1. Introduction
Anal squamous cell carcinoma (ASCC) is a relatively rare disease, accounting for approximately 2.6% of all digestive cancers, with around 27,000 estimated cases worldwide in 2008 [
1]. Its incidence is however rising, mainly in high-income countries including the USA, France, Australia and the UK, possibly due to changes in environmental risk factors [
2]. The main known risk factor is human papilloma virus (HPV) infection, in particular HPV16, as well as usually correlated risk factors such as sexual behavior, concomitant human immune-deficiency virus infection via immunosuppression allowing HPV replication, and probably tobacco smoking. Women and patients older than 65 years old are more at risk [
3].
With an indolent natural history and low rate of distant metastases at diagnosis, ASCC is usually amenable to loco-regional treatment. During recent decades, the standard of care for non-metastatic disease has evolved from non-conservative surgery, namely abdomino-perineal resection (APR), to concomitant chemo-radiotherapy (CRT), based on the results of phase III trials performed in the 1990s [
4,
5]. This is because, apart from modest improvements such as better chemotherapy management [
6,
7] and modern intensity-modulated radiation therapy (IMRT) techniques, no major therapeutic progress has been made. Currently, the standard of care for non-metastatic ASCC thus relies on a combination of mitomycin and 5-fluorouracil-based chemotherapy (CT) and up to 59.4 Gy radiation doses to the tumor volume, with salvage APR saved in case of loco-regional relapse. This association allows for organ preservation and achieves good curative results with around 80% 5-year overall survival (OS) and excellent local control (LC) for T1-T2, N0 localized tumors. However, cure rates are lower for more advanced cases (T3-T4 or N-positive), with up to 30%–40% loco-regional or metastatic relapses, justifying the need to explore ways to improve these outcomes [
8,
9].
Numerous trials have explored different hypotheses, for example, RT dose escalation to more than 60 Gy or neo-adjuvant CT in the ACCORD 03 trial [
10], but all failed to demonstrate efficacy on unselected non-metastatic patients. Recent efforts aim to escalate the treatment by adding targeted therapies, epidermal growth factor receptor (EGFR) inhibitors, to classic CRT, but proof of efficacy is still lacking [
11,
12,
13]. One of the foreseeable solutions is to better identify patient subgroups, in order to individualize the treatment by escalating or de-escalating it following estimated risk stratification. To this matter, large trials are being conducted like the PLATO trial (ISRCTN88455282), that aims to personalize the RT dose in three separate groups for low, intermediate and high-risk disease (ACT3, ACT4 and ACT5).
To this day, there is no reliable way to predict which patients will experience disease recurrence following CRT. This ability to foretell response to CRT at baseline would be of significant clinical benefit, as it would allow personalized treatment and adjusted follow-up. It is trusted that novel areas of study, like radiomics and machine learning (ML), could be of critical help in better identifying upstream which patients would benefit most from treatment adjustment, justifying this critical review of the literature [
14].
2. Materials and Methods
The authors conducted a literature review in February 2020 using PubMed/Medline, Scopus and Google Scholar. The terms ‘machine learning’, ‘radiomics’, ‘anal squamous cell cancer’ and/or ‘prediction’ were included, as well as other associated technical ML keywords. Articles were selected based on relevance to the subject and reference lists of said articles were hand searched in order to investigate novel articles. Selected articles were published between 2010 and 2020, with a surge in the 2018–2020 time period. Several articles were excluded for the following reasons: no mention of radiomics or machine learning techniques, main focus on rectal cancers, no description of methodology.
