Discrimination of Potato (Solanum tuberosum L.) Accessions Collected in Majella National Park (Abruzzo, Italy) Using Mid-Infrared Spectroscopy and Chemometrics Combined with Morphological and Molecular Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Potato Samples
2.2. Potato Cropping
2.3. Morpho-Agronomic Characterization of Potato Cultivars
2.4. Genotyping
2.5. ATR-FTIR Measurements
2.6. Multivariate Statistical Analysis
3. Results and Discussion
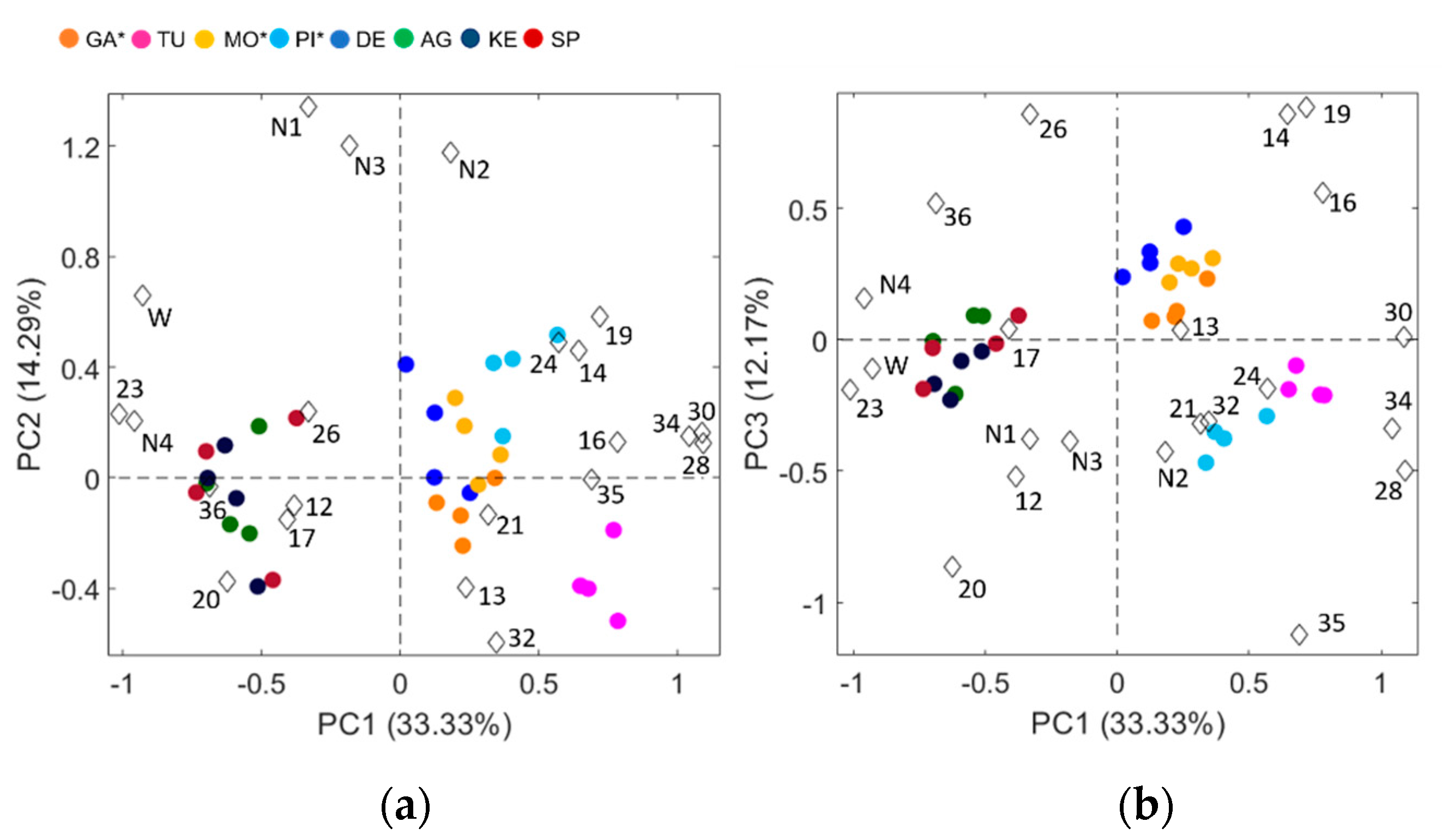
3.1. Morpho-Agronomic Characterization of Potato Accessions
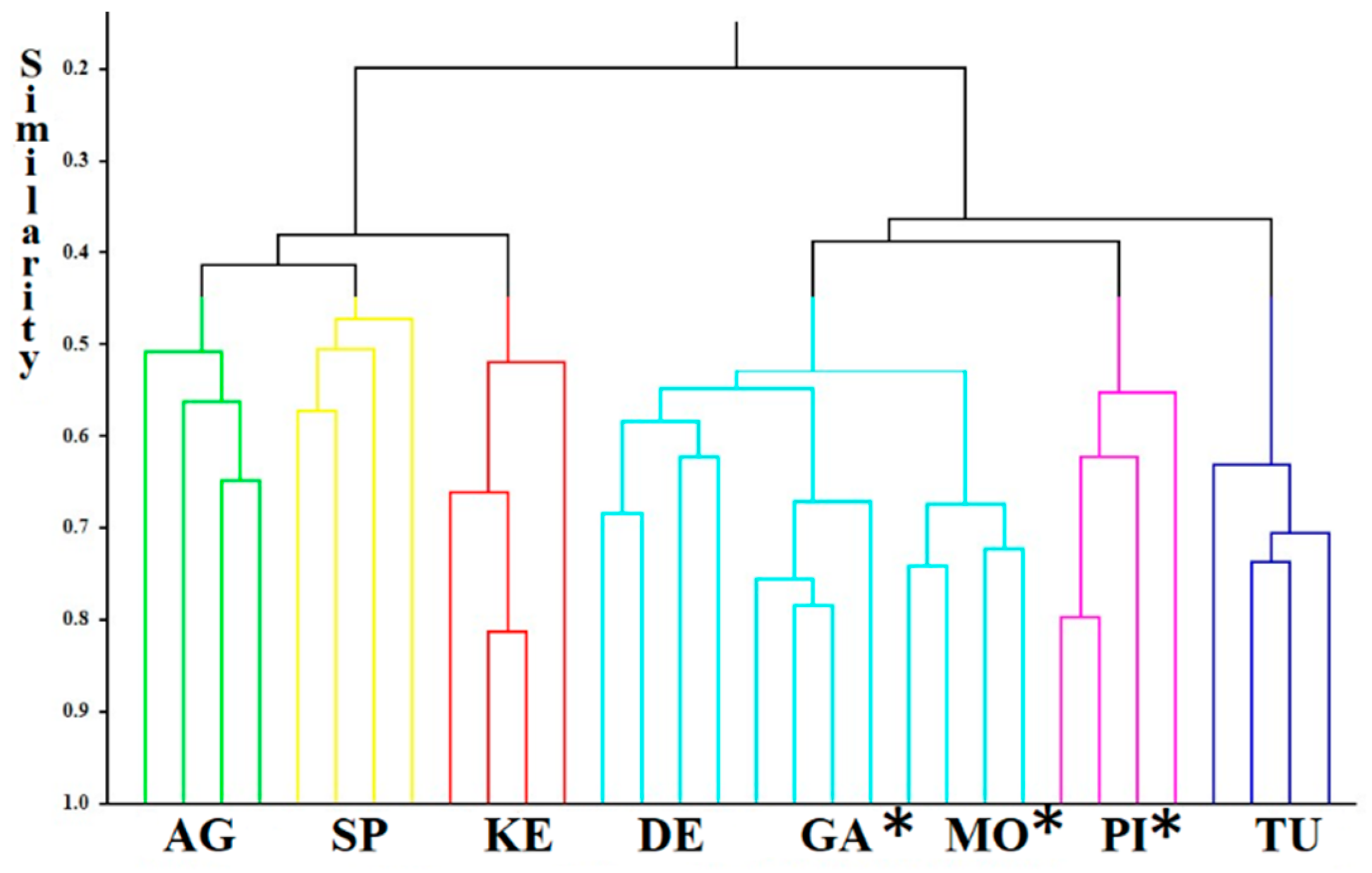
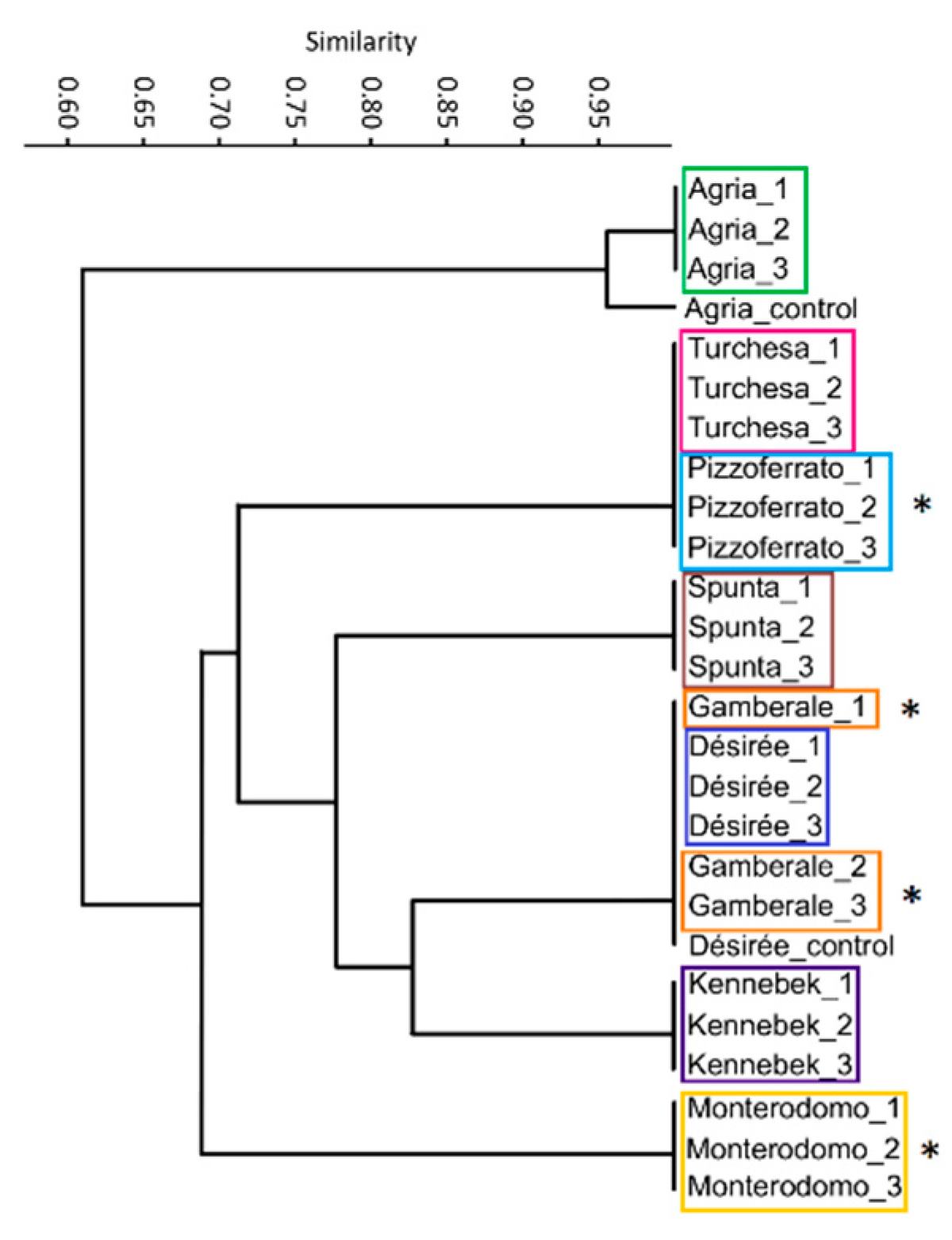
3.2. DNA Fingerprinting
3.3. Characterization of Potatoes Using ATR-FTIR Spectroscopy
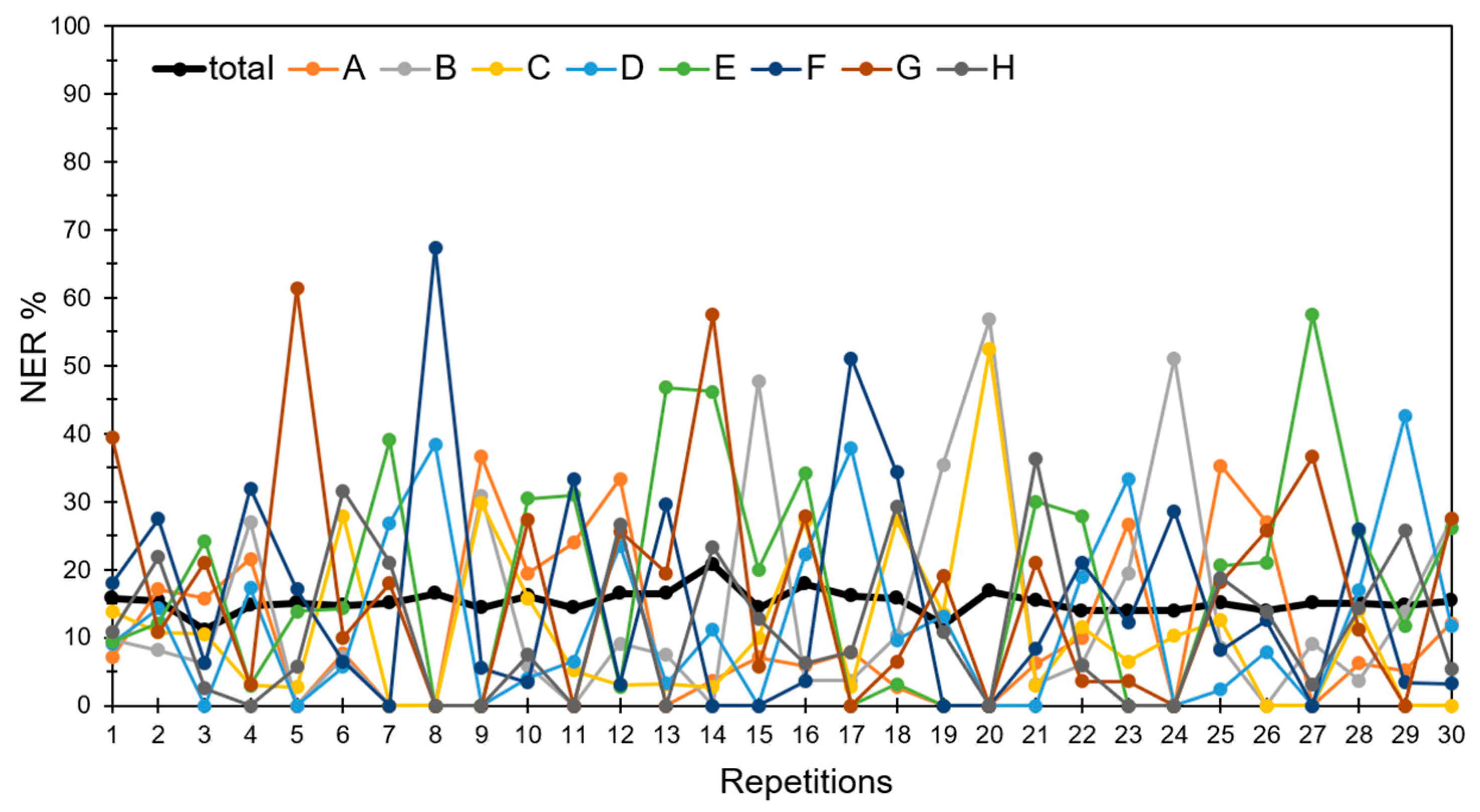
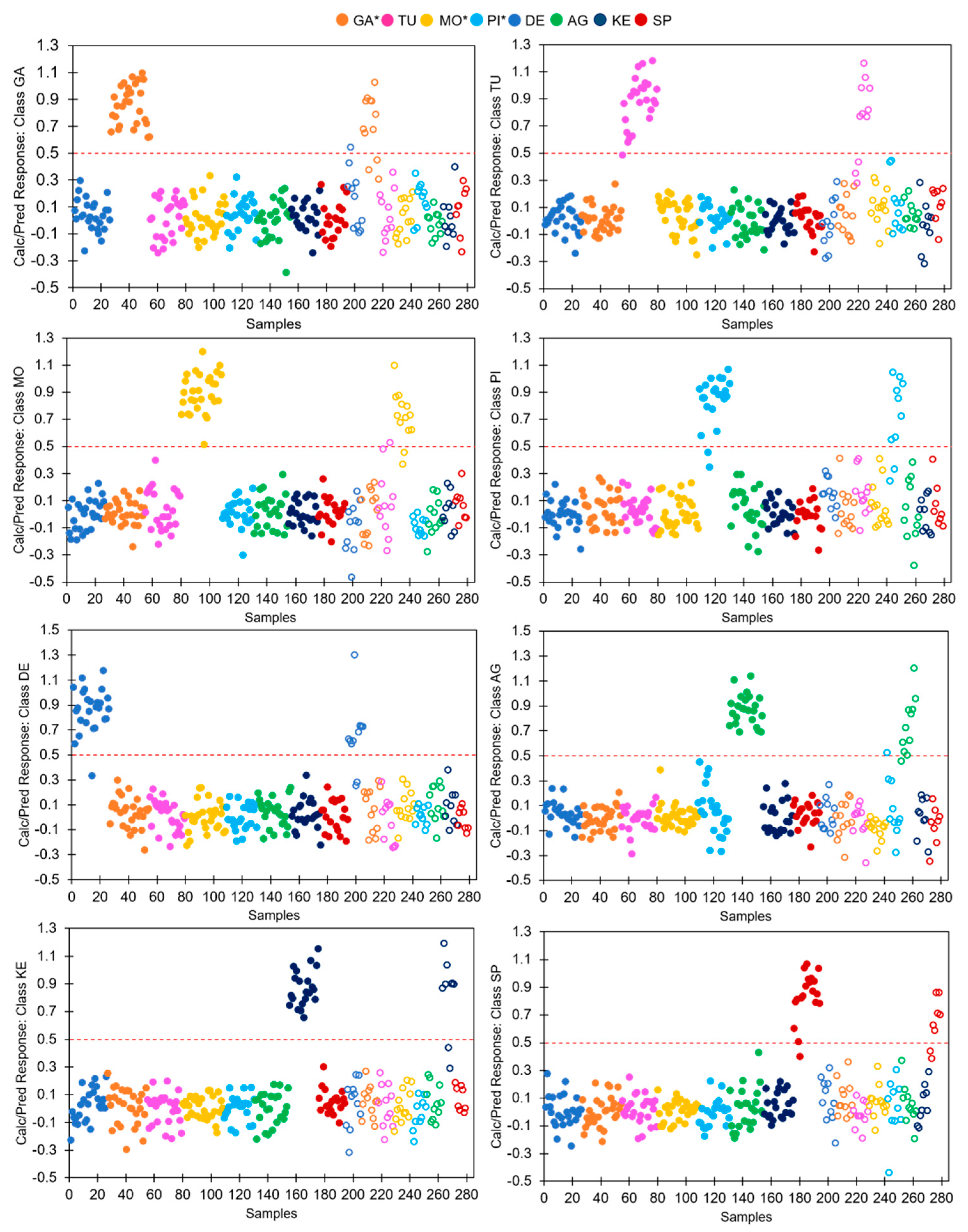
3.4. Discrimination of Potato Varieties Using the PLS-DA of ATR-FTIR Spectra
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A

| Accession | 12 | 13 | 14 | 16 | 17 | 19 | 20 | 21 | 23 | 24 | 26 | 28 | 30 | 32 | 34 | 35 | 36 | N1 | W | N2 | N3 | N4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AG | 1 | 2 | 1 | 3 | 5 | 1 | 7 | 5 | 6 | 7 | 7 | 1 | 5 | 4 | 2 | 2 | 4 | 81 | 14.7 | 14 | 21 | 46 |
| 2 | 5 | 1 | 2 | 5 | 1 | 7 | 5 | 6 | 7 | 3 | 1 | 1 | 4 | 2 | 2 | 4 | 105 | 15.8 | 17 | 46 | 42 | |
| 2 | 5 | 3 | 5.8 | 5 | 3 | 7 | 5 | 6 | 7 | 7 | 1 | 1 | 4 | 2 | 2 | 4 | 149 | 18.0 | 59 | 45 | 45 | |
| 2 | 6 | 3 | 2 | 5 | 1 | 7 | 5 | 6 | 7 | 7 | 1 | 1 | 4 | 2 | 2 | 4 | 122 | 18.6 | 24 | 49 | 49 | |
| DE | 1 | 6 | 7 | 5 | 3.6 | 5 | 5 | 1 | 3 | 7 | 7 | 4 | 7 | 4 | 3 | 2 | 2 | 76 | 12.2 | 16 | 29 | 31 |
| 1 | 5 | 7 | 4.4 | 3.6 | 5 | 5 | 1 | 3 | 7 | 7 | 4.8 | 7 | 5 | 3 | 2 | 2 | 129 | 18.0 | 30 | 58 | 41 | |
| 2 | 6.2 | 7 | 3 | 3 | 7 | 5 | 5 | 3 | 7 | 7 | 3.9 | 6.3 | 4 | 3 | 2 | 2 | 86 | 16.8 | 12 | 31 | 43 | |
| 2 | 5 | 7 | 3 | 3 | 6 | 5 | 3 | 3 | 7 | 7 | 4 | 5 | 3 | 3 | 2 | 2 | 152 | 16.4 | 73 | 40 | 39 | |
