A Deep-Learning Approach for Diagnosis of Metastatic Breast Cancer in Bones from Whole-Body Scans
Abstract
1. Introduction
1.1. Related Research in Breast Cancer Diagnosis Using Convolutional Neural Networks (CNNs)
1.2. Literature Review in Bone Metastasis Diagnosis from Breast Cancer Patients in Nuclear Medicine
1.3. Literature Review in Bone Metastasis Classification Using CNNs in Nuclear Medicine
1.4. Motivation and Aim of This Research Study
- The creation and demonstration of a customized, robust CNN-based classification tool for identification of metastatic breast cancer in bones from whole-body scans.
- The meticulous exploration of CNN hyper-parameter selection to define the best architecture and select the RGB mode selection that could lead to enhanced classification performance.
- The comparative experimental analysis performed for utilizing popular image classification CNN architectures, like ResNet50, VGG16, GoogleNET, etc.
2. Materials and Methods
2.1. Breast Cancer Patient Images
2.2. Main Aspects of Convolutional Neural Networks
2.3. Methodology
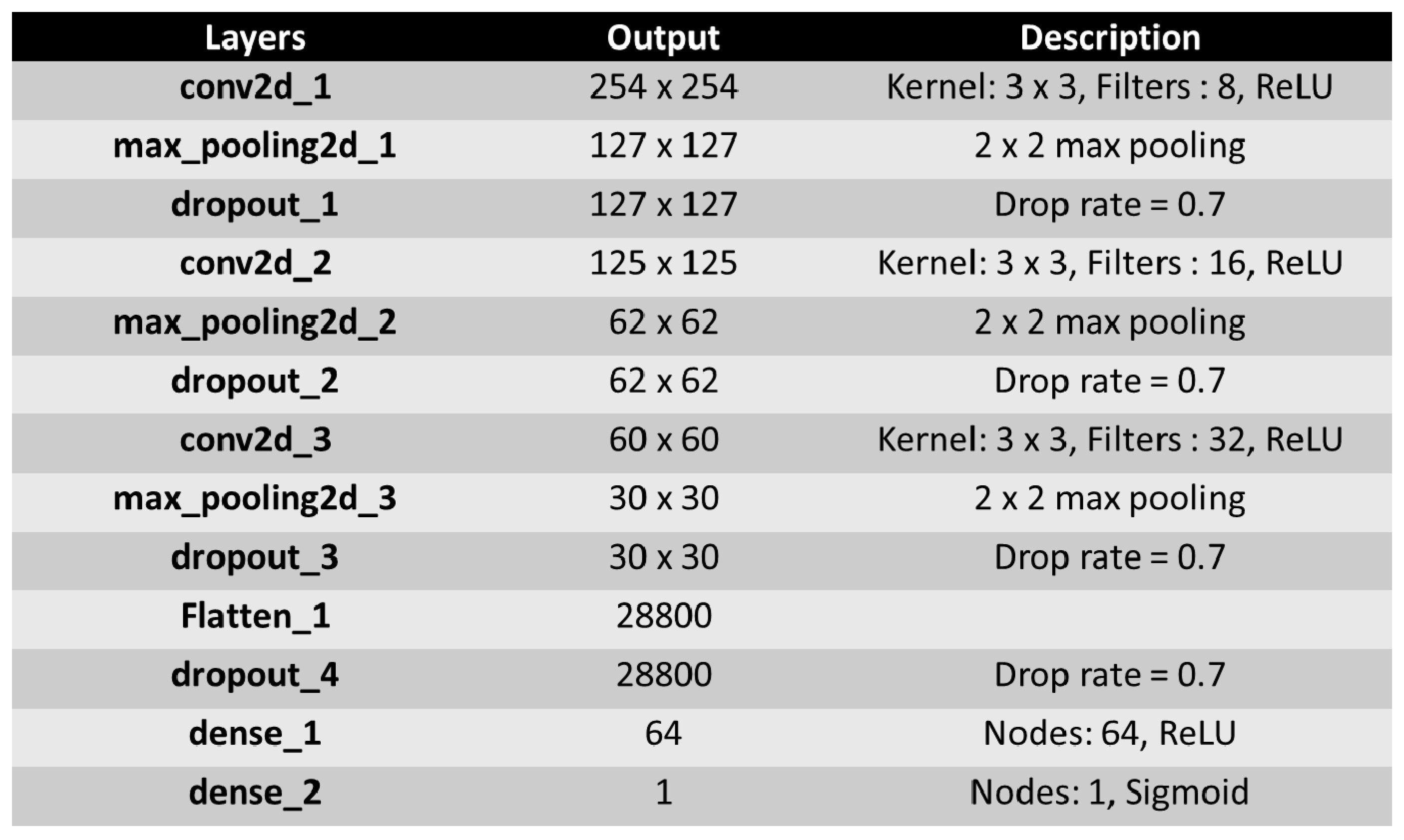
3. The Proposed CNN Architecture
4. Results
4.1. Classification Performance Evaluation
4.2. Confusion Matrix
4.3. Presentation of Results
4.4. Comparison with Benchmark CNNs
- ▪
- Best CNN: batch-size = 8, dropout = 0.7, nodes (3 conv layers) = 8,16,32, dense nodes: 64, epochs:200, pixel = (256,256,3)
- ▪
- VGG16: pixel size (200 × 200), batch size = 32, dropout = 0.2, dense nodes 2 × 512, epochs = 200
- ▪
- ResNet50: pixel size (224 × 224), batch size = 32, dropout = 0.5, dense nodes 32 × 32, epochs = 200
- ▪
- MobileNet: pixel size (250 × 250), batch size = 8, dropout = 0.5, global average pooling, dense = 1500 × 1500, epochs = 200
- ▪
- DenseNet: pixel size (200X200), batch size = 8, dropout = 0.9, global average pooling, dense nodes = (128 × 128), epochs = 200.
5. Discussion
- ▪
- The proposed CNN method exhibits outstanding performance when RGB analysis is performed, for the examined images of this case study. This can emanate from the fact that the results produced from the application of the CNN architecture are based on distinct features that appear specifically on the bone metastasis presence scans, compared to the healthy scans (no malignant spots).
- ▪
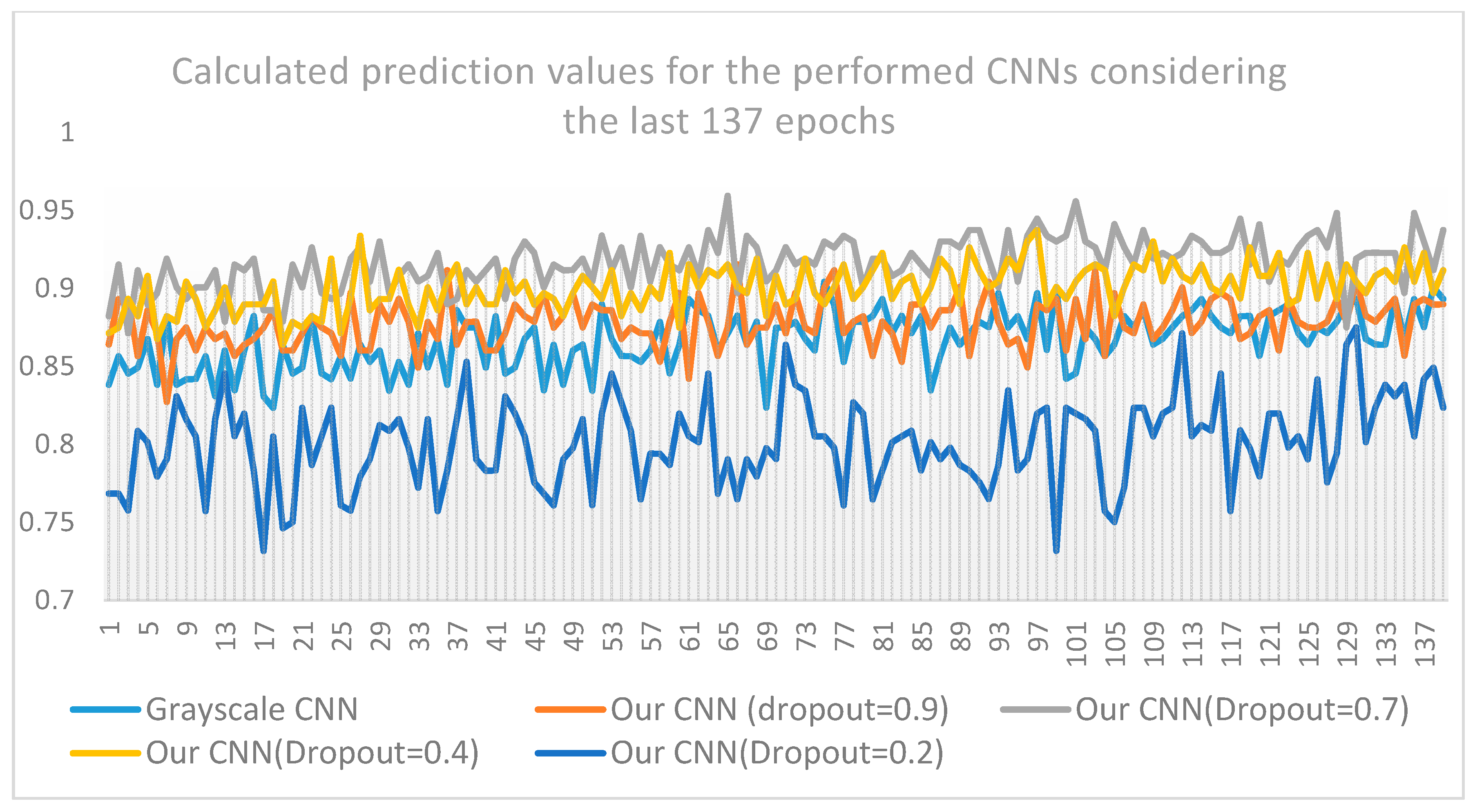
- The proposed CNN architecture is superior compared to three out of four benchmarks and well-known CNN architectures (ResNet50, VGG16, MobileNet, and DenseNet) which have been previously and efficiently used in medical image-processing problems. As observed from the aforementioned tables and figures, the results deriving from the application of the CNN method, are better in terms of classification accuracy, prediction, sensitivity, specificity and f1 score, than those coming from popular and well-known CNN approaches found in the relevant literature. Even if CNN has an inferior performance compared to DenseNet, the accuracy and loss values of the proposed CNN show a quite low variability whereas the corresponding DenseNet values present substantial variations among predicted outcomes (see Figure A3).
