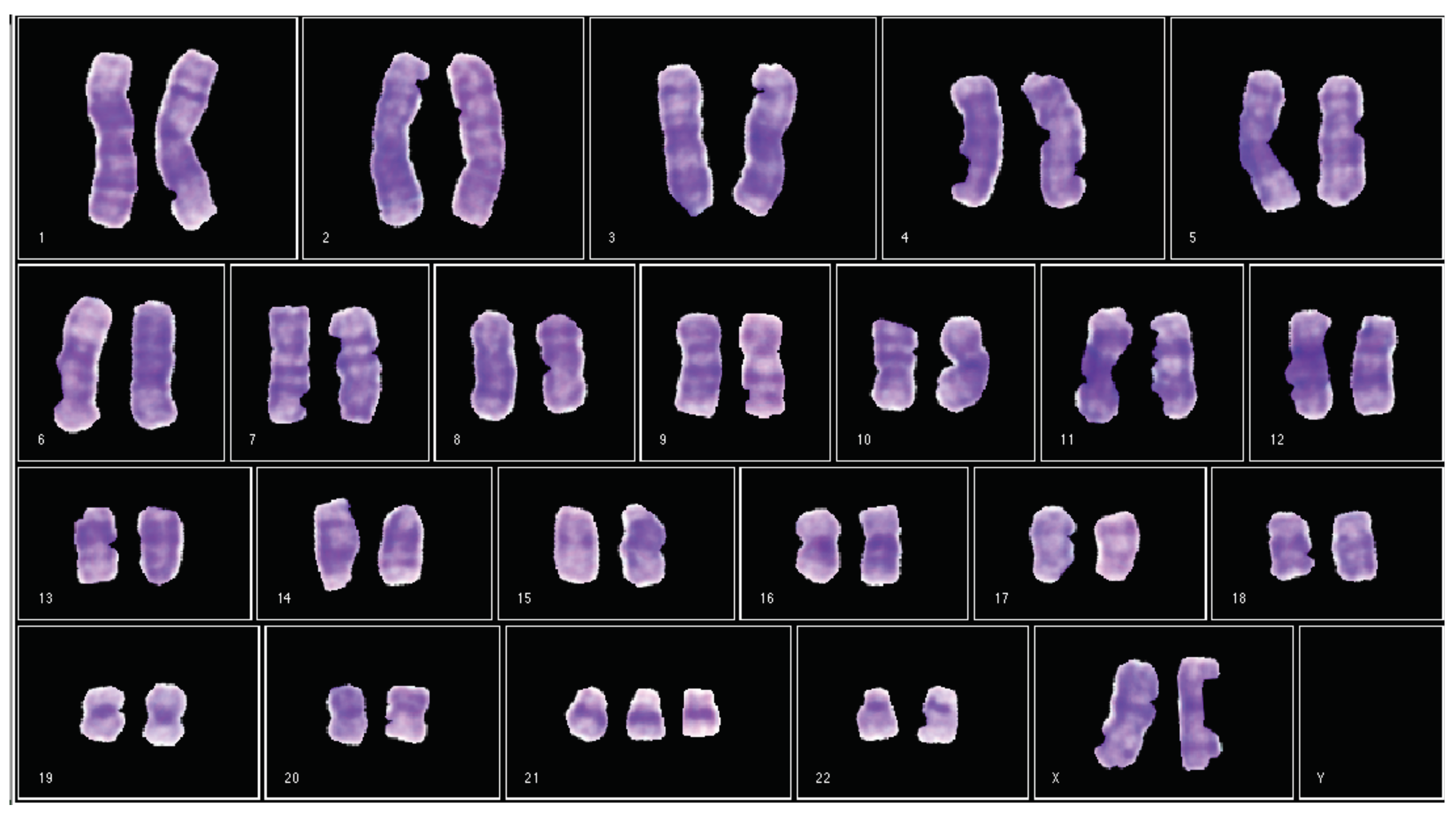
- It is a GUI that generates a karyogram using the classified chromosomes obtained in module B. This karyogram is interactive, since the module allows the user to change the chromosome polarity and its membership class. In addition, this module generates a cytogenetic report in the format defined by the cytogeneticist.