| GA | 2 | 5.2 | 7 | 4.4 | 3 | 7 | 5.8 | 4 | 1 | 5 | 5 | 3.7 | 6.4 | 4 | 3 | 2 | 2 | 103 | 9.4 | 52 | 25 | 26 |
| 2 | 5.8 | 5.4 | 3 | 3 | 3 | 5.8 | 3 | 1 | 5 | 5 | 4.6 | 7 | 4 | 3 | 2 | 2 | 101 | 12.6 | 34 | 33 | 34 | |
| 2 | 5.8 | 5 | 4.2 | 3 | 3.6 | 7 | 3.8 | 1 | 5 | 7 | 4 | 5 | 5 | 3 | 2 | 2 | 79 | 7.7 | 29 | 36 | 14 | |
| 2 | 5 | 5 | 4 | 3 | 5 | 7 | 3 | 1 | 5 | 5 | 3.9 | 7 | 4 | 3 | 2 | 2 | 86 | 10.0 | 33 | 32 | 21 | |
| KE | 3 | 5 | 1 | 2.2 | 3 | 1 | 7 | 3 | 4 | 3 | 7 | 1 | 1 | 4 | 1 | 2 | 1 | 68 | 11.8 | 12 | 28 | 28 |
| 3 | 5 | 1 | 2 | 6 | 1 | 7 | 1 | 4 | 3 | 7 | 1 | 1 | 3 | 3 | 3 | 1 | 155 | 18.7 | 73 | 36 | 46 | |
| 3 | 4.2 | 1 | 1 | 3 | 1 | 7 | 1 | 4 | 3 | 7 | 1 | 1 | 4 | 2 | 2 | 1 | 133 | 18.7 | 41 | 47 | 45 | |
| 2 | 4.2 | 1 | 1.6 | 3 | 1 | 7 | 1 | 4 | 3 | 7 | 1 | 1 | 4 | 2 | 2 | 1 | 120 | 16.9 | 42 | 36 | 42 | |
| MO | 1 | 4.4 | 5 | 3 | 5 | 4.4 | 5 | 1 | 2 | 5 | 7 | 5 | 9 | 4 | 3 | 2 | 2 | 87 | 6.9 | 34 | 46 | 7 |
| 1 | 3 | 5 | 4.1 | 5 | 5 | 5 | 3 | 2 | 5 | 7 | 3.8 | 7 | 4 | 3 | 2 | 2 | 142 | 9.5 | 72 | 55 | 15 | |
| 1 | 3.8 | 5 | 5 | 3 | 5 | 5 | 1 | 2 | 5 | 7 | 5 | 7 | 4 | 3 | 2 | 2 | 101 | 6.5 | 47 | 46 | 8 | |
| 2 | 3 | 3 | 5 | 5 | 7 | 5 | 2 | 2 | 5 | 7 | 5.7 | 7 | 4 | 3 | 1 | 2 | 122 | 8.65 | 72 | 40 | 10 | |
| PI | 2 | 5 | 5 | 3 | 3 | 3.6 | 7 | 1 | 3 | 9 | 5 | 8.3 | 9 | 4 | 4 | 4 | 1 | 169 | 12.8 | 89 | 64 | 16 |
| 2 | 5 | 3 | 3 | 3 | 3 | 7 | 3.4 | 3 | 9 | 5 | 9 | 9 | 4 | 4 | 4 | 1 | 173 | 14.0 | 72 | 77 | 24 | |
| 1 | 3 | 3 | 3 | 3 | 3 | 7 | 5 | 3 | 7 | 5 | 9 | 7 | 4 | 4 | 4 | 1 | 123 | 12.3 | 51 | 56 | 16 | |
| 2 | 3.8 | 5 | 5 | 4 | 5 | 5 | 6.2 | 3 | 7 | 5 | 9 | 9 | 5 | 4 | 4 | 1 | 183 | 15.1 | 93 | 73 | 17 | |
| SP | 2 | 4 | 3 | 3 | 6.4 | 1 | 7 | 1 | 5 | 3 | 5 | 1 | 1 | 5 | 3 | 2 | 3 | 76 | 12.2 | 16 | 29 | 31 |
| 1 | 2.2 | 1 | 1.8 | 3 | 1 | 7 | 1 | 5 | 3 | 7 | 1 | 5 | 5 | 1 | 2 | 3 | 141 | 20.2 | 38 | 62 | 41 | |
| 2 | 5 | 7 | 4.4 | 3.6 | 5 | 7 | 1 | 5 | 3 | 5 | 1 | 1 | 5 | 2 | 2 | 3 | 152 | 19.5 | 60 | 59 | 33 | |