3. Radiomics and ML as Powerful Clinical Tools
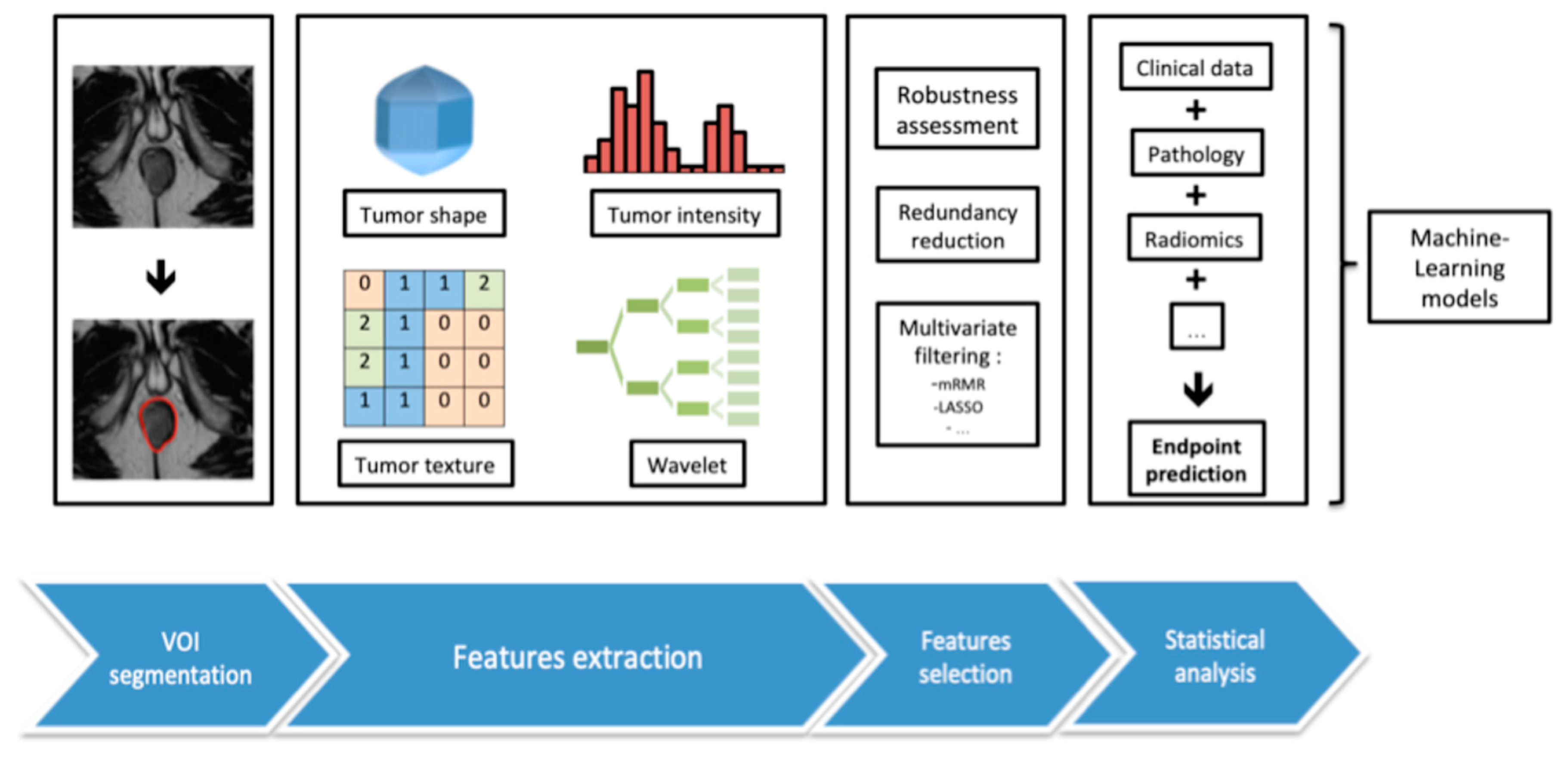
Radiomics is a relatively recent area of study, defined by the use of data-characterization algorithms and mathematical tools to extract large amounts of features from manually or automatically segmented volumes of radiographic medical images (magnetic resonance imaging (MRI), positron emission tomography-computed tomography (PET-CT), CT). These features, mostly inaccessible to the human eye, can be classified into different categories: first-order features describing voxels’ intensities or spatial distribution, second-order features comparing relationships between adjacent voxels and third-order features exploring relationships between more than two voxels.
Several steps are conducted during the radiomic process, each bearing their own specifications, including image acquisition, data standardization, segmentation, features extraction and qualification, feature selection by stability and filtering, and finally, exploration of association with a selected clinical or paraclinical endpoint (
Figure 1). These techniques already display promising results in the field of oncology, for a wide range of diseases [
15,
16]. Nevertheless, several limitations prevent radiomic tools to be extensively used in clinical routine, mostly due to the lack of technical standardization and need of validation on independent cohorts.
ML is a subset of artificial intelligence in which an algorithm, supervised with labeled data or unsupervised, learns by pattern-recognition and inference from a dataset. ML algorithms are already used in a wide range of applications, and offer significant hopes in the medical field. The goal is to produce a model capable of prediction, prediction endpoints including but not limited to diagnosis, prognosis, treatment decision, evaluation of efficacy and management [
17,
18,
19,
20]. As such techniques can simultaneously encompass a large number of variables, they could surpass human decision-making abilities and lead to better patient care as well as to the discovery of hitherto unused parameters [
21]. In fact, similar detection rates, or even those that surpass experts, have already been achieved by ML for some applications [
22,
23,
24]. Conventional ML techniques include logical regression algorithms, support vector machine (SVM), decisional trees (DT) and Bayesian methods such as naive Bayes classifiers and Bayesian networks. These algorithms can be combined in ensemble learning methods, such as random decision forests (RF), designed to obtain better predictive performance over any of their constituent learning algorithms alone. The development of deep neural networks (DNN), using multiple layers of connected perceptrons, is a more recent step in ML and they are some of the most popular network architectures in use today. While conventional ML techniques require, as input, manually extracted engineered features that quantify predictive information of an image, DNN are able to directly learn such features from the data without specifying them as input. The feature extraction stage thus vanishes, merged with the classification step of the algorithm into a single stage, in which a hierarchical representation is subsequently constructed over the layers of the network. The common limitations for ML techniques lie in the lack of data quality (missing data, duplicated data, labeling errors, etc.) and need of a large quantity of input, in order to better perform and avoid a loss of robustness (especially in the field of radiomics with the large number of features extracted). Each ML method holds respective strengths and weaknesses, taken into account during the selection process for the right approach, depending on the desired outcome.