3. Results and Discussion
3.1. Experiment 1. Feature Selection
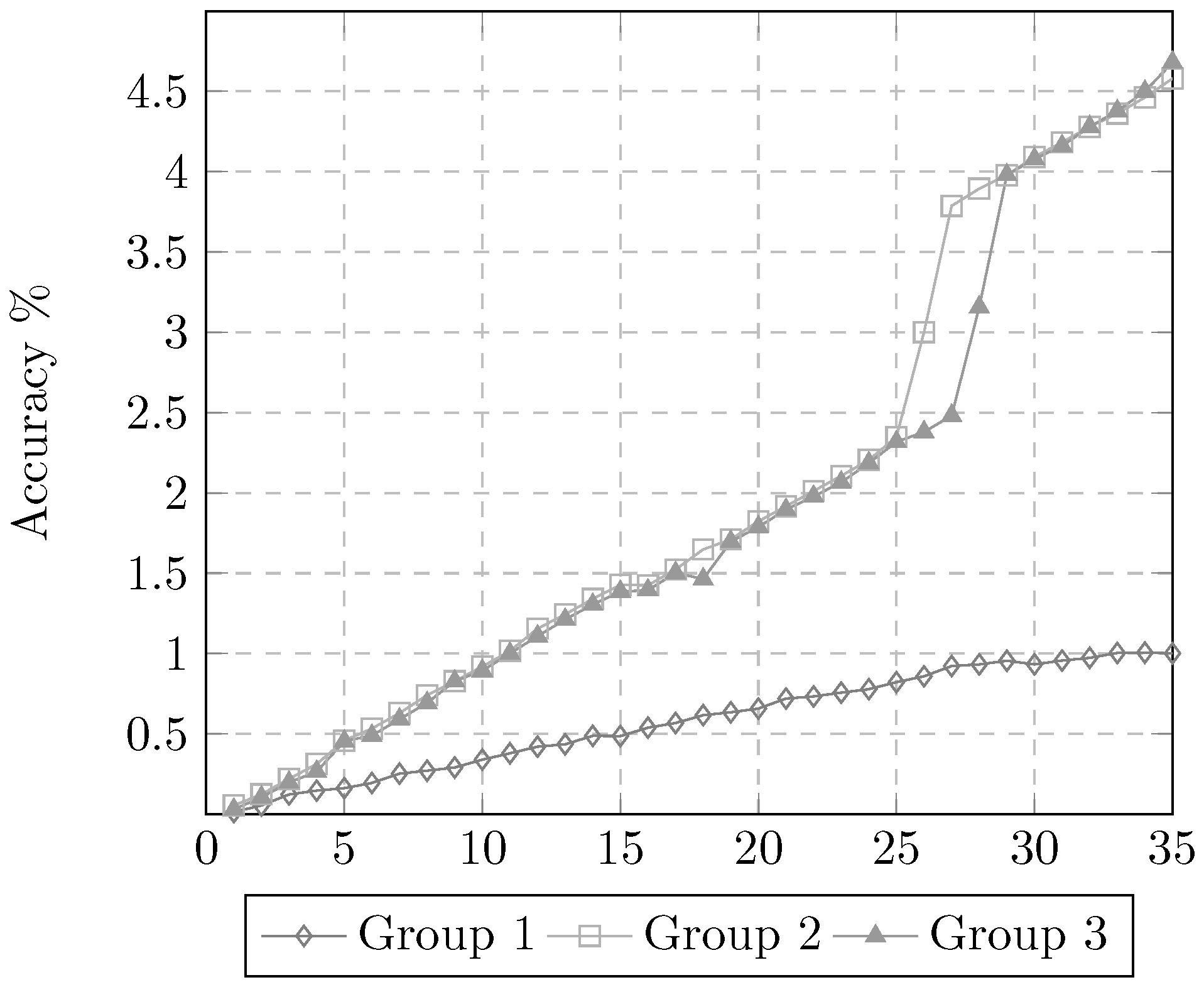
3.2. Experiment 2. Training Time
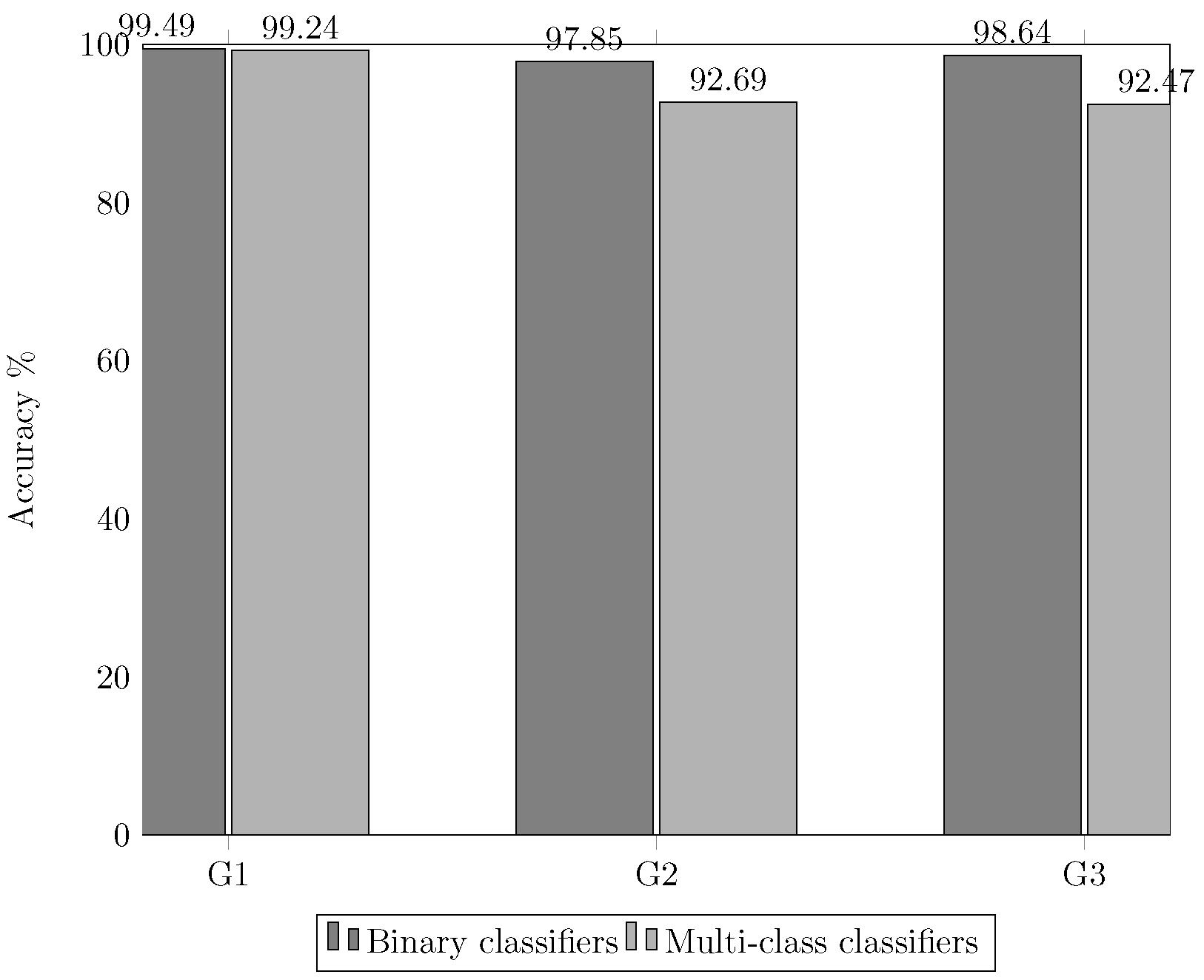
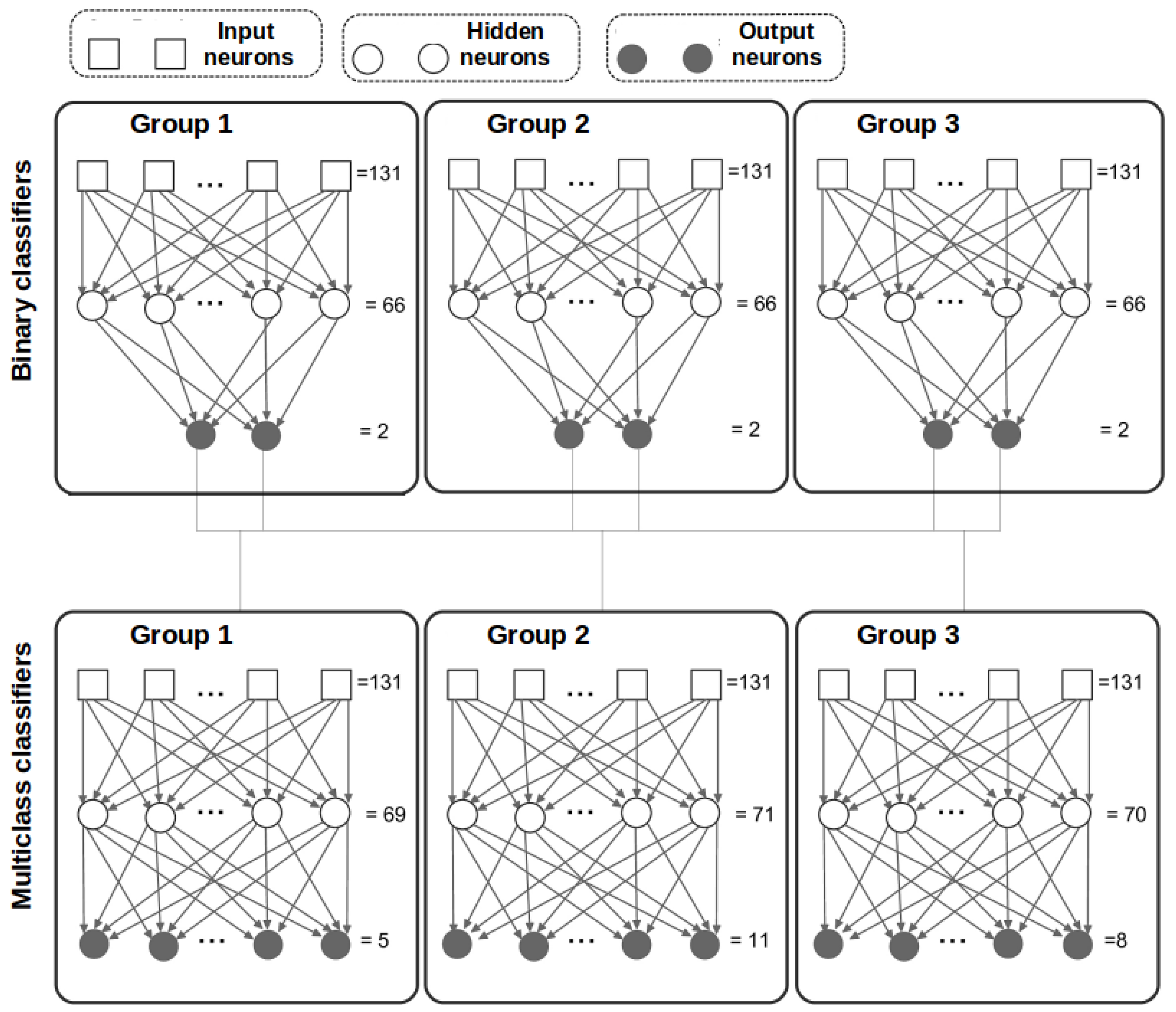
3.3. Experiment 3. Two Stage Classification
- Group 1: Chromosomes 1 to 7.
- Group 2: Chromosomes 8 to 15.
- Group 3: Chromosomes 16 to 23.
3.4. Pre-Classification Stage
- 1.
- Multi-class ( architecture). Two groups of features, and , are used to decide if the current chromosome belongs to one of the three groups.
- 2.
- Binary ( architecture). The groups of features, and , are used to decide if a chromosome:
- (a)
- Belongs to group 1 (G1) or does not belong to group 1 (NOTG1).
- (b)
- Belongs to group 2 (G2) or does not belong to group 2 (NOTG2).
- (c)
- Belongs to group 3 (G3) or does not belong to group 3 (NOTG3).