| 2 | 5 | 1 | 1 | 5 | 1 | 7 | 1 | 5 | 3 | 5 | 1 | 1 | 5 | 1 | 2 | 3 | 136 | 19.0 | 60 | 48 | 28 | |
| TU | 1 | 7 | 3 | 4.5 | 3.5 | 3 | 7 | 5 | 1 | 7 | 5 | 9 | 9 | 6 | 4 | 4 | 1 | 43 | 5.5 | 16 | 20 | 7 |
| 2 | 4.6 | 3 | 4.2 | 3.4 | 3 | 5 | 5 | 1 | 7 | 5 | 9 | 9 | 5 | 4 | 4 | 1 | 81 | 4.5 | 52 | 25 | 4 | |
| 2 | 5 | 4 | 4.7 | 3 | 3 | 7 | 3 | 1 | 5 | 5 | 9 | 9 | 5 | 4 | 4 | 1 | 54 | 5.9 | 24 | 19 | 11 | |
| 2 | 5 | 3.7 | 4 | 5 | 3 | 5 | 3 | 1 | 5 | 5 | 9 | 9 | 5 | 4 | 4 | 1 | 53 | 4.4 | 34 | 8 | 11 |
| Locus | Motif | Primer | Ta | Map | Total Found Alleles |
|---|---|---|---|---|---|
| STM5121 | (TGT)5 | FW: CACCGGAATAAGCGGATCT | 48 | XII | 301, 305, 308 |
| RW: TCTTCCCTTCCATTTGTCA | |||||
| STI0001 | (AAT)n | FW: CAGCAAAATCAGAACCCGAT | 60 | IV | 196, 199, 202, 205, 207, 210 |
| RW: GGATCATCAAATTCACCGCT | |||||
| STM1064 | (TA)12..(TG)4 GT (TG)5 | FW: GTTCTTTTGGTGGTTTTCCT | 55 | II | 207, 210, 212 |
| RW: TTATTTCTCTGTTGTTGCTG | |||||
| STG0016 | (AGA)8 | FW: AGCTGCTCAGCATCAAGAGA | 55 | I | 142, 149, 152, 155, 173 |
| RW: ACCACCTCAGGCACTTCATC | |||||
| STPoAc58 | (TA)13 | FW: TTGATGAAAGGAATGCAGCTTGTG | 55 | V | 250, 264 |
| RW: ACGTTAAAGAAGTGAGAGTACGAC |
| DE_1 | DE_2 | DE_3 | KE_1 | KE_2 | KE_3 | GA_1 | GA_2 | GA_3 | MO_1 | MO_2 | MO_3 | TU_1 | TU_2 | TU_3 | SP_1 | SP_2 | SP_3 | PI_1 | PI_2 | PI_3 | AG_1 | AG_2 | AG_3 | AG_control | DE_control | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DE_1 | 1 | 1.000 | 1.000 | 0.828 | 0.828 | 0.828 | 1.000 | 1.000 | 1.000 | 0.692 | 0.692 | 0.692 | 0.720 | 0.720 | 0.720 | 0.786 | 0.786 | 0.786 | 0.720 | 0.720 | 0.720 | 0.615 | 0.615 | 0.615 | 0.640 | 1.000 |
| DE_2 | 1 | 1.000 | 0.828 | 0.828 | 0.828 | 1.000 | 1.000 | 1.000 | 0.692 | 0.692 | 0.692 | 0.720 | 0.720 | 0.720 | 0.786 | 0.786 | 0.786 | 0.720 | 0.720 | 0.720 | 0.615 | 0.615 | 0.615 | 0.640 | 1.000 | |
| DE_3 | 1 | 0.828 | 0.828 | 0.828 | 1.000 | 1.000 | 1.000 | 0.692 | 0.692 | 0.692 | 0.720 | 0.720 | 0.720 | 0.786 | 0.786 | 0.786 | 0.720 | 0.720 | 0.720 | 0.615 | 0.615 | 0.615 | 0.640 | 1.000 | ||