Typically, the development of new ML tools relies on the separation of the original dataset into two independent cohorts: one used by the ML algorithm to learn (training set) and the other used to evaluate the performances on a separate population (testing set). To reduce variability in the evaluation, these two sets may be iteratively rotated over the whole dataset to perform a cross-validation. Moreover, each ML model, as well as feature extraction methods, display hyperparameters (for example the number of layers for DNN, number of decisional trees for RF), tuned in order to enhance performance. The optimal values for those parameters should not be determined based on the whole test set performance, as this amounts to a form of overfitting on the test set (data leakage). Hence, a sizable part of the training set is split off to validate the model’s performance for a certain set of hyperparameters in a third set, called the validation set. Prediction performance is then evaluated based on several indicators on the testing set; mostly sensitivity, specificity, accuracy of the prediction and area under the curve of the receiver-operating characteristic curve (AUC ROC), used to optimize the algorithm.
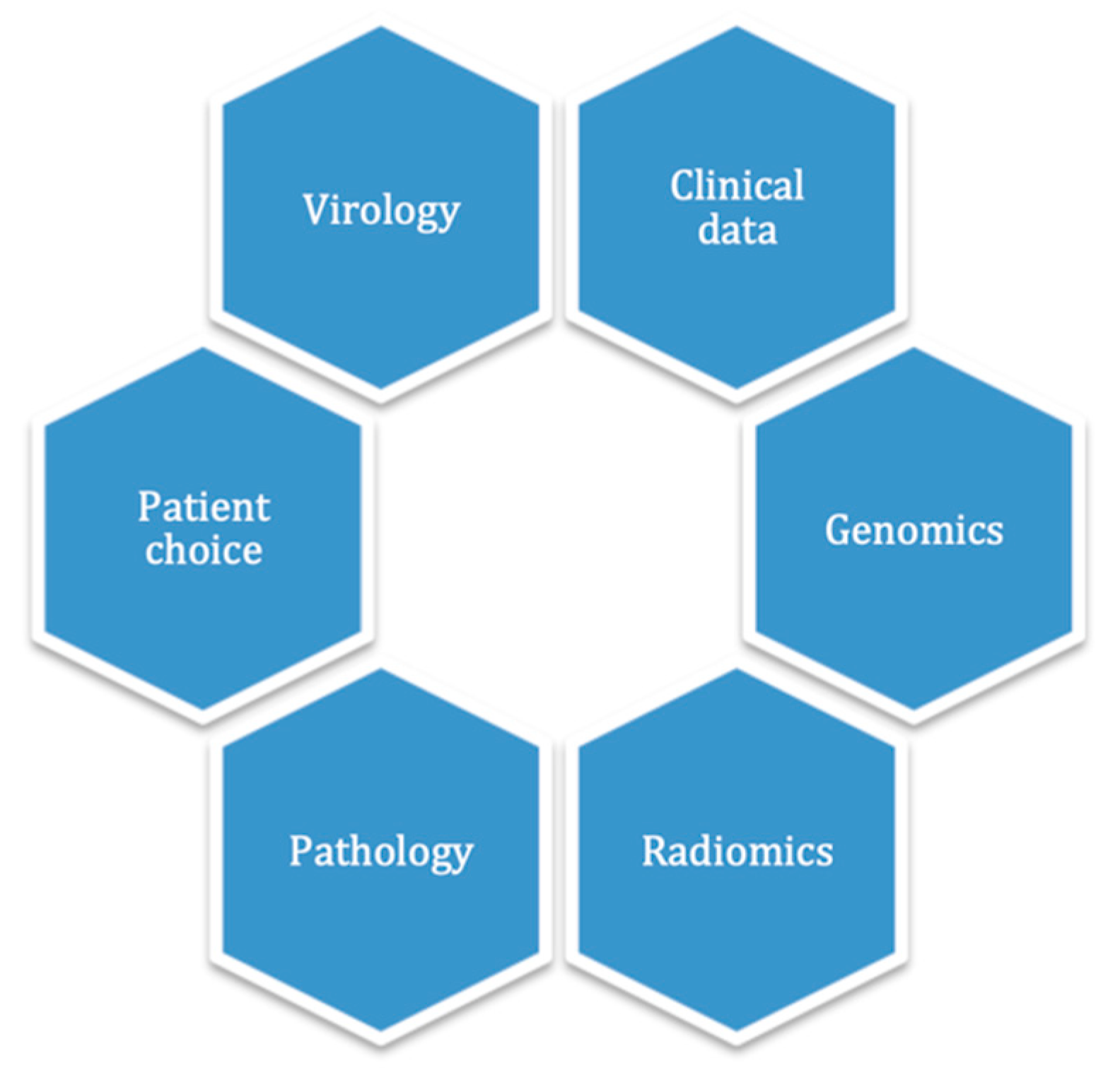
In the medical field, information originates from a large variety of sources, such as clinical, biology, genomics, radiology and radiomics, pathology, metabolomics and proteomics [
14,
25]. Theoretically, all of this data could be centered in a unique ML clinical decision support system, to allow individual patient-centered decision-making (
Figure 2).