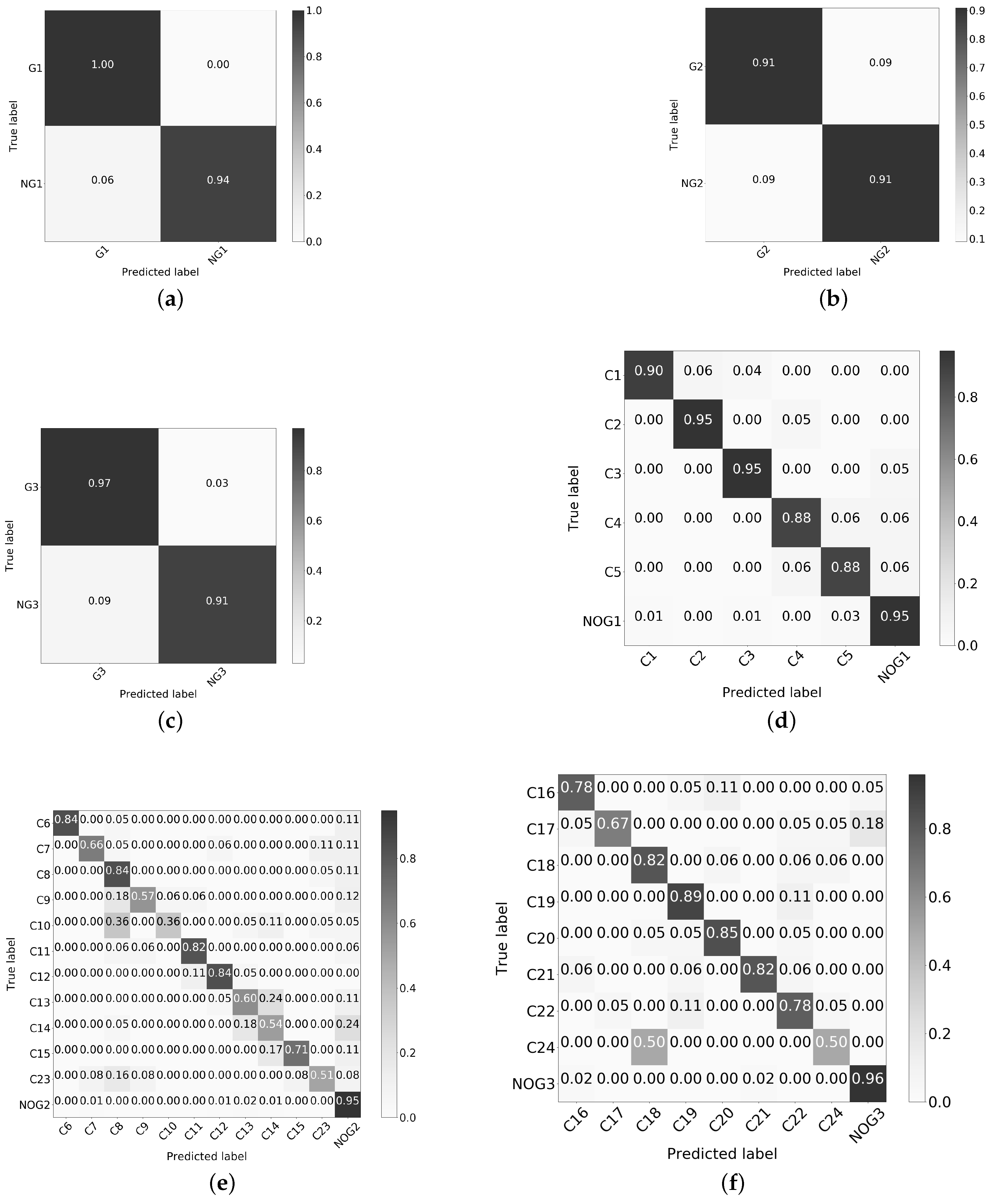
3.5. Post-Classification Stage
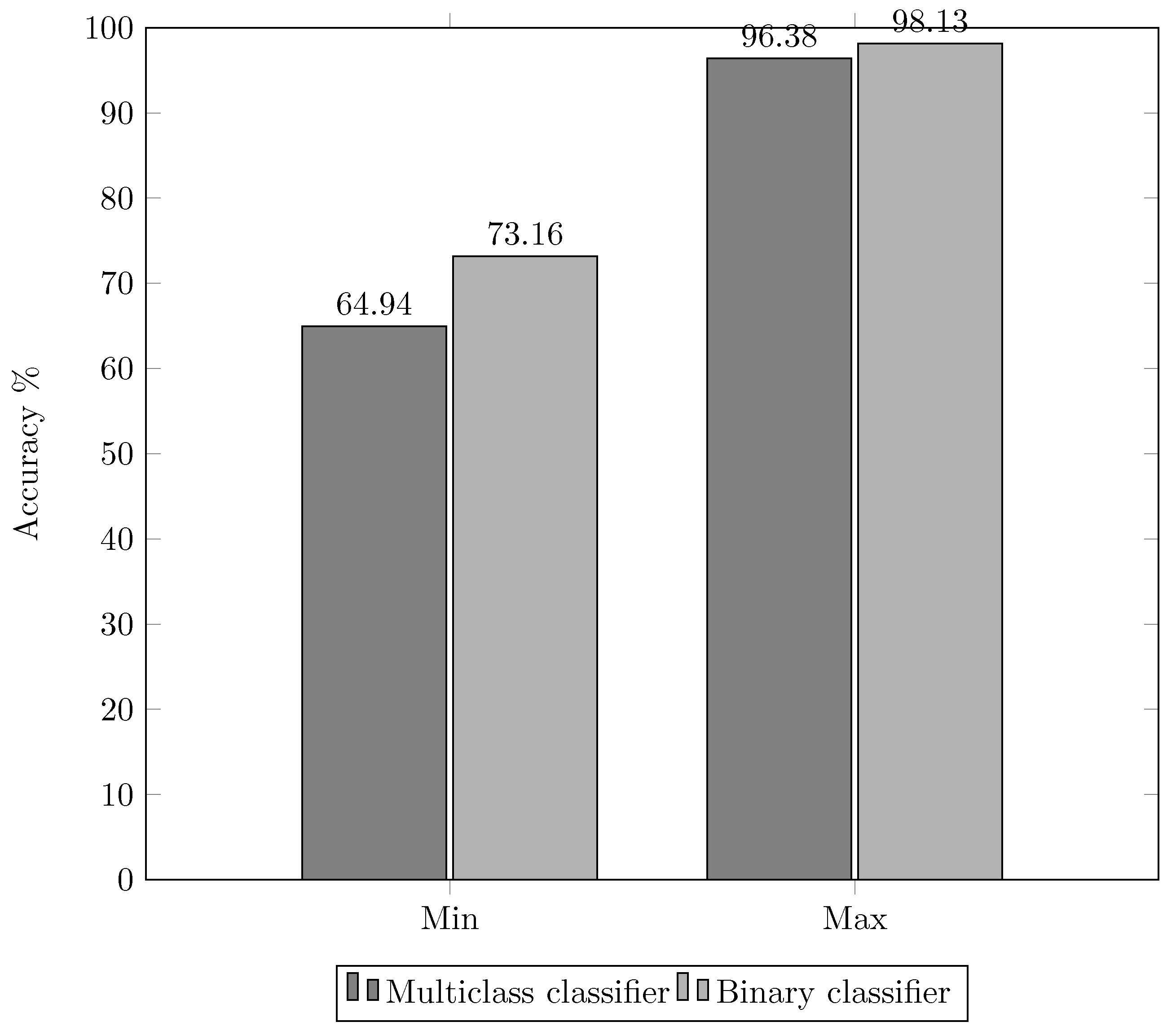
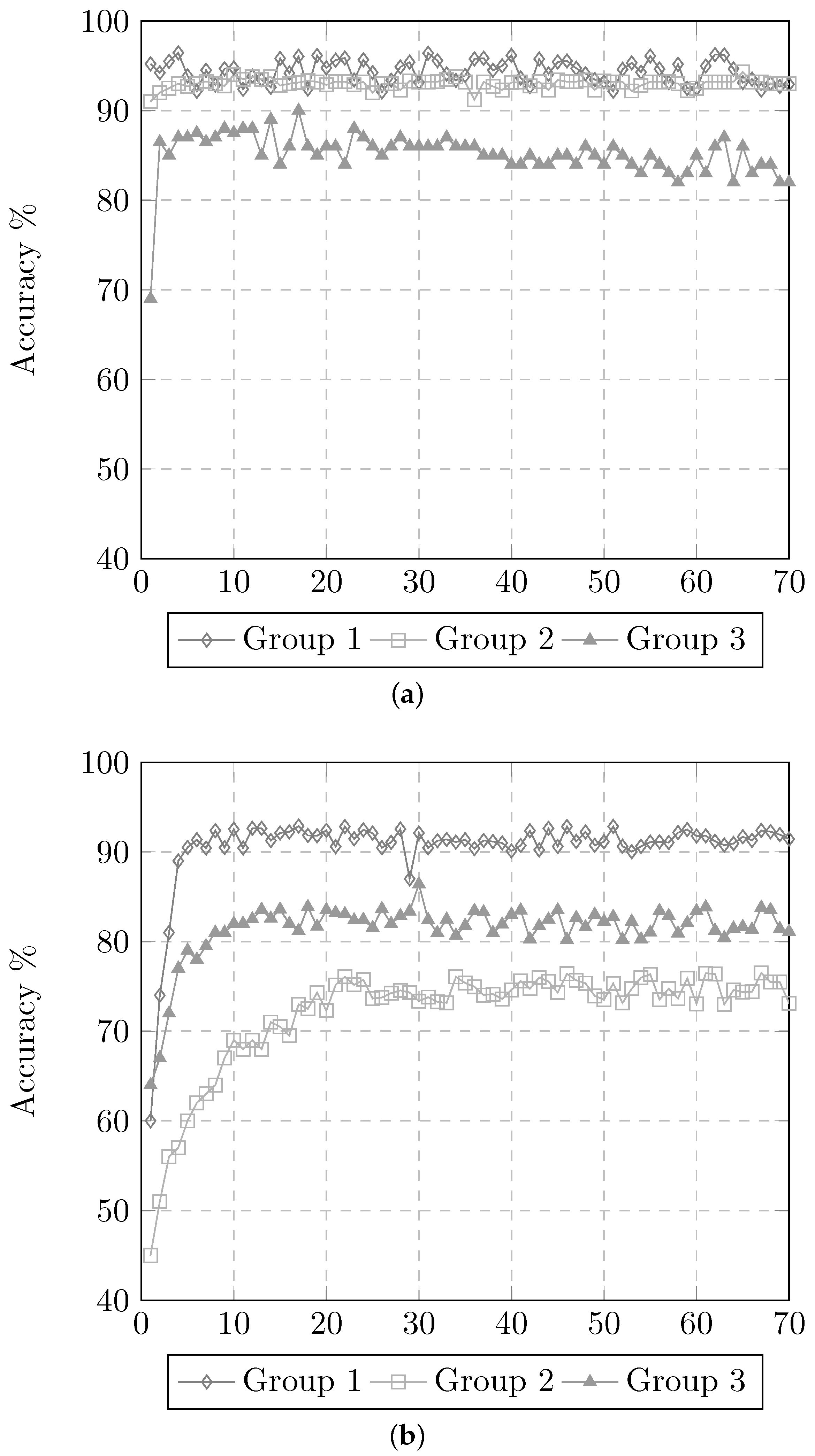
3.6. Experiment 4. Redefinition of the 2 Stage Classification
- Feature group ;
- MLP classifier; and
- (Phase 1, binary classifiers) and (Phase 2, multi-class classifiers) architectures.