- ▪
- The proposed bone scan deep learning performs efficiently despite the fact that it was trained on a small number of images.
- ▪
- Overall, the deployed process seemingly improves the diagnostic effect of the deep-learning method, making it more efficient compared to other benchmark CNN architectures for image analysis. For the purpose of medical image analysis and classification, the CNN approach can be used effectively in the classification of whole-body images in nuclear medicine, outweighing the popular CNN architectures for medical image analysis.
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| CNN (100 × 100 × 3) | CNN (200 × 200 × 3) | CNN (300 × 300 × 3) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Batch size = 8 | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test |
| Run 1 | 82.81 | 0.76 | 73.21 | 1.04 | 85.94 | 0.17 | 92.86 | 0.17 | 87.5 | 0.296 | 96.428 | 0.157 |
| Run 2 | 82.81 | 0.76 | 87.5 | 0.3 | 92.19 | 0.2 | 91.1 | 0.29 | 89.06 | 0.247 | 94,64 | 0.167 |
| Run 3 | 81.25 | 0.39 | 80.36 | 0.64 | 81.25 | 0.28 | 87.5 | 0.24 | 87.5 | 0.27 | 89.28 | 0.206 |
| Run 4 | 84.37 | 0.37 | 87.5 | 0.45 | 84.37 | 0.25 | 83.93 | 0.27 | 92.187 | 0.21 | 89.28 | 0.255 |
| Run 5 | 81.25 | 0.39 | 76.78 | 0.54 | 92.19 | 0.2 | 92.86 | 0.15 | 87.5 | 0.276 | 89.28 | 0.235 |
| Average | 82.5 | 0.53 | 81.07 | 0.59 | 87.19 | 0.22 | 89.65 | 0.22 | 88.75 | 0.26 | 91.78 | 0.20 |
| Dropout = 0.2 | Dropout = 0.4 | Dropout = 0.9 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Batch size = 8 | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test |
| Run 1 | 93.75 | 0.22 | 91.1 | 0.21 | 87.5 | 0.44 | 92.86 | 0.18 | 71.87 | 0.55 | 71.43 | 0.57 |
| Run 2 | 84.37 | 0.45 | 83.93 | 0.29 | 89.1 | 0.47 | 85.71 | 0.34 | 81.25 | 0.41 | 87.5 | 0.37 |
| Run 3 | 83.37 | 0.38 | 85.67 | 0.28 | 87.5 | 0.21 | 89.28 | 0.17 | 92.19 | 0.36 | 85.71 | 0.41 |
| Run 4 | 95.31 | 0.7 | 89.29 | 0.17 | 87.5 | 0.25 | 85.71 | 0.4 | 79.69 | 0.43 | 87.5 | 0.46 |
| Run 5 | 90.62 | 0.21 | 92.86 | 0.23 | 82.1 | 0.36 | 87.25 | 0.43 | 84.37 | 0.4 | 78.57 | 0.43 |
| Average | 89.48 | 0.39 | 85.57 | 0.24 | 86.74 | 0.34 | 88.16 | 0.3 | 81.87 | 0.43 | 82.14 | 0.45 |
| VGG16 (Batch Size=32) | ||||
|---|---|---|---|---|
| Acc. Val | Loss Val | Acc Test | Loss Test | |
| Run 1 | 84.37 | 0.3 | 81.25 | 0.36 |
| Run 2 | 73.44 | 0.53 | 78.12 | 0.26 |
| Run 3 | 75 | 0.46 | 84.37 | 0.38 |
| Run 4 | 89.06 | 0.31 | 90.62 | 0.22 |
| Run 5 | 90.62 | 0.26 | 84.37 | 0.51 |
| Average | 82.49 | 0.37 | 83.74 | 0.34 |
Appendix B




References