4. Radiomics and ML in ASCC
In ASCC, physical examination remains the cornerstone of local response assessment. Imaging however also bears an important role in pre-therapeutic and follow-up modalities, MRI and PET-CT being the most informative imaging techniques available [
26,
27]. Post-therapeutic clinical and imaging assessments determine treatment response, histologic proof being only required for doubtful cases. Thus, they act as the gold-standard to which ML tools predictions are compared. Radiomics have been tested for both MRI and PET-CT for ASCC, and have the potential to uncover novel clinical information parameters (
Table 1).
4.1. MRI
Disregarding radiomics, MRI analysis struggles to provide valuable prognostic help [
28]. Nonetheless, it is worth noting that the use of MRI-determined tumor regression grading (TRG) to predict local relapse at three and six months post-CRT has been associated with an almost 100% negative predictive value for TRG 1/2 scores at three and six months, and almost 100% positive predictive value for TRG 4/5 at six months [
29].
Three published studies have explored various complexity levels of MRI radiomics in ASCC. In a monocentric retrospective study, conducted by Hocquelet et al., 28 non-metastatic ASCC patients were included and pre-CRT tumor volumes were manually segmented on axial T2-weighted (T2w) sequences. First-order radiomics features and second-order statistical features derived from the grey-level co-occurrence matrix (GLCM) were performed, using an in-house ITK library-based Python script. Inter-observer variability in manual segmentation was assessed by case-to-case consensus between 2 experts, and all MRI acquisitions were performed on the same 1.5-T machine, thus limiting results extrapolation to other centers. After adjusting for age, gender and tumor grade, two radiomics features were associated with event (disease progression or death) occurrence: skewness (HR = 0.131,
p = 0.005) and cluster shade_d1 (HR = 0.601,
p = 0.027). The corresponding Harrell C-indices were respectively 0.846 and 0.851 [
30].
Similarly, in a retrospective study published by Owczarczyk et al. in 2019, pre and post-CRT (median of 15 weeks from the start of radiotherapy with 90% of scans performed within 12 weeks of completion of treatment) MRI heterogeneity was evaluated. T2w but also functional diffusion-weighted (DW) imaging sequences were analyzed to predict post-treatment recurrence (locoregional or metastatic). Three 1.5-T MRI machines were used, all with the same acquisition protocol, and 40 patients were included. The tumor volume was manually delineated on all slices of axial T2w and ADC parametric maps on pre and post-CRT acquisitions, generating four separate whole tumor 3D volumes-of-interest (VOI) per case. Tumor maximum size, volume, extent and TNM stage were recorded by a consensus of two experts. Tumor volume and seventy-eight first-order, second-order (based on GLCM statistics) and fractal features were extracted from the VOI using in-house software. A ML random forest method was used to select variables with highest discriminatory value in predicting the two selected outcomes: disease recurrence and two-year disease-free survival (DFS). A baseline multivariate clinical-only model (age, gender, T stage, N stage) and an extended multiparametric model (addition of top performing imaging features) were developed and compared. Two radiomics features were found to be of high informative value: baseline T2w “energy” and DWI “coefficient of variation” appeared to be predictive of CRT outcome, independently of clinical characteristics alone. The addition of these two imaging features to multivariate logistic regression models based on clinical characteristics yielded an increase in the predictive accuracy for both endpoints when using C-statistic and net reclassification improvement (
p < 0.001) algorithms. This extended model demonstrated 34.8% error reduction beyond baseline clinical model in terms of disease recurrence prediction and 18.1% error reduction in terms of 2y-DFS post CRT in an independent cross validation analysis [
31].
Finally, in a recent prospective study led by Jones et al., 25 patients with non-metastatic ASCC underwent multiparametric 3-T MRI incorporating diffusion-weighted magnetic resonance imaging (DW-MRI) and dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) sequences at baseline, weeks two and four of treatment, and eight weeks after the end of standard CRT. Standard radiomics and delta-radiomics, a measure of change in radiomics between several imagings, were performed on manually segmented volumes, using only first-order statistical-based features (histogram analysis of the multi-parametric maps). Local recurrence was correlated with a few apparent diffusion coefficient (ADC) metrics extracted from DW-MRI: baseline skewness (
p = 0.04, ROC AUC 0.90) and standard deviation (SD) (
p = 0.02, ROC AUC 0.90), week two skewness (
p = 0.02, ROC AUC 0.91) and SD (
p = 0.01, ROC AUC 0.94), week four kurtosis (
p = 0.01, AUC 0.92) and SD (
p = 0.01, ROC AUC 0.96). Delta-radiomics changes in minimum ADC between baseline and week two (
p = 0.02, ROC AUC 0.94), and between baseline and week four (
p = 0.02, ROC AUC 0.94) were also prognostic of local recurrence. K-trans min at second follow-up (
p = 0.05, AUC 0.84) was the only DCE-MRI feature associated with local recurrence. For any recurrence, minimum ADC (
p = 0.02, ROC AUC 0.87) and SD (
p = 0.01, ROC AUC 0.85) at baseline, and maximum ADC (
p = 0.03, ROC AUC 0.77) and SD (
p = 0.02, ROC AUC 0.81) at week four were found to be of interest. After least absolute shrinkage and selection operator (LASSO) logistic regression was performed, minimum ADC and SD at baseline were retained for any recurrence. No other machine-learning algorithm was tested. The authors argue that use of higher-order radiomics is warranted and could further investigate the highlighted associations with disease recurrence [
32].