| KE_1 | 1 | 1.000 | 1.000 | 0.828 | 0.828 | 0.828 | 0.667 | 0.667 | 0.667 | 0.769 | 0.769 | 0.769 | 0.759 | 0.759 | 0.759 | 0.769 | 0.769 | 0.769 | 0.593 | 0.593 | 0.593 | 0.615 | 0.828 | |||
| KE_2 | 1 | 1.000 | 0.828 | 0.828 | 0.828 | 0.667 | 0.667 | 0.667 | 0.769 | 0.769 | 0.769 | 0.759 | 0.759 | 0.759 | 0.769 | 0.769 | 0.769 | 0.593 | 0.593 | 0.593 | 0.615 | 0.828 | ||||
| KE_3 | 1 | 0.828 | 0.828 | 0.828 | 0.667 | 0.667 | 0.667 | 0.769 | 0.769 | 0.769 | 0.759 | 0.759 | 0.759 | 0.769 | 0.769 | 0.769 | 0.593 | 0.593 | 0.593 | 0.615 | 0.828 | |||||
| GA_1 | 1 | 1.000 | 1.000 | 0.692 | 0.692 | 0.692 | 0.720 | 0.720 | 0.720 | 0.786 | 0.786 | 0.786 | 0.720 | 0.720 | 0.720 | 0.615 | 0.615 | 0.615 | 0.640 | 1.000 | ||||||
| GA_2 | 1 | 1.000 | 0.692 | 0.692 | 0.692 | 0.720 | 0.720 | 0.720 | 0.786 | 0.786 | 0.786 | 0.720 | 0.720 | 0.720 | 0.615 | 0.615 | 0.615 | 0.640 | 1.000 | |||||||
| GA_3 | 1 | 0.692 | 0.692 | 0.692 | 0.720 | 0.720 | 0.720 | 0.786 | 0.786 | 0.786 | 0.720 | 0.720 | 0.720 | 0.615 | 0.615 | 0.615 | 0.640 | 1.000 | ||||||||
| MO_1 | 1 | 1.000 | 1.000 | 0.696 | 0.696 | 0.696 | 0.692 | 0.692 | 0.692 | 0.696 | 0.696 | 0.696 | 0.583 | 0.583 | 0.583 | 0.609 | 0.692 | |||||||||
| MO_2 | 1 | 1.000 | 0.696 | 0.696 | 0.696 | 0.692 | 0.692 | 0.692 | 0.696 | 0.696 | 0.696 | 0.583 | 0.583 | 0.583 | 0.609 | 0.692 | ||||||||||
| MO_3 | 1 | 0.696 | 0.696 | 0.696 | 0.692 | 0.692 | 0.692 | 0.696 | 0.696 | 0.696 | 0.583 | 0.583 | 0.583 | 0.609 | 0.692 | |||||||||||
| TU_1 | 1 | 1.000 | 1.000 | 0.640 | 0.640 | 0.640 | 1.000 | 1.000 | 1.000 | 0.522 | 0.522 | 0.522 | 0.545 | 0.720 | ||||||||||||
| TU_2 | 1 | 1.000 | 0.640 | 0.640 | 0.640 | 1.000 | 1.000 | 1.000 | 0.522 | 0.522 | 0.522 | 0.545 | 0.720 | |||||||||||||
| TU_3 | 1 | 0.640 | 0.640 | 0.640 | 1.000 | 1.000 | 1.000 | 0.522 | 0.522 | 0.522 | 0.545 | 0.720 | ||||||||||||||
| SP_1 | 1 | 1.000 | 1.000 | 0.640 | 0.640 | 0.640 | 0.769 | 0.769 | 0.769 | 0.800 | 0.786 | |||||||||||||||