- Roodman, G.D. Mechanisms of bone metastasis. N. Engl. J. Med. 2004, 350, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.E. Metastatic bone disease: Clinical features, pathophysiology and treatment strategies. Cancer Treat. Rev. 2001, 27, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Macedo, F.; Ladeira, K.; Pinho, F.; Saraiva, N.; Bonito, N.; Pinto, L.; Goncalves, F. Bone Metastases: An Overview. Oncol. Rev. 2017, 11, 321. [Google Scholar] [CrossRef] [PubMed]
- Battafarano, G.; Rossi, M.; Marampon, F.; Del Fattore, A. Management of bone metastases in cancer: A review. Crit. Rev. Oncol. Hematol. Int. J. Mol. Sci. 2005. [Google Scholar] [CrossRef]
- Manders, K.; van de Poll-Franse, L.V.; Creemers, G.J.; Vreugdenhil, G.; van der Sangen, M.J.; Nieuwenhuijzen, G.A.; Roumen, R.M.; Voogd, A.C. Clinical management of women with metastatic breast cancer: A descriptive study according to age group. BMC Cancer 2006, 6, 179. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, A.; Dorri, S.; Atashi, A.; Shirafkan, H.; Zabolinezhad, H. Bone Metastasis Prognostic Factors in Breast Cancer. Breast Cancer 2019, 13. [Google Scholar] [CrossRef]
- Coleman, R.E. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin. Cancer Res. 2006, 12, 6243s–6249s. [Google Scholar] [CrossRef]
- Muhammad, A.; Mehwish, I.; Muhammad, D.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef]
- Talbot, J.N.; Paycha, F.; Balogova, S. Diagnosis of bone metastasis: Recent comparative studies of imaging modalities. Q. J. Nucl. Med. Mol. Imaging 2011, 55, 374–410. [Google Scholar]
- O’Sullivan, G.J.; Carty, F.L.; Cronin, C.G. Imaging of bone metastasis: An update. World J. Radiol. 2015, 7, 202–211. [Google Scholar] [CrossRef]
- Chang, C.Y.; Gill, C.M.; Simeone, J.F.; Taneja, A.K.; Huang, A.J.; Torriani, M.; Bredella, M.A. Comparison of the diagnostic accuracy of 99 m-Tc-MDP bone scintigraphy and 18 F-FDG PET/CT for the detection of skeletal metastases. Acta Radiol. 2016, 57, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Savelli, G.; Maffioli, L.; Maccauro, M.; De Deckere, E.; Bombardieri, E. Bone scintigraphy and the added value of SPECT (single photon emission tomography) in detecting skeletal lesions. Q. J. Nucl. Med. 2001, 45, 27–37. [Google Scholar] [PubMed]
- Ghosh, P. The role of SPECT/CT in skeletal malignancies. Semin. Musculoskelet. Radiol. 2014, 18, 175–193. [Google Scholar] [CrossRef]
- Rieden, K. Conventional imaging and computerized tomography in diagnosis of skeletal metastases. Radiologe 1995, 35, 15–20. [Google Scholar]
- Hamaoka, T.; Madewell, J.E.; Podoloff, D.A.; Hortobagyi, G.N.; Ueno, N.T. Review—Bone imaging in metastatic breast cancer. J. Clin. Oncol. 2004, 22, 2942–2953. [Google Scholar] [CrossRef]
- Wyngaert, T.; Strobel, K.; Kampen, W.U.; Kuwert, T.; Bruggen, W.; Mohan, H.K.; Gnanasegaran, G.; Delgado-Bolton, R.; Weber, W.A.; Beheshti, M.; et al. The EANM practice guidelines for bone scintigraphy. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 1723–1738. [Google Scholar] [CrossRef]