4.2. PET-CT
For PET-CT, several functional parameters, alone or in combination with clinical parameters, have previously been linked to recurrence. For instance, in a monocentric trial, Rusten et al. included 93 ASCC patients from the prospective ANCARAD trial (NCT01937780) who benefited from baseline PET-CT imaging (and at two weeks of treatment for 39 patients). Alongside available clinical data, several PET parameters were investigated: standard uptake volume (SUV) max/peak/mean, metabolic tumor volume (MTV), total lesion glycolysis (TLG) and a proposed Z-normalized combination of MTV and SUV peaks (ZMP). In the bivariate analysis, HPV status was the most independent predictor in combinations with N3 stage, ZMP, TLG, and MTV (
p < 0.02). The 18-F-fluoro-2-deoxyglucose (FDG)-PET parameters at two weeks into radiotherapy decreased by 30%–40% of the initial values, but this decrease failed to improve the prediction models, thus questioning the utility of an intermediary PET-CT after two weeks of treatment when using classical PET parameters [
33].
Jones et al. prospectively included 19 ASCC patients and performed PET-CT assessments before and 12 weeks following CRT. Six VOI were extracted and analyzed: five based on SUV (maximum, mean, median, standard deviation and peak), MTV and TLG. Exact logistic regression and ROC AUC analyses were completed. Two PET-CT parameters were found to be associated with recurrence: MTV bounded by a threshold of 41% maximum SUV on the pre-CRT PET-CT predicted for any recurrence (
p = 0.03, ROC AUC 0.89), and median SUV within a VOI bounded by an SUV of three on post-CRT PET-CT correlated with local recurrence (
p < 0.01, ROC AUC 1.00 with a median SUV threshold of 3.38) [
34].
The only found research using radiomics for PET-CT in ASCC patients was a monocentric retrospective trial trying to predict PFS, that included 189 non-metastatic ASCC patients [
35]. A single operator semi-autonomously segmented the primary tumor and associated lymph nodes on baseline PET-CT, and voxels with an SUV greater than 1.5 times the mean liver SUV were included in the final VOI. First, second and third-order features were extracted using the LifeX software [
36]. Elastic net regularization and feature selection were used for logistic regression model generation on a randomly selected training cohort and applied to a validation cohort. Three models were created and evaluated using ROC-AUC analysis: a clinical prognostic factors model (age, sex, tumor and nodal stage), a radiomics model and a combined model. GLCM “entropy”, neighborhood grey-level different matrix (NGLDM) “busyness”, minimum CT value (lowest Hounsfield unit within the lesion) and standardized MTV were selected for inclusion in the prognostic model, alongside tumor and nodal stage. The combined model performed best for PFS prediction comparatively with the clinical and radiomics model, with AUCs of respectively 0.738, 0.602 and 0.660 on the validation set. This highlights the usefulness of combining both imaging and clinical parameters for increased prediction performance.
5. Discussion
Male gender, N-positive stage and tumor length greater than five cm are recognized as ASCC clinical prognostic factors, associated with worse clinical outcomes [
37,
38]. A few other prognostic factors have been suggested throughout the years, like the baseline neutrophil to lymphocyte ratio for LR recurrence [
39], or an age greater than 55, increased circumferential tumor spread, skin ulceration, inguinal node development and a total RT dose of more than 60 Gy for worse colostomy-free survival [
40]. Nomograms have even been created with promising results in predicting cancer specific survival and overall survival as well as risk-stratifying patients, but were only based on clinical data [
41].
Yet, radiomics features extracted from medical imaging have the potential to bring additional informative data, as showcased above in both PET and MRI, even if used alone. Adding these novel parameters to previously used clinical parameters seems to offer additive performance, paving the way to build more accurate predictive models.
Given the potentially high number of radiomics features created on top of a rising number of clinical variables, powerful algorithms are needed to encompass and make all available data flourish. This is where ML algorithms thrive and have already shown tremendous results for a number of malignancies [
42,
43,
44], but have, for now, barely been explored in ASCC. As an exception, using various ML algorithms including random forest and J48 decision trees, De Bari et al. created a model predicting inguinal relapse with respective sensitivity, specificity and accuracy of 86.4%, 50% and 83.1% on the validation dataset (and superior results compared to logistic regression), highlighting the potential of such algorithms for ASCC care [
45].
However, the available data are still sparse, as all published studies remain exploratory and the created models are unfit for extensive clinical use. Indeed, several limitations curb the expansion of radiomics: inter-observer segmentation variability, lack of a harmonized process for image normalization and acquisition (especially for MRI with high signal inconsistencies), as well as the absence of standardized features’ extraction methods. As for ML, large ASCC patients’ cohorts are needed in order to fulfill their prerequisites and allow complex ML algorithms to prove superiority over simpler predictive models such as standard logistic regression, but are difficult to gather, given the relative rarity of the disease.