| SP_2 | 1 | 1.000 | 0.640 | 0.640 | 0.640 | 0.769 | 0.769 | 0.769 | 0.800 | 0.786 | ||||||||||||||||
| SP_3 | 1 | 0.640 | 0.640 | 0.640 | 0.769 | 0.769 | 0.769 | 0.800 | 0.786 | |||||||||||||||||
| PI_1 | 1 | 1.000 | 1.000 | 0.522 | 0.522 | 0.522 | 0.545 | 0.720 | ||||||||||||||||||
| PI_2 | 1 | 1.000 | 0.522 | 0.522 | 0.522 | 0.545 | 0.720 | |||||||||||||||||||
| PI_3 | 1 | 0.522 | 0.522 | 0.522 | 0.545 | 0.720 | ||||||||||||||||||||
| AG_1 | 1 | 1.000 | 1.000 | 0.957 | 0.615 | |||||||||||||||||||||
| AG_2 | 1 | 1.000 | 0.957 | 0.615 | ||||||||||||||||||||||
| AG_3 | 1 | 0.957 | 0.615 | |||||||||||||||||||||||
| AG_control | 1 | 0.640 | ||||||||||||||||||||||||
| DE_control | 1 |
References
- FAOSTAT Database. Available online: http://www.fao.org/faostat/en (accessed on 29 November 2019).
- Pacifico, D.; Casciani, L.; Ritota, M.; Mandolino, G.; Onofri, C.; Moschella, A.; Parisi, B.; Cafiero, C.; Valentini, M. NMR-based metabolomics for organic farming traceability of early potatoes. J. Agric. Food Chem. 2013, 61, 11201–11211. [Google Scholar] [CrossRef]
- Manzelli, M.; Romagnoli, S.; Ghiselli, L.; Benedettelli, S.; Palchetti, E.; Andrenelli, L.; Vecchio, V. Typicity in potato: Characterization of geographic origin. Ital. J. Agron. 2010, 5, 61–67. [Google Scholar] [CrossRef]
- European Commission. Agricultural and Rural Development DOOR Database. Available online: https://ec.europa.eu/agriculture/quality/door/list.html (accessed on 29 November 2019).
- Negri, V. Landraces in central Italy: Where and why they are conserved and perspectives for their on-farm conservation. Genet. Resour. Crop Evol. 2003, 50, 871–885. [Google Scholar] [CrossRef]
- UPOV TG/23/06, Potato (Solanum tuberosum L.)- Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability, International Union for the Protection of New Varieties of Plants. Available online: https://www.upov.int (accessed on 29 November 2019).