- Even-Sapir, E.; Metser, U.; Mishani, E.; Lievshitz, G.; Lerman, H.; Leibovitch, I. The detection of bone metastases in patients with high-risk prostate cancer: 99mTc-MDP Planar bone scintigraphy, single- and multi-field-of-view SPECT, 18F-fluoride PET, and 18F-fluoride PET/CT. J. Nucl. Med. 2006, 47, 287–297. [Google Scholar]
- Hahn, S.; Heusner, T.; Kümmel, S.; Köninger, A.; Nagarajah, J.; Müller, S.; Boy, C.; Forsting, M.; Bockisch, A.; Antoch, G. Comparison of FDG-PET/CT and bone scintigraphy for detection of bone metastases in breast cancer. Acta Radiol. 2011, 52, 1009–1014. [Google Scholar] [CrossRef]
- Drakopoulos, M.; Vejdani, K.; Osman, M. Comparison of diagnostic certainty and accuracy of 18F-NaF PET/CT and planar 99mTc-MDP bone scan in patients with prostate cancer. J. Nucl. Med. 2014, 55, 1669. [Google Scholar]
- Nemoto, M.; Masutani, Y.; Nomura, Y.; Hanaoka, S.; Miki, S.; Yoshikawa, T.; Hayashi, N.; Ootomo, K. Machine Learning for Computer-aided Diagnosis. Igaku Butsuri 2016, 36, 29–34. [Google Scholar] [CrossRef]
- Suzuki, K. Machine Learning in Computer-Aided Diagnosis: Medical Imaging Intelligence and Analysis: 9781466600591: Medicine & Healthcare Books; University of Chicago: Chicago, IL, USA, 2012. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.; Setio, A.; Ciompi, F.; Ghafoorian, M.; Laak, J.; Ginneken, B.; Sánchez, C. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Cuadrado-Godia, E.; Laird, J.R.; Marinhoe, R.T.; Sanches, J.M.; Nicolaides, A.; et al. State-of-the-art review on deep learning in medical imaging. Front. Biosci. 2019, 24, 392–426. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46. [Google Scholar] [CrossRef] [PubMed]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, X.; Li, P.; Zhang, D.; Zhang, L. Part-based convolutional neural network for visual recognition. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017. [Google Scholar]
- Komeda, Y.; Handa, H.; Watanabe, T.; Nomura, T.; Kitahashi, M.; Sakurai, T.; Okamoto, A.; Minami, T.; Kono, M.; Arizumi, T.; et al. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology 2017, 93 (Suppl. S1), 30–34. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef]
- Gao, J.; Jiang, Q.; Zhou, B.; Chen, D. Convolutional neural networks for computer-aided detection or diagnosis in medical image analysis: An overview. Math. Biosci. Eng. 2019, 16, 6536–6561. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In European Conference on Computer Vision; Springer: Berlin, Germany, 2014; pp. 818–833. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks; Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Dutchess County, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, Hawaii, 21–26 July 2016. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. arXiv 2016, arXiv:1606.00915. [Google Scholar] [CrossRef]
- Nahid, A.; Kong, Y. Involvement of Machine Learning for Breast Cancer Image Classification: A Survey. Comput. Math. Methods Med. 2017, 2017, 29. [Google Scholar] [CrossRef]