6. Conclusions
As the standard of care in non-metastatic ASCC remains unchanged since the 1990s, with stalling 30%–40% recurrence rates for locally advanced stages, along with not uncommon treatment-induced side effects, being able to predict treatment outcome is of paramount clinical interest. Recent research is still struggling to bring alternatives to the fore, although hypotheses such as EGFR inhibitors, immunotherapy or RT dose adjustment are under investigation.
Radiomics have the potential to increase prognostic information and help distinguish risk groups, but are confronted with previously described arduously movable barriers. Ideally, these novel parameters can be integrated alongside all other medical information (such as clinical and environmental data, virology, pathology and biology) in a common clinical individualized decision-making model, used for precision medicine. Given the high number of parameters, ML algorithms should be the best suited to fulfill this task, and are bound to ineluctably evolve in the upcoming years.
Yet, for this to work, there is a crucial need for a high number of quality clinical data, preferably multicentric and prospective. A few research projects are underway in this matter, for example collecting imaging data from national trials such as the French phase I-II multicenter FFCD-0904 (NCT01581840), which tested panitumumab, an anti-EGFR targeted therapy, in addition to standard RCT, to identify and predict which patients would benefit most from this association, or using prospective data from the English PLATO trial (ISRCTN 88455282) [
31]. Joint efforts are warranted, and this should involve international or national collaboration, such as the French ANABASE cohort, vowing to nationally congregate all ASCC cases [
46].
Author Contributions
Conceptualization, N.G. and P.S.; methodology, N.G.; validation, P.S., N.L., O.S., T.V. and V.V.; writing—original draft preparation, N.G.; writing—review and editing, P.S., N.L., O.S., T.V. and V.V.; supervision, P.S. and V.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Islami, F.; Ferlay, J.; Lortet-Tieulent, J.; Bray, F.; Jemal, A. International trends in anal cancer incidence rates. Int. J. Epidemiol. 2017, 46, 924–938. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, A.R.; Nyitray, A.G.; Kreimer, A.R.; Pierce Campbell, C.M.; Goodman, M.T.; Sudenga, S.L.; Monsonego, J.; Franceschi, S. EUROGIN 2014 roadmap: Differences in human papillomavirus infection natural history, transmission and human papillomavirus-related cancer incidence by gender and anatomic site of infection. Int. J. Cancer 2015, 136, 2752–2760. [Google Scholar] [CrossRef] [PubMed]
- Bosset, J.F.; Pavy, J.J.; Roelofsen, F.; Bartelink, H. Combined radiotherapy and chemotherapy for anal cancer EORTC Radiotherapy and Gastrointestinal Cooperative Groups. Lancet 1997, 349, 205–206. [Google Scholar] [CrossRef]
- UKCCCR. Epidermoid anal cancer: Results from the UKCCCR randomised trial of radiotherapy alone versus radiotherapy, 5-fluorouracil, and mitomycin. UKCCCR Anal Cancer Trial Working Party. UK Coordinating Committee on Cancer Research. Lancet 1996, 348, 1049–1054. [Google Scholar] [CrossRef]
- Ajani, J.A.; Winter, K.A.; Gunderson, L.L.; Pedersen, J.; Benson, A.B.; Thomas, C.R.; Mayer, R.J.; Haddock, M.G.; Rich, T.A.; Willett, C. Fluorouracil, mitomycin, and radiotherapy vs. fluorouracil, cisplat- in, and radiotherapy for carcinoma of the anal canal: A randomized controlled trial. JAMA 2008, 299, 1914–1921. [Google Scholar] [CrossRef] [PubMed]
- James, R.D.; Glynne-Jones, R.; Meadows, H.M.; Cunningham, D.; Myint, A.S.; Saunders, M.P.; Maughan, T.; McDonald, A.; Essapen, S.; Leslie, M.; et al. Mitomycin or cisplatin chemoradiation with or without maintenance chemotherapy for treatment of squamous-cell carcinoma of the anus (ACT II): A randomised, phase 3, open-label, 2 × 2 factorial trial. Lancet Oncol. 2013, 14, 516–524. [Google Scholar] [CrossRef]
- Shridhar, R.; Shibata, D.; Chan, E.; Thomas, C.R. Anal cancer: Current standards in care and recent changes in practice. CA Cancer J. Clin. 2015, 65, 139–162. [Google Scholar] [CrossRef]
- Renehan, A.G.; Saunders, M.P.; Schofield, P.F.; O’Dwyer, S.T. Patterns of local disease failure and outcome after salvage surgery in patients with anal cancer. Br. J. Surg. 2005, 92, 605–612. [Google Scholar] [CrossRef]
- Peiffert, D.; Tournier-Rangeard, L.; Gérard, J.-P.; Lemanski, C.; Francois, E.; Giovannini, M.; Cvitkovic, F.; Mirabel, X.; Bouche, O.; Luporsi, E.; et al. Induction chemotherapy and dose intensification of the radiation boost in locally advanced anal canal carcinoma: Final analysis of the randomized UNICANCER ACCORD 03 trial. J. Clin. Oncol. 2012, 30, 1941–1948. [Google Scholar] [CrossRef]