- Ferradini, N.; Torricelli, R.; Terzaroli, N.; Albertini, E.; Russi, L. The genetic structure of the Field Pea Landrace “Roveja di Civita di Cascia”. Sustainability 2019, 11, 6493. [Google Scholar] [CrossRef]
- Torricelli, R.; Silveri, D.D.; Ferradini, N.; Venora, G.; Veronesi, F.; Russi, L. Characterization of the lentil landrace Santo Stefano di Sessanio from Abruzzo, Italy. Genet. Resour. Crop Evol. 2012, 59, 261–276. [Google Scholar] [CrossRef]
- Di Giacomo, F.; Del Signore, A.; Giaccio, M. Determining the geographic origin of potatoes using mineral and trace element content. J. Agric. Food Chem. 2007, 55, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.C.; Hernández, P.S.; Rodríguez, E.M.R.; Martín, J.D.; Romero, C.D. Mineral concentrations in cultivars of potatoes. Food Chem. 2003, 83, 247–253. [Google Scholar] [CrossRef]
- Galdón, B.R.; Rodríguez, L.H.; Mesa, D.R.; León, H.L.; Pérez, N.L.; Rodríguez Rodríguez, E.M.; Romero, C.D. Differentiation of potato cultivars experimentally cultivated based on their chemical composition and by applying linear discriminant analysis. Food Chem. 2012, 133, 1241–1248. [Google Scholar] [CrossRef]
- Adamo, P.; Zampella, M.; Quétel, C.R.; Aversano, R.; Dal Piaz, F.; De Tommasi, N.; Frusciante, L.; Iorizzo, M.; Lepore, L.; Carputo, D. Biological and geochemical markers of the geographical origin and genetic identity of potatoes. J. Geochemical Explor. 2012, 121, 62–68. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, J.K.; Jin, Y.I.; Oh, Y.T.; Prabakaran, M.; Youn, K.J.; Kim, S.H. Discriminative study of a potato (Solanum tuberosum L.) cultivation region by measuring the stable isotope ratios of bio-elements. Food Chem. 2016, 212, 48–57. [Google Scholar] [CrossRef]
- Camin, F.; Moschella, A.; Miselli, F.; Parisi, B.; Versini, G.; Ranalli, P.; Bagnaresi, P. Evaluation of markers for the traceability of potato tubers grown in an organic versus conventional regime. J. Sci. Food Agric. 2007, 87, 1330–1336. [Google Scholar] [CrossRef]
- Mahne Opatić, A.; Nečemer, M.; Budič, B.; Lojen, S. Stable isotope analysis of major bioelements, multi-element profiling, and discriminant analysis for geographical origins of organically grown potato. J. Food Compos. Anal. 2018, 71, 17–24. [Google Scholar] [CrossRef]
- Claassen, C.; Kuballa, J.; Rohn, S. Metabolomics-Based Approach for the Discrimination of Potato Varieties (Solanum tuberosum) using UPLC-IMS-QToF. J. Agric. Food Chem. 2019, 67, 5700–5709. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, F.; Casiello, G.; Sacco, D.; Tedone, L.; Sacco, A. Characterisation of the geographical origin of Italian potatoes, based on stable isotope and volatile compound analyses. Food Chem. 2011, 124, 1708–1713. [Google Scholar] [CrossRef]
- Firmani, P.; De Luca, S.; Bucci, R.; Marini, F.; Biancolillo, A. Near infrared (NIR) spectroscopy-based classification for the authentication of Darjeeling black tea. Food Control 2019, 100, 292–299. [Google Scholar] [CrossRef]
- Biancolillo, A.; Marini, F.; D’Archivio, A.A. Geographical discrimination of red garlic (Allium sativum L.) using fast and non-invasive Attenuated Total Reflectance-Fourier Transformed Infrared (ATR-FTIR) spectroscopy combined with chemometrics. J. Food Compos. Anal. 2020, 86, 103351. [Google Scholar] [CrossRef]
- Biancolillo, A.; De Luca, S.; Bassi, S.; Roudier, L.; Bucci, R.; Magrì, A.D.; Marini, F. Authentication of an Italian PDO hazelnut (“Nocciola Romana”) by NIR spectroscopy. Environ. Sci. Pollut. Res. 2018, 25, 28780–28786. [Google Scholar] [CrossRef]
- Kouvoutsakis, G.; Mitsi, C.; Tarantilis, P.A.; Polissiou, M.G.; Pappas, C.S. Geographical differentiation of dried lentil seed (Lens culinaris) samples using Diffuse Reflectance Fourier Transform Infrared Spectroscopy (DRIFTS) and discriminant analysis. Food Chem. 2014, 145, 1011–1014. [Google Scholar] [CrossRef]
- Dominguez-Vidal, A.; Pantoja-De La Rosa, J.; Cuadros-Rodríguez, L.; Ayora-Cañada, M.J. Authentication of canned fish packing oils by means of Fourier transform infrared spectroscopy. Food Chem. 2016, 190, 122–127. [Google Scholar] [CrossRef]
- Santha, N.; Sudha, K.G.; Vijayakumari, K.P.; Nayar, V.U.; Moorthy, S.N. Raman and infrared spectra of starch samples of sweet potato and cassava. J. Chem. Sci. 1990, 102, 705–712. [Google Scholar] [CrossRef]
- Xu, G.-Y.; Liao, A.-M.; Huang, J.-H.; Zhang, J.-G.; Thakur, K.; Wei, Z.-J. Evaluation of structural, functional, and anti-oxidant potential of differentially extracted polysaccharides from potatoes peels. Int. J. Biol. Macromol. 2019, 129, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Reddy, C.K.; Xu, B. A systematic comparative study on morphological, crystallinity, pasting, thermal and functional characteristics of starches resources utilized in China. Food Chem. 2018, 259, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Warren, F.J.; Gidley, M.J.; Flanagan, B.M. Infrared spectroscopy as a tool to characterise starch ordered structure—A joint FTIR–ATR, NMR, XRD and DSC study. Carbohydr. Polym. 2016, 139, 35–42. [Google Scholar] [CrossRef]