- Cheng, H.; Shi, X.; Min, R.; Hu, L.; Cai, X.; Du, H. Approaches for automated detection and classification of masses in mammograms. Pattern Recognit. 2006, 39, 646–668. [Google Scholar] [CrossRef]
- Ponraj, D.N.; Jenifer, M.E.; Poongodi, D.P.; Manoharan, J.S. A survey on the preprocessing techniques of mammogram for the detection of breast cancer. J. Emerg. Trends Comput. Inf. Sci. 2011, 2, 656–664. [Google Scholar]
- Jiang, Y.; Chen, L.; Zhang, H.; Xiao, X. Breast cancer histopathological image classification using convolutional neural networks with small SE-ResNet module. PLoS ONE. 2019, 14, e0214587. [Google Scholar] [CrossRef]
- Sert, E.; Ertekin, S.; Halici, U. Ensemble of convolutional neural networks for classification of breast microcalcification from mammograms. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2017, 2017, 689–692. [Google Scholar]
- Rangayyan, R.M.; Ayres, F.J.; Desautels, J.L. A review of computer-aided diagnosis of breast cancer: Toward the detection of subtle signs. J. Frankl. Inst. 2007, 344, 312–348. [Google Scholar] [CrossRef]
- Magna, G.; Casti, P.; Jayaraman, S.V.; Salmeri, M.; Mencattini, A.; Martinelli, E.; Natale, C.D. Identification of mammography anomalies for breast cancer detection by an ensemble of classification models based on artificial immune system. Knowl. Based Syst. 2016, 101, 60–70. [Google Scholar] [CrossRef]
- Yassin, N.I.R.; Omran, S.; El Houby, E.M.F.; Allam, H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review. Comput. Methods Programs Biomed. 2018, 156, 25–45. [Google Scholar] [CrossRef]
- Gardezi, S.J.S.; Elazab, A.; Lei, B.; Wang, T. Breast Cancer Detection and Diagnosis Using Mammographic Data: Systematic Review. J. Med. Internet Res. 2019, 21, e14464. [Google Scholar] [CrossRef]
- Munir, K.; Elahi, H.; Ayub, A.; Frezza, F.; Rizzi, A. Cancer Diagnosis Using Deep Learning: A Bibliographic Review. Cancers 2019, 11, 1235. [Google Scholar] [CrossRef]
- Chougrad, H.; Zouaki, H.; Alheyane, O. Deep Convolutional Neural Networks for breast cancer screening. Comput. Methods Programs Biomed. 2018, 157, 19–30. [Google Scholar] [CrossRef]
- Abdelhafiz, D.; Yang, C.; Ammar, R.; Nabavi, S. Deep convolutional neural networks for mammography: Advances, challenges and applications. BMC Bioinform. 2019, 20 (Suppl. S11), 481. [Google Scholar] [CrossRef]
- CNNs Applied in Breast Cancer Classification. Available online: https://towardsdatascience.com/convolutional-neural-network-for-breast-cancer-classification-52f1213dcc9 (accessed on 10 November 2019).
- Kumar, K.; Chandra Sekhara Rao, A. Breast cancer classification of image using convolutional neural network. In Proceedings of the 2018 4th International Conference on Recent Advances in Information Technology (RAIT), Dhanbad, India, 15–17 March 2018; Available online: https://ieeexplore.ieee.org/abstract/document/8389034 (accessed on 6 January 2020).