- Garg, M.K.; Zhao, F.; Sparano, J.A.; Palefsky, J.; Whittington, R.; Mitchell, E.P.; Mulcahy, M.F.; Armstrong, K.I.; Nabbout, N.H.; Kalnicki, S.; et al. Cetuximab Plus Chemoradiotherapy in Immunocompetent Patients with Anal Carcinoma: A Phase II Eastern Cooperative Oncology Group–American College of Radiology Imaging Network Cancer Research Group Trial (E3205). J. Clin. Oncol. 2017, 35, 718. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Lee, J.Y.; Palefsky, J.; Henry, D.H.; Wachsman, W.; Rajdev, L.; Aboulafia, D.; Ratner, L.; Fitzgerald, T.J.; Kachnic, L.; et al. Cetuximab Plus Chemoradiotherapy for HIV-Associated Anal Carcinoma: A Phase II AIDS Malignancy Consortium Trial. J. Clin. Oncol. 2017, 35, 727. [Google Scholar] [CrossRef] [PubMed]
- Sclafani, F.; Rao, S. Systemic Therapies for Advanced Squamous Cell Anal Cancer. Curr. Oncol. Rep. 2018, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Larue, R.T.H.M.; Defraene, G.; De Ruysscher, D.; Lambin, P.; van Elmpt, W. Quantitative radiomics studies for tissue characterization: A review of technology and methodological procedures. Br. J. Radiol. 2017, 90, 20160665. [Google Scholar] [CrossRef]
- Kourou, K. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Montagnon, E.; Cerny, M.; Cadrin-Chênevert, A.; Hamilton, V.; Derennes, T.; Ilinca, A.; Vandenbroucke-Menu, F.; Turcotte, S.; Kadoury, S.; Tang, A. Deep learning workflow in radiology: A primer. Insights Imaging 2020, 11, 22. [Google Scholar] [CrossRef]
- Cuocolo, R.; Perillo, T.; De Rosa, E.; Ugga, L.; Petretta, M. Current applications of big data and machine learning in cardiology. J. Geriatr. Cardiol. 2019, 16, 601–607. [Google Scholar]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Ugga, L.; Romeo, V.; Radice, L.; Brunetti, A.; Imbriaco, M. Machine learning applications in prostate cancer magnetic resonance imaging. Eur. Radiol. Exp. 2019, 3, 35. [Google Scholar] [CrossRef]
- Abernethy, A.P.; Etheredge, L.M.; Ganz, P.A.; Wallace, P.; German, R.R.; Neti, C.; Bach, P.B.; Murphy, S.B. Rapid-Learning System for Cancer Care. J. Clin. Oncol. 2010, 28, 4268–4274. [Google Scholar] [CrossRef] [PubMed]
- Ehteshami Bejnordi, B.; Veta, M.; van Johannes Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; the CAMELYON16 Consortium. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017, 318, 2199. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Linares Perez, M.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94. [Google Scholar] [CrossRef] [PubMed]
- De Groof, A.J.; Struyvenberg, M.R.; van der Putten, J.; van der Sommen, F.; Fockens, K.N.; Curvers, W.L.; Zinger, S.; Pouw, R.E.; Coron, E.; Baldaque-Silva, F.; et al. Deep-Learning System Detects Neoplasia in Patients With Barrett’s Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology 2020, 158, 915–929.e4. [Google Scholar] [CrossRef] [PubMed]
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards clinical application of image mining: A systematic review on artificial intelligence and radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672. [Google Scholar] [CrossRef]
- Glynne-Jones, R.; Nilsson, P.J.; Aschele, C.; Goh, V.; Peiffert, D.; Cervantes, A.; Arnold, D. Anal cancer: ESMO-ESSO-ESTRO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, 10–20. [Google Scholar] [CrossRef]
- Jones, M.; Hruby, G.; Solomon, M.; Rutherford, N.; Martin, J. The role of FDG-PET in the initial staging and response assessment of anal cancer: A systematic review and meta-analysis. Ann. Surg. Oncol. 2015, 22, 3574–3581. [Google Scholar] [CrossRef]
- Goh, V.; Gollub, F.K.; Liaw, J.; Wellsted, D.; Przybytniak, I.; Padhani, A.R.; Glynne-Jones, R. Magnetic resonance imaging assessment of squamous cell carcinoma of the anal canal before and after chemoradiation: Can MRI predict for eventual clinical out- come? Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, 715–721. [Google Scholar] [CrossRef]
- Kochhar, R.; Renehan, A.G.; Mullan, D.; Chakrabarty, B.; Saunders, M.P.; Carrington, B.M. The assessment of local response using magnetic resonance imaging at 3- and 6-month post chemoradiotherapy in patients with anal cancer. Eur. Radiol. 2017, 27, 607–617. [Google Scholar] [CrossRef]
- Hocquelet, A.; Auriac, T.; Perier, C.; Dromain, C.; Meyer, M.; Pinaquy, J.B.; Denys, A.; Trillaud, H.; Denis de Seneville, B.; Vendrely, V. Pre-treatment magnetic resonance-based texture features as potential imaging biomarkers for predicting event free survival in anal cancer treated by chemoradiotherapy. Eur. Radiol. 2018, 28, 2801. [Google Scholar] [CrossRef]