- Shi, M.; Jing, Y.; Yang, L.; Huang, X.; Wang, H.; Yan, Y.; Liu, Y. Structure and Physicochemical Properties of Malate Starches from Corn, Potato, and Wrinkled Pea Starches. Polymers 2019, 11, 1523. [Google Scholar] [CrossRef]
- Su, W.-H.; Bakalis, S.; Sun, D.-W. Fourier transform mid-infrared-attenuated total reflectance (FTMIR-ATR) microspectroscopy for determining textural property of microwave baked tuber. J. Food Eng. 2018, 218, 1–13. [Google Scholar] [CrossRef]
- Kizil, R.; Irudayaraj, J.; Seetharaman, K. Characterization of irradiated starches by using FT-Raman and FTIR spectroscopy. J. Agric. Food Chem. 2002, 50, 3912–3918. [Google Scholar] [CrossRef]
- Ding, X.; Ni, Y.; Kokot, S. NIR spectroscopy and chemometrics for the discrimination of pure, powdered, purple sweet potatoes and their samples adulterated with the white sweet potato flour. Chemom. Intell. Lab. Syst. 2015, 144, 17–23. [Google Scholar] [CrossRef]
- Dankar, I.; Haddarah, A.; Omar, F.E.L.; Pujolà, M.; Sepulcre, F. Characterization of food additive-potato starch complexes by FTIR and X-ray diffraction. Food Chem. 2018, 260, 7–12. [Google Scholar] [CrossRef]
- López, A.; Arazuri, S.; García, I.; Mangado, J.; Jarén, C. A review of the application of near-infrared spectroscopy for the analysis of potatoes. J. Agric. Food Chem. 2013, 61, 5413–5424. [Google Scholar] [CrossRef]
- Su, W.H.; Bakalis, S.; Sun, D.W. Chemometrics in tandem with near infrared (NIR) hyperspectral imaging and Fourier transform mid infrared (FT-MIR) microspectroscopy for variety identification and cooking loss determination of sweet potato. Biosyst. Eng. 2019, 180, 70–86. [Google Scholar] [CrossRef]
- Tierno, R.; López, A.; Riga, P.; Arazuri, S.; Jarén, C.; Benedicto, L.; Ruiz de Galarreta, J.I. Phytochemicals determination and classification in purple and red fleshed potato tubers by analytical methods and near infrared spectroscopy. J. Sci. Food Agric. 2016, 96, 1888–1899. [Google Scholar] [CrossRef] [PubMed]
- Di Cecco, V.; Di Musciano, M.; D’Archivio, A.A.; Frattaroli, A.R.; Di Martino, L. Analysis of intraspecific seed diversity in Astragalus aquilanus (Fabaceae), an endemic species of Central Apennine. Plant Biol. 2019, 21, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, L.; Del Vecchio, S.; Di Cecco, V.; Di Santo, M.; Stanisci, A.; Frattaroli, A.R. The role of GA3 in the germination process of high-mountain endemic and threatened species: Leontopodium nivalePinguicula fiorii and Soldanella minima subsp. samnitica (central Apennines, Italy). Plant Biosyst. 2014, 148, 1231–1238. [Google Scholar] [CrossRef]
- Aversano, R.; Caruso, I.; Aronne, G.; De Micco, V.; Scognamiglio, N.; Carputo, D. Stochastic changes affect Solanum wild species following autopolyploidization. J. Exp. Bot. 2013, 64, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Masi, P.; Aversano, R.; Carucci, F.; Palomba, S.; Carputo, D. Microstructure and tuber properties of potato varieties with different genetic profiles. Food Chem. 2018, 239, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Bontempo, P.; De Masi, L.; Carafa, V.; Rigano, D.; Scisciola, L.; Iside, C.; Grassi, R.; Molinari, A.M.; Aversano, R.; Nebbioso, A.; et al. Anticancer activities of anthocyanin extract from genotyped Solanum tuberosum L. “Vitelotte.”. J. Funct. Foods 2015, 19, 584–593. [Google Scholar] [CrossRef]
- Corliss, J.O.; Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification. Trans. Am. Microsc. Soc. 1974, 93, 303. [Google Scholar] [CrossRef]
- Garcia-Vallvé, S.; Palau, J.; Romeu, A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol. Biol. Evol. 1999, 16, 1125–1134. [Google Scholar] [CrossRef]
- Wold, S. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Brereton, R.G. Applied Chemometrics for Scientists; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2007; ISBN 0470016868. [Google Scholar]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Brereton, R.G.; Lloyd, G.R. Partial least squares discriminant analysis: Taking the magic away. J. Chemom. 2014, 28, 213–225. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Ghislain, M.; Núñez, J.; Del Rosario Herrera, M.; Pignataro, J.; Guzman, F.; Bonierbale, M.; Spooner, D.M. Robust and highly informative microsatellite-based genetic identity kit for potato. Mol. Breed. 2009, 23, 377–388. [Google Scholar] [CrossRef]
- Spooner, D.; van Treuren, R.C.; de Vicente, M.C. Molecular markers for genebank management. In IPGRI Tech. Bull. No. 10; IPGRI: Rome, Italy, 2005; ISBN 9789290436843. [Google Scholar]
- Leonel, M.; do Carmo, E.L.; Fernandes, A.M.; Soratto, R.P.; Ebúrneo, J.A.M.; Garcia, É.L.; dos Santos, T.P.R. Chemical composition of potato tubers: The effect of cultivars and growth conditions. J. Food Sci. Technol. 2017, 54, 2372–2378. [Google Scholar] [CrossRef]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef]