- Suzuki, S.; Zhang, X.; Homma, N.; Ichiji, K.; Sugita, N.; Kawasumi, Y.; Ishibashi, T.; Yoshizawa, M. Mass detection using deep convolutional neural networks for mammoghraphic computer-aided diagnosis. In Proceedings of the 55th Annual Conference of the Society of Intruments and Control Engineers of Japan (SICE), Tsukuba, Japan, 20–23 September 2016; pp. 1382–1386. [Google Scholar]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using convolutional neural networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2560–2567. [Google Scholar]
- Wichakam, I.; Vateekul, P. Combining deep convolutional networks and SVMs for mass detection on digital mammograms. In Proceedings of the 8th International Conference on Knowledge and Smart Technology (KST), Bangkok, Thailand, 3–6 February 2016; pp. 239–244. [Google Scholar]
- Swiderski, B.; Kurek, J.; Osowski, S.; Kruk, M.; Barhoumi, W. Deep learning and non-negative matrix factorization in recognition of mammograms. In Proceedings of the Eighth International Conference on Graphic and Image Processing, International Society of Optics and Photonics, Tokyo, Japan, 8 February 2017; Volume 10225, p. 102250B. [Google Scholar]
- Kallenberg, M.; Petersen, K.; Nielsen, M.; Ng, A.Y.; Diao, P.; Igel, C.; Vachon, C.M.; Holland, K.; Winkel, R.R.; Karssemeijer, N.; et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans. Med. Imaging 2016, 35, 1322–1331. [Google Scholar] [CrossRef]
- Giger, M.L.; Vybomy, C.L.; Huo, Z.; Kupinski, M.A. Computer-aided diagnosis in mammography. In Handbook of Medical Imaging, 2nd ed.; Breast Cancer Detection and Diagnosis Using Mammographic Data: Systematic Review; SPIE Digital Library: Cardiff, Wales, 2000; pp. 915–1004. [Google Scholar]
- Fenton, J.J.; Taplin, S.H.; Carney, P.A.; Abraham, L.; Sickles, E.A.; D’Orsi, C.; Berns, E.A.; Cutter, G.; Hendrick, R.E.; Barlow, W.E.; et al. Influence of computer-aided detection on performance of screening mammography. N. Engl. J. Med. 2017, 356, 1399–1409. [Google Scholar] [CrossRef]
- Zhou, L.Q.; Wu, X.L.; Huang, S.Y.; Wu, G.G.; Ye, H.R.; Wei, Q.; Bao, L.Y.; Deng, Y.B.; Li, X.R.; Cui, X.W.; et al. Lymph Node Metastasis Prediction from Primary Breast Cancer US Images Using Deep Learning. Radiology 2020, 294, 19–28. [Google Scholar] [CrossRef]
- Steiner, D.F.; MacDonald, R.; Liu, Y.; Truszkowski, P.; Hipp, J.D.; Gammage, C.; Thng, F.; Peng, L.; Stumpe, M.C. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am. J. Surg. Pathol. 2018, 42, 1636–1646. [Google Scholar] [CrossRef]
- Takayoshi, U.; Sachiko, Y.; Seigo, Y.; Takeshi, A.; Naoki, M.; Masahiro, E.; Hiroyoshi, F.; Yoshihiro, U.; An Junichiro, W. Comparison of FDG PET and SPECT for Detection of Bone Metastases in Breast Cancer. Breast Imaging Am. J. Roentgenol. Diagn. Adv. Search 2005, 184, 1266–1273. [Google Scholar]
- Soyeon, P.; Yoon, J.K.; Lee, S.J.; Kang, S.Y.; Yim, H.; An, Y.S. Prognostic utility of FDG PET/CT and bone scintigraphy in breast cancer patients with bone-only metastasis. Medicine 2017, 96, e8985. [Google Scholar]