3.7. Application of the Proposed Model to the G-Band Images Dataset
3.7.1. Changes in the Number of Neurons in the Hidden Layer
3.7.2. Validation of the ANN Models
3.7.3. Desktop Application for Semi-Automatic Chromosome Classification
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nair, R.M.; Remya, R.; Sabeena, K. Karyotyping Techniques of Chromosomes: A Survey. Int. J. Comput. Trends Technol. 2015, 22, 30–34. [Google Scholar] [CrossRef]
- Kannan, T.P. Cytogenetics: Past, Present And Future. Malays. J. Med. Sci. 2009, 16, 4–9. [Google Scholar] [PubMed]
- Lahmiri, S.; Shmuel, A. Performance of machine learning methods applied to structural MRI and ADAS cognitive scores in diagnosing Alzheimer’s disease. Biomed. Signal Process. Control 2019, 52, 414–419. [Google Scholar] [CrossRef]
- Lahmiri, S.; Shmuel, A. Detection of Parkinson’s disease based on voice patterns ranking and optimized support vector machine. Biomed. Signal Process. Control 2019, 49, 427–433. [Google Scholar] [CrossRef]
- Lahmiri, S.; Dawson, D.A.; Shmuel, A. Performance of machine learning methods in diagnosing Parkinson’s disease based on dysphonia measures. Biomed. Eng. Lett. 2018, 8, 29–39. [Google Scholar] [CrossRef]
- Chantrapornchai, C.; Navapanitch, S.; Choksuchat, C. Parallel Patient Karyotype Information System using Multi-threads. Appl. Med. Inform. 2015, 37, 39–48. [Google Scholar]
- Zhang, H.; Albitar, M. Computer-Assisted Karyotyping. U.S. Patent 9,336,430, 10 May 2016. [Google Scholar]
- Markou, C.; Maramis, C.; Delopoulo, A.; Daiou, C.; Lambropoulos, A. Automatic Chromosome Classification using Support Vector Machines. In Pattern Recognition: Methods and Applications; Hosny, K., de la Calleja, J., Eds.; CreateSpace Independent Publishing Platform: Scotts Valley, CA, USA, 2013; Chapter 13. [Google Scholar]
- Arora, T.; Dhir, R. A review of metaphase chromosome image selection techniques for automatic karyotype generation. Med. Biol. Eng. Comput. 2016, 54, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Silla, C.N., Jr.; Freitas, A.A. A Survey of Hierarchical Classification Across Different Application Domains. Data Min. Knowl. Discov. 2011, 22, 31–72. [Google Scholar] [CrossRef]
- Xiong, Z.; Wu, Q.; Castlemen, K.R. Enhancement, Classification And Compression Of Chromosome Images. In Proceedings of the Workshop on Genomic Signal Processing and Statistics (GENSIPS), Raleigh, NC, USA, 12–13 October 2002. [Google Scholar]
- Qiu, Y.; Song, J.; Lu, X.; Li, Y.; Zheng, B.; Li, S.; Liu, H. Feature Selection for the Automated Detection of Metaphase Chromosomes: Performance Comparison Using a Receiver Operating Characteristic Method. Anal. Cell. Pathol. 2014. [Google Scholar] [CrossRef]
- Emary, I.M.M.E. On the Application of Artificial Neural Networks in Analyzing and Classifying the Human Chromosomes. J. Comput. Sci. 2006, 2, 72–75. [Google Scholar] [CrossRef][Green Version]
- Mashadi, N.T.; Seyedin, S.A. Direct classification of human G-banded chromosome images using support vector machines. In Proceedings of the 2007 9th International Symposium on Signal Processing and Its Applications, Sharjah, United Arab Emirates, 12–15 February 2007; pp. 1–4. [Google Scholar] [CrossRef]
- Kusakci, A.O.; Gagula-Palalic, S. Human Chromosome Classification Using Competitive Support Vector Machine Teams. Southeast Eur. J. Soft Comput. 2014. [Google Scholar] [CrossRef][Green Version]
- Kou, Z.; Ji, L.; Zhang, X. Karyotyping of comparative genomic hybridization human metaphases by using support vector machines. Cytometry 2002, 47, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Hadziabdic, K. Classification of chromosomes using nearest neighbor classifier. South. Eur. J. Soft Comput. 2012. [Google Scholar] [CrossRef][Green Version]
- Sethakulvichai, W.; Manitpornsut, S.; Wiboonrat, M.; Lilakiatsakun, W.; Assawamakin, A.; Tongsima, S. Estimation of band level resolutions of human chromosome images. In Proceedings of the 2012 Ninth International Conference on Computer Science and Software Engineering (JCSSE), Bangkok, Thailand, 30 May–1 June 2012; pp. 276–282. [Google Scholar] [CrossRef]
- Shah, P. Automatic Karyotyping of Human Chromosomes Using Band Patterns. Bangladesh J. Sci. Res. 2013, 2, 154–156. [Google Scholar] [CrossRef]