- Owczarczyk, K.; Prezzi, D.; Cascino, M.; Kozarski, R.; Gaya, A.; Siddique, M.; Cook, G.J.; Glynne-Jones, R.; Goh, V. MRI heterogeneity analysis for prediction of recurrence and disease free survival in anal cancer. Radiother. Oncol. 2019, 134, 119. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Hruby, G.; Coolens, C.; Driscoll, B.; Stanwell, P.; Kumar, M.; Capp, A.; Sridharan, S.; Arm, J.; Gallagher, S.; et al. A prospective, multi-centre trial of multi-parametric MRI as a biomarker in anal carcinoma. Radiother. Oncol. 2020, 144, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Rusten, E.; Rekstad, B.L.; Undseth, C.; Klotz, D.; Hernes, E.; Guren, M.G.; Malinen, E. Anal cancer chemoradiotherapy outcome prediction using 18F-fluorodeoxyglucose positron emission tomography and clinicopathological factors. BJR 2019, 92, 20181006. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.P.; Hruby, G.; Metser, U.; Sridharan, S.; Capp, A.; Kumar, M.; Gallagher, S.; Rutherford, N.; Holder, C.; Oldmeadow, C.; et al. FDG-PET parameters predict for recurrence in anal cancer—Results from a prospective, multicentre clinical trial. Radiat. Oncol. 2019, 14, 140. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.J.; Zhong, J.; Frood, R.; Currie, S.; Gilbert, A.; Appelt, A.L.; Sebag-Montefiore, D.; Scarsbrook, A. Prediction of outcome in anal squamous cell carcinoma using radiomic feature analysis of pre-treatment FDG PET-CT. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2790–2799. [Google Scholar] [CrossRef] [PubMed]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuze, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterization of Tumor Heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef]
- Bartelink, H.; Roelofsen, F.; Eschwege, F.; Rougier, P.; Bosset, J.F.; Gonzalez, D.G.; Peiffert, D.; van Glabbeke, M.; Pierart, M. Concomitant radiotherapy and chemotherapy is superior to radiotherapy alone in the treatment of locally advanced anal cancer: Results of a phase III randomized trial of the European Organization for Research and Treatment of Cancer Radiotherapy and Gastrointestinal Cooperative Groups. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1997, 15, 2040–2049. [Google Scholar]
- Myerson, R.J.; Kong, F.; Birnbaum, E.H.; Fleshman, J.W.; Kodner, I.J.; Picus, J.; Ratkin, G.A.; Read, T.E.; Walz, B.J. Radiation therapy for epidermoid carcinoma of the anal canal, clinical and treatment factors associated with outcome. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2001, 61, 15–22. [Google Scholar] [CrossRef]
- Toh, E.; Wilson, J.; Sebag-Montefiore, D.; Botterill, I. Neutrophil:lymphocyte ratio as a simple and novel biomarker for prediction of locoregional recurrence after chemoradiotherapy for squamous cell carcinoma of the anus. Colorectal Dis. 2014, 16, O90–O97. [Google Scholar] [CrossRef]
- Faivre, J.-C.; Peiffert, D.; Vendrely, V.; Lemanski, C.; Hannoun-Levi, J.M.; Mirabel, X.; Stanbury, T.; Salleron, J.; Guillemin, F. Prognostic factors of colostomy free survival in patients presenting with locally advanced anal canal carcinoma: A pooled analysis of two prospective trials (KANAL 2 and ACCORD 03). Radiother. Oncol. 2018, 129, 463–470. [Google Scholar] [CrossRef]
- Wu, Y.; Han, X.; Li, Y.; Zhu, K.; Yu, J. Survival prediction models for patients with anal carcinoma receiving definitive chemoradiation: A population-based study. Oncol. Lett. 2019, 19, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Brinker, T.; Hekler, A.; Enk, A.; Berking, C.; Haferkamp, S.; Hauschild, A.; Weichenthal, M.; Klode, J.; Schadendorf, D.; Holland-Letz, T.; et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer. 2019, 119, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Arcadu, F.; Benmansour, F.; Maunz, A.; Willis, J.; Haskova, Z.; Prunotto, M. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit. Med. 2019, 2, 92. [Google Scholar] [CrossRef] [PubMed]
- Munir, K.; Elahi, H.; Ayub, A.; Frezza, F.; Rizzi, A. Cancer Diagnosis Using Deep Learning: A Bibliographic Review. Cancers 2019, 11, 1235. [Google Scholar] [CrossRef]
- De Bari, B.; Vallati, M.; Gatta, R.; Lestrade, L.; Manfrida, S.; Carrie, C.; Valentini, V. Development and validation of a machine learning-based predictive model to improve the prediction of inguinal status of anal cancer patients: A preliminary report. Oncotarget 2017, 8, 108509. [Google Scholar] [CrossRef]
- Vendrely, V.; Lemanski, C.; Francois, E.; Barbier, E.; Baba Hamed, N.; Bonichon-Lamichhane, N.; de la Rochefordiere, A.; Bouche, O.; Toucheron, D.; Diaz, D.; et al. OC-0284: First results of the French cohort ANABASE: Treatment and outcome in non-metastatic anal cancer. Radiother. Oncol. 2018, 127, S146. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).