- Łukaszewski, B.; Nazar, J.; Goch, M.; Łukaszewska, M.; Stępiński, A.; Jurczyk, M.U. Diagnostic methods for detection of bone metastases. Contemp. Oncol. 2017, 21, 98–103. [Google Scholar] [CrossRef]
- Aslantas, A.; Dandil, E.; Saǧlam, S.; Çakiroǧlu, M. CADBOSS: A computer-aided diagnosis system for whole-body bone scintigraphy scans. J. Can. Res. Ther. 2016, 12, 787–792. [Google Scholar] [CrossRef]
- Sadik, M. Computer-Assisted Diagnosis for the Interpretation of Bone Scintigraphy: A New Approach to Improve Diagnostic Accuracy. Ph.D. Thesis, University of Gothenburg, Gothenburg, Sweden, June 2019. [Google Scholar]
- Fogelman, I.; Cook, G.; Israel, O.; Van der Wall, H. Positron emission tomography and bone metastases. Semin. Nucl. Med. 2005, 35, 135–142. [Google Scholar] [CrossRef]
- Pianou, N.K.; Stavrou, P.Z.; Vlontzou, E.; Rondogianni, P.; Exarhos, D.N.; Datseris, I.E. More advantages in detecting bone and soft tissue metastases from prostate cancer using 18F-PSMA PET/CT. Hell. J. Nucl. Med. 2019, 22, 6–9. [Google Scholar] [CrossRef]
- Newberg, A. Bone Scans. In Radiology Secrets Plus, 3rd ed.; Elsevier: Cambridge, MA, USA, 2011; pp. 376–381. ISBN 978-0-323-06794-2. [Google Scholar] [CrossRef]
- Dang, J. Classification in Bone Scintigraphy Images Using Convolutional Neural Networks. Master’s Thesis, Lund University, Lund, Sweden, 2016. [Google Scholar]
- Bradshaw, T.; Perk, T.; Chen, S.; Hyung-Jun, I.; Cho, S.; Perlman, S.; Jeraj, R. Deep learning for classification of benign and malignant bone lesions in [F-18]NaF PET/CT images. J. Nucl. Med. 2018, 59 (Suppl. S1), 327. [Google Scholar]
- Furuya, S.; Kawauchi, K.; Hirata, K.; Manabe, O.; Watanabe, S.; Kobayashi, K.; Ichikawa, S.; Katoh, C.; Shiga, T.J. A convolutional neural network-based system to detect malignant findings in FDG PET-CT examinations. Nucl. Med. 2019, 60, 1210. [Google Scholar]
- Furuya, S.; Kawauchi, K.; Hirata, K.; Manabe, O.; Watanabe, S.; Kobayashi, K.; Ichikawa, S.; Katoh, S.; Shiga, T.J. Can CNN detect the location of malignant uptake on FDG PET-CT? Nucl. Med. 2019, 60 (Suppl. S1), 285. [Google Scholar]
- Kawauchi, K.; Hirata, K.; Katoh, C.; Ichikawa, S.; Manabe, O.; Kobayashi, K.; Watanabe, S.; Furuya, S.; Shiga, T. A convolutional neural network based system to prevent patient misidentification in FDG-PET examinations. Sci. Rep. 2019, 9, 7192. [Google Scholar] [CrossRef]
- Kawauchi, K.; Hirata, K.; Ichikawa, S.; Manabe, O.; Kobayashi, K.; Watanabe, S.; Haseyama, M.; Ogawa, T.; Togo, R.; Shiga, T.; et al. Strategy to develop convolutional neural network-based classifier for diagnosis of whole-body FDG PET images. Nucl. Med. 2018, 59 (Suppl. S1), 326. [Google Scholar]
- Gjertsson, K. Segmentation in Skeletal Scintigraphy Images Using CNNs. Master’s Thesis, Lund University, Lund, Sweden, 2017. [Google Scholar]
- Weiner, M.G.; Jenicke, L.; Mller, V.; Bohuslavizki, H.K. Artifacts and nonosseous, uptake in bone scintigraphy. Imaging reports of 20 cases. Radiol. Oncol. 2001, 35, 185–191. [Google Scholar]