- Legrand, B.; Chang, C.; Ong, S.; Neo, S.Y.; Palanisamy, N. Chromosome classification using dynamic time warping. Pattern Recognit. Lett. 2008, 29, 215–222. [Google Scholar] [CrossRef]
- Ritter, G.; Pesch, C. Polarity-free automatic classification of chromosomes. Comput. Stat. Data Anal. 2001, 35, 351–372. [Google Scholar] [CrossRef]
- Lerner, B.; Guterman, H.; Dinstein, I. A classification-driven partially occluded object segmentation (CPOOS) method with application to chromosome analysis. IEEE Trans. Signal Process. 1998, 46, 2841–2847. [Google Scholar] [CrossRef]
- Abid, F.; Hamami, L. A survey of neural network based automated systems for human chromosome classification. Artif. Intell. Rev. 2016, 49, 41–56. [Google Scholar] [CrossRef]
- Errington, P.A.; Graham, J. Application of artificial neural networks to chromosome classification. Cytometry 1993, 14, 627–639. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, B.; Li, S.; Mulvihill, J.J.; Wood, M.C.; Liu, H. Automated Classification of Metaphase Chromosomes: Optimization of an Adaptive Computerized Scheme. J. Biomed. Inform. 2009, 42, 22–31. [Google Scholar] [CrossRef]
- Poletti, E.; Grisan, E.; Ruggeri, A. A modular framework for the automatic classification of chromosomes in Q-band images. Comput. Methods Programs Biomed. 2012, 105, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Nabil, A.; Sarra, F. Q-Banding. In Reference Module in Life Sciences; Elsevier: Oxford, UK, 2017; pp. 1–3. ISBN 978-0-12-809633-8. [Google Scholar] [CrossRef]
- Yang, X.; Wen, D.; Cui, Y.; Cao, X.; Lacny, J.; Tseng, C. Computer Based Karyotyping. In Proceedings of the 2009 Third International Conference on Digital Society, Cancun, Mexico, 1–7 February 2009; pp. 310–315. [Google Scholar] [CrossRef]
- Balaji, V.S.; Vidhya, S. A novel and maximum-likelihood segmentation algorithm for touching and overlapping human chromosome images. ARPN J. Eng. Appl. Sci. 2015, 10, 2777–2781. [Google Scholar]
- Gagula-Palalic, S.; Can, M. Automatic Segmentation of Human Chromosomes. South. Eur. J. Soft Comput. 2012. [Google Scholar] [CrossRef][Green Version]
- Moradi, M.; Setarehdan, S.K.; Ghaffari, S.R. Automatic Locating the Centromere on Human Chromosome Pictures. In Proceedings of the 16th IEEE Conference on Computer-based Medical Systems, New York, NY, USA, 26–27 June 2003; IEEE Computer Society: Washington, DC, USA, 2003. CBMS’03. pp. 56–61. [Google Scholar]
- Ritter, G.; Gao, L. Automatic segmentation of metaphase cells based on global context and variant analysis. Pattern Recognit. 2008, 41, 38–55. [Google Scholar] [CrossRef]
- Kao, J.-H.; Chuang, J.-H.; Wang, T. Chromosome classification based on the band profile similarity along approximate medial axis. Pattern Recognit. 2008, 41, 77–89. [Google Scholar] [CrossRef]
- Gagula-Palalic, S.; Can, M. Extracting Gray Level Profiles of Human Chromosomes by Curve Fitting. South. Eur. J. Soft Comput. 2012. [Google Scholar] [CrossRef]
- Somasundaram, D.; Kumar, V.V. Separation of overlapped chromosomes and pairing of similar chromosomes for karyotyping analysis. Measurement 2014, 48, 274–281. [Google Scholar] [CrossRef]
- Moradi, M.; Setarehdan, S.K. New Features for Automatic Classification of Human Chromosomes: A Feasibility Study. Pattern Recognit. Lett. 2006, 27, 19–28. [Google Scholar] [CrossRef]
- Badawi, A.M.; Hassan, K.; Aly, E.; Messiha, R.A. Chromosomes classification based on neural networks, fuzzy rule based, and template matching classifiers. In Proceedings of the 2003 46th Midwest Symposium on Circuits and Systems, Cairo, Egypt, 27–30 December 2003; Volume 46, p. 383. [Google Scholar]
- Poletti, E.; Grisan, E.; Ruggeri, A. Automatic classification of chromosomes in Q-band images. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 1911–1914. [Google Scholar]
- Markou, C.; Maramis, C.; Delopoulos, A.; Daiou, C.; Lambropoulos, A. Automatic Chromosome Classification Using Support Vector Machines; iConceptPress: Hong Kong, China, 2012. [Google Scholar]
- Shaffer, L.G.; McGowan-Jordan, J.; Schmid, M. ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013); Karger Medical and Scientific Publishers: Basel, Switzerland, 2013. [Google Scholar]




| Set | Description | Normalized | |
|---|---|---|---|
 | 32 intensity values along medial axis | NO | |
 | 32 intensity values along medial axis | YES | |