- Schulz, H.; Baranska, M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Rinnan, Å.; Van Den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]

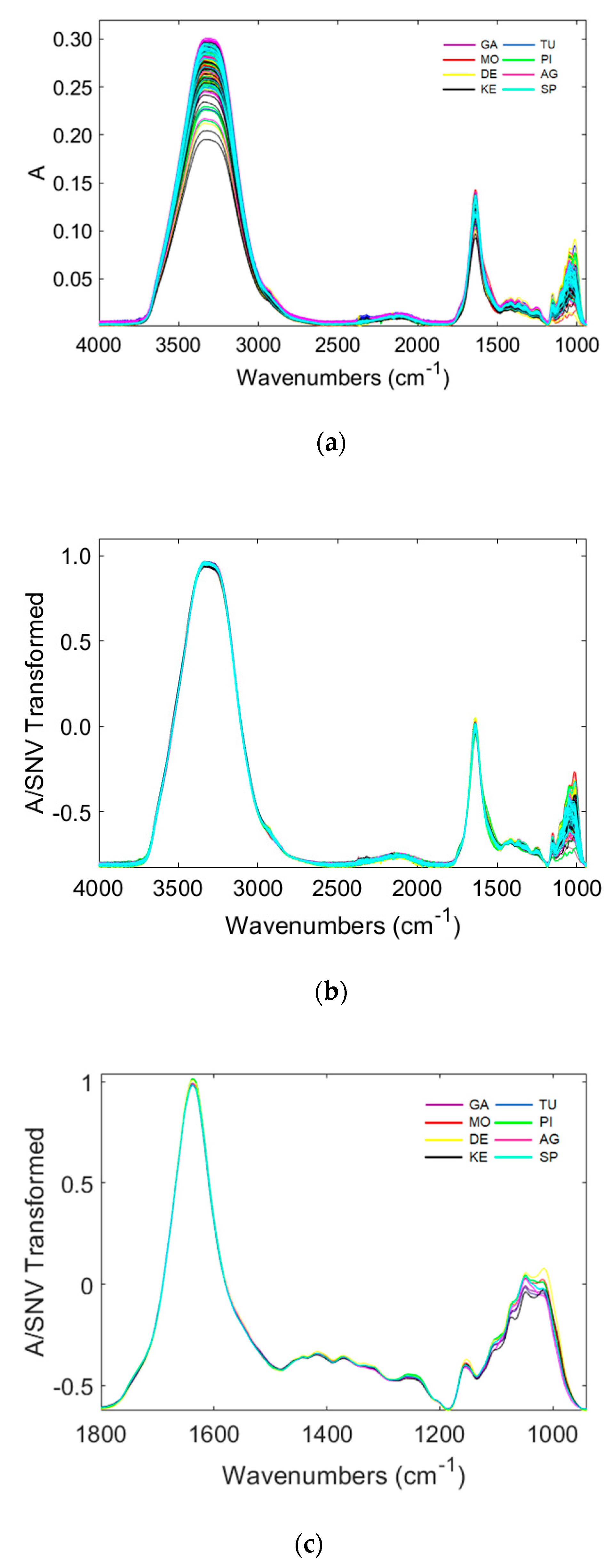
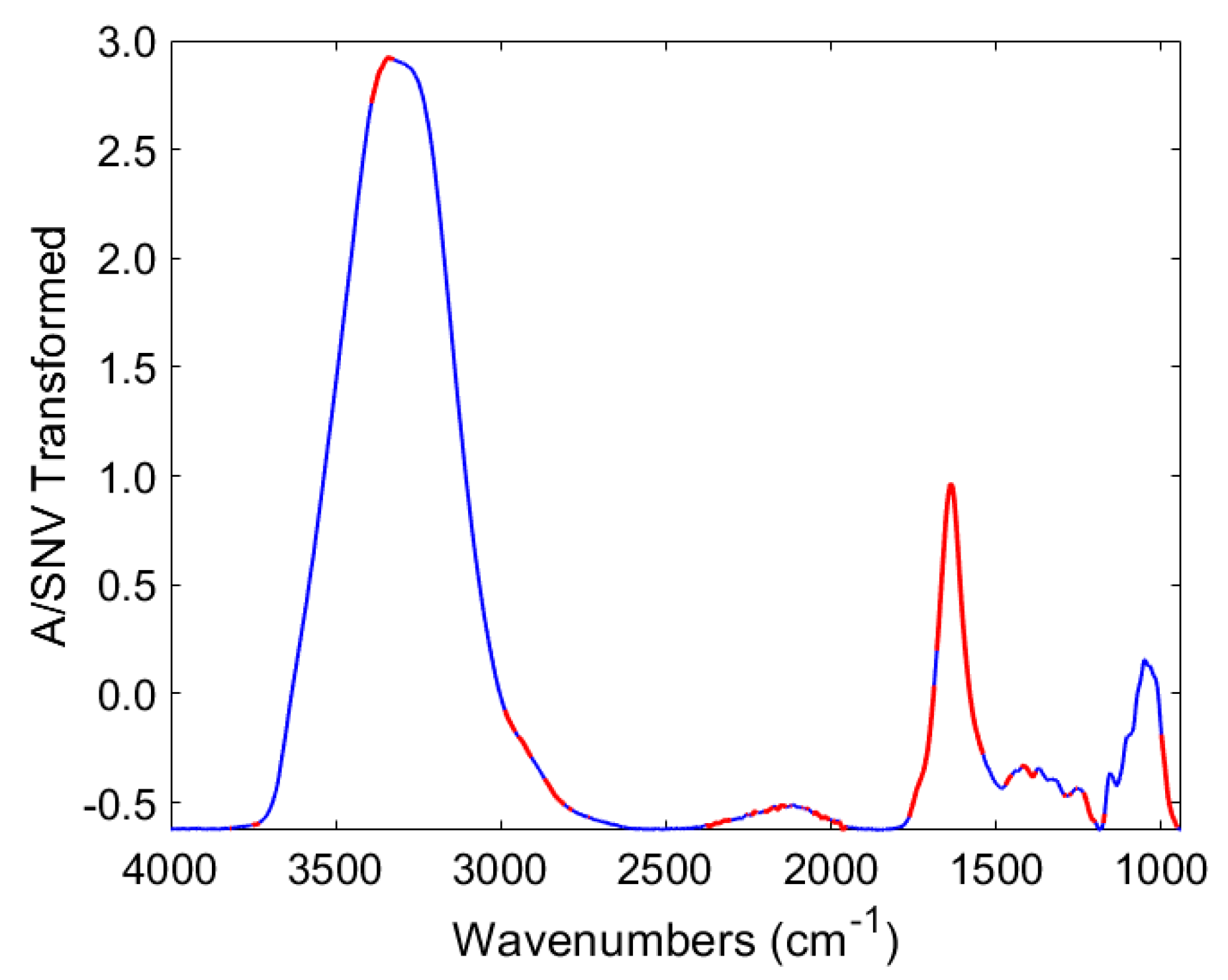
| Plant Part | Descriptors | Expressions | Abbreviation |
|---|---|---|---|
| Plant | Height | 1 = very short, 3 = short; 5 = medium; 7 = tall; 9 = very tall | 23 1 |
| Plant | Growth habit | 3 = upright; 5 = semi-upright; 7 = spreading | 13 |
| Plant | Anthocyanin coloration of stem | 1 = absent or very weak; 3 = weak; 5 = medium; 7 = strong; 9 = very strong | 14 |
| Plant | Foliage structure | 1 = stem type; 2 = intermediate type; 3 = leaf type | 12 |
| Plant | Flower frequency | 1 = absent or very low; 3 = low; 5 = medium; 7 = high; 9 = very high | 24 |
| Leaf | Openness | 1 = closed; 3 = intermediate; 5 = open | 16 |
| Leaf | Presence of secondary leaflets | 3 = weak; 5 = medium; 7 = strong | 17 |
| Leaf | Anthocyanin coloration on midrib of upper side | 1 = absent or very weak; 3 = weak; 5 = medium; 7 = strong; 9 = very strong | 19 |
| Leaf | Width in relation to length of lateral leaflets | 3 = narrow; 5 = medium; 7 = broad | 20 |
| Leaf | Frequency of coalescence of lateral leaflets | 1 = absent or very low; 3 = low; 5 = medium; 7 = high; 9 = very high | 21 |
| Flower | Anthocyanin coloration on peduncle of inflorescence | 1 = absent or very weak; 3 = weak; 5 = medium; 7 = strong; 9 = very strong | 26 |
| Flower | Intensity of anthocyanin coloration on inner side of corolla | 1 = absent or very weak; 3 = weak; 5 = medium; 7 = strong; 9 = very strong | 28 |
| Flower | Extent of anthocyanin coloration on inner side of corolla | 1 = absent or very small; 3 = small; 5 = medium; 7 = large; 9 = very large | 30 |
| Tuber | Shape | 1 = round; 2 = short oval; 3 = oval; 4 = long-oval; 5 = long; 6 = very long | 32 |
| Tuber | Color of skin | 1 = light beige; 2 = yellow; 3 = red; 4 = red parti-colored; 5 = blue; 6 = blue parti-colored; 7 = reddish brown | 34 |
| Tuber | Color of base of eye | 1 = white; 2 = yellow; 3 = red; 4 = blue | 35 |
| Tuber | Color of flesh | 1 = white; 2 = cream; 3 = light yellow; 4 = medium yellow; 5 = dark yellow; 6 = red; 7 = red parti-colored; 8 = blue; 9 = blue parti-colored | 36 |
| Tuber | Average tuber number | Number | N1 |
| Tuber | Average tuber weight | Weight (kg) | W |
| Tuber | Average tuber number (<40 mm) | Number | N2 |
| Tuber | Average tuber number (40–60 mm) | Number | N3 |
| Tuber | Average tuber number (>60 mm) | Number | N4 |
| Pre-Processing of ATR-FTIR Spectra | NER% in Cross-Validation |
|---|---|
| None | 87.1 |
| First derivative | 86.1 |
| Second derivative | 81.4 |
| SNV 1 | 97.4 |
| SNV + first derivative | 88.7 |
| SNV + second derivative | 79.9 |
| Class | GA | TU | MO | PI | DE | AG | KE | SP |
|---|---|---|---|---|---|---|---|---|
| Computed classes | 100.0 | 96.0 | 100.0 | 90.9 | 96.2 | 100.0 | 100.0 | 94.7 |
| Predicted classes | 75.0 | 72.7 | 84.6 | 80.0 | 81.8 | 90.9 | 77.8 | 75.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Donato, F.; Di Cecco, V.; Torricelli, R.; D’Archivio, A.A.; Di Santo, M.; Albertini, E.; Veronesi, F.; Garramone, R.; Aversano, R.; Marcantonio, G.; et al. Discrimination of Potato (Solanum tuberosum L.) Accessions Collected in Majella National Park (Abruzzo, Italy) Using Mid-Infrared Spectroscopy and Chemometrics Combined with Morphological and Molecular Analysis. Appl. Sci. 2020, 10, 1630. https://doi.org/10.3390/app10051630
Di Donato F, Di Cecco V, Torricelli R, D’Archivio AA, Di Santo M, Albertini E, Veronesi F, Garramone R, Aversano R, Marcantonio G, et al. Discrimination of Potato (Solanum tuberosum L.) Accessions Collected in Majella National Park (Abruzzo, Italy) Using Mid-Infrared Spectroscopy and Chemometrics Combined with Morphological and Molecular Analysis. Applied Sciences. 2020; 10(5):1630. https://doi.org/10.3390/app10051630
Chicago/Turabian StyleDi Donato, Francesca, Valter Di Cecco, Renzo Torricelli, Angelo Antonio D’Archivio, Marco Di Santo, Emidio Albertini, Fabio Veronesi, Raffaele Garramone, Riccardo Aversano, Giuseppe Marcantonio, and et al. 2020. "Discrimination of Potato (Solanum tuberosum L.) Accessions Collected in Majella National Park (Abruzzo, Italy) Using Mid-Infrared Spectroscopy and Chemometrics Combined with Morphological and Molecular Analysis" Applied Sciences 10, no. 5: 1630. https://doi.org/10.3390/app10051630
APA StyleDi Donato, F., Di Cecco, V., Torricelli, R., D’Archivio, A. A., Di Santo, M., Albertini, E., Veronesi, F., Garramone, R., Aversano, R., Marcantonio, G., & Di Martino, L. (2020). Discrimination of Potato (Solanum tuberosum L.) Accessions Collected in Majella National Park (Abruzzo, Italy) Using Mid-Infrared Spectroscopy and Chemometrics Combined with Morphological and Molecular Analysis. Applied Sciences, 10(5), 1630. https://doi.org/10.3390/app10051630