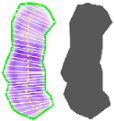 | Perimeter Area Medial axis length Intensity levels along 64 traversal lines touching the medial axis Length of 64 traversal lines touching the medial axis | YES | |
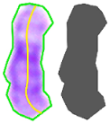 | Perimeter Area Medial axis length | YES | |
| 1 BayesNet | 27 NNge (Non-Nested generalized exemplars) |
| 2 DMNBtext (Discriminative Multinomial Naive Bayes) | 28 OneR (1-R classifier) |
| 3 NaiveBayes | 29 PART (Partial decision trees) |
| 4 NaiveBayesMultinomial | 30 Ridor (RIpple-DOwn Rule) |
| 5 NaiveBayesMultinomialUpdateable | 31 ZeroR (0-R classifier) |
| 6 NaiveBayesSimple | 32 BFTree (Best-First decision tree) |
| 7 NaiveBayesUpdateable | 33 DecisionStump |
| 8 IB1 (Instance-based classifier) | 34 FT (Functional Trees) |
| 9 KStar | 35 J48 (C4.5 Decision Tree) |
| 10 LWL (Locally Weighted Learning) | 36 J48graft (Decision Tree Grafting) |
| 11 AdaBoostM1 | 37 LADTree (LogitBoost Alternating Decision Tree ) |
| 12 AttributeSelectedClassifier | 38 RandomForest |
| 13 Bagging | 39 RandomTree |
| 14 ClassificationViaClustering | 40 REPTree (Reduced-Error Pruning) |
| 15 ClassificationViaRegression | 41 SimpleCart |
| 16 CVParameterSelection (Cross-Validation) | 42 Logistic |
| 17 END (Ensembles of Balanced Nested Dichotomies) | 43 MultilayerPerceptron |
| 18 FilteredClassifier | 44 RBFNetwork (Radial Basis Function Network) |
| 19 Grading | 45 SimpleLogistic |
| 20 LogitBoost (Additive Logistic Regression) | 46 SMO (Sequential Minimal Optimization) |
| 21 MultiBoostAB (Ada Boost) | 47 DTNB (Decision Table/Naive Bayes hybrid) |
| 22 MultiClassClassifier | 48 Dagging |
| 23 MultiScheme | 49 Decorate (Diverse Ensemble Creation) |
| 24 HyperPipes | 50 LMT (Logistic Model Trees) |
| 25 VFI (Voting Feature Intervals) | 51 NBTree (Naive-Bayes Tree hybrid) |
| 26 JRip (Repeated Incremental Pruning) |
| 1 IB1 (Instance-based classifier) |
| 2 KStar |
| 3 Random Forest |
| 4 Multilayer perceptron |
| 5 SMO (Sequential Minimal Optimization) |
| Set | Type | Outputs | Description |
|---|---|---|---|
| Multiclass | 24 | Classify chromosomes into 24 classes | |
| Multiclass | 3 | Classify chromosomes into 3 groups | |
| G1: Chromosomes from 1 to 7 | |||
| G2: Chromosomes from 8 to 15 | |||
| G3: Chromosomes from 16 to 24 | |||
| Binary | 6 | 3 binary classifiers are included: | |
| BG1: Chromosomes belongs to G1 or not | |||
| BG2: Chromosomes belongs to G2 or not | |||
| BG3: Chromosomes belongs to G3 or not | |||
| Multiclass | 24 | 3 multiclass classifiers are included: | |
| MG1: Chromosomes from 1 to 7 | |||
| MG2: Chromosomes from 8 to 15 | |||
| MG3: Chromosomes from 16 to 24 |
| New Class | Chromosomes | |
|---|---|---|
| Model 1 | Model 2 | |
| Group 1 | 1 to 7 | 1 to 5 |
| Group 2 | 8 to 15 | 6 to 15 and 23 |
| Group 3 | 16 to 24 | 16 to 22 and 24 |
| Original Configuration | Modified Configuration | ||||
|---|---|---|---|---|---|
| Binary Classification | |||||
| Group | Neurons | Accuracy | Neurons | Accuracy | |
| 1 | 66 | 95.26 | 4 | 96.44 | |
| 2 | 66 | 90.56 | 20 | 90.29 | |
| 3 | 66 | 93.60 | 34 | 93.60 | |
| Multiclass Classification | |||||
| 1 | 68 | 91.12 | 17 | 92.89 | |
| 2 | 71 | 77.08 | 67 | 79.24 | |
| 3 | 70 | 84.40 | 30 | 86.40 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Mier, Y.; Nuño-Maganda, M.A.; Polanco-Martagón, S.; García-Chávez, M.d.R. Machine Learning Classifiers Evaluation for Automatic Karyogram Generation from G-Banded Metaphase Images. Appl. Sci. 2020, 10, 2758. https://doi.org/10.3390/app10082758
Hernández-Mier Y, Nuño-Maganda MA, Polanco-Martagón S, García-Chávez MdR. Machine Learning Classifiers Evaluation for Automatic Karyogram Generation from G-Banded Metaphase Images. Applied Sciences. 2020; 10(8):2758. https://doi.org/10.3390/app10082758
Chicago/Turabian StyleHernández-Mier, Yahir, Marco Aurelio Nuño-Maganda, Said Polanco-Martagón, and María del Refugio García-Chávez. 2020. "Machine Learning Classifiers Evaluation for Automatic Karyogram Generation from G-Banded Metaphase Images" Applied Sciences 10, no. 8: 2758. https://doi.org/10.3390/app10082758
APA StyleHernández-Mier, Y., Nuño-Maganda, M. A., Polanco-Martagón, S., & García-Chávez, M. d. R. (2020). Machine Learning Classifiers Evaluation for Automatic Karyogram Generation from G-Banded Metaphase Images. Applied Sciences, 10(8), 2758. https://doi.org/10.3390/app10082758