- O’Shea, K.T.; Nash, R. An Introduction to Convolutional Neural Networks. 2015. Available online: https://arxiv.org/abs/1511.08458 (accessed on 12 November 2019).
- Albelwi, S.; Mahmood, A. A Framework for Designing the Architectures of Deep Convolutional Neural Networks. Entropy 2017, 19, 242. [Google Scholar] [CrossRef]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net, Proceedings of ICLR-2015. arXiv 2014, arXiv:1412.6806. [Google Scholar]
- Yamashita, R.; Nishio, M.; Kinh, R.; Togashi, G. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Fully Connected Layers in Convolutional Neural Networks: The Complete Guide. Available online: https://missinglink.ai/guides/convolutional-neural-networks/fully-connected-layers-convolutional-neural-networks-complete-guide/ (accessed on 5 January 2020).
- Jayalakshmi, T.; Santhakumaran, A. Statistical normalization and back propagation for classification. Int. J. Comput. Theory Eng. 2011, 3, 89–93. [Google Scholar] [CrossRef]
- Bishop, C.M.; Hart, P.E.; Stork, D.G. Pattern Recognition and Machine Learning; Springer: Berlin, Germany, 2006. [Google Scholar]
- Moustakidis, S.; Christodoulo, E.; Papageorgiou, E.; Kokkotis, C.; Papandrianos, N.; Tsaopoulos, D. Application of machine intelligence for osteoarthritis classification: A classical implementation and a quantum perspective. Quantum Mach. Intell. 2019. [Google Scholar] [CrossRef]
- Theodoridis, S.; Koutroumbas, K.; Stork, D.G. Pattern Recognition; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Labatut, V.; Cherifi, H. Accuracy measures for the comparison of classifiers. In Proceedings of the 5th International Conference on Information Technology, Amman, Jordan, 11–13 May 2011. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Loffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Flatten Layer What Does. Available online: https://www.google.com/search?q=flatten+layer+what+does&oq=flatten+layer+what+does&aqs=chrome.69i57j0l3.5821j0j7&sourceid=chrome&ie=UTF-8 (accessed on 3 January 2020).
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Chollet, F. “Keras.” GitHub Repository. 2015. Available online: https://github.com/fchollet/keras (accessed on 9 December 2019).
- Google Colab, Colaboratory Cloud Environment Supported by Google. Available online: https://colab.research.google.com/ (accessed on 25 January 2020).
- Jia, D.; Wei, D.; Richard, S.; Li-Jia, L.; Kai, L.; Li, F.-F. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in Network. Proceedings in ICLR 2013. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Densely Connected Convolutional Networks. Available online: https://arthurdouillard.com/post/densenet/ (accessed on 3 January 2020).
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, J.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. (IJCV) 2015, 115, 211–252. [Google Scholar] [CrossRef]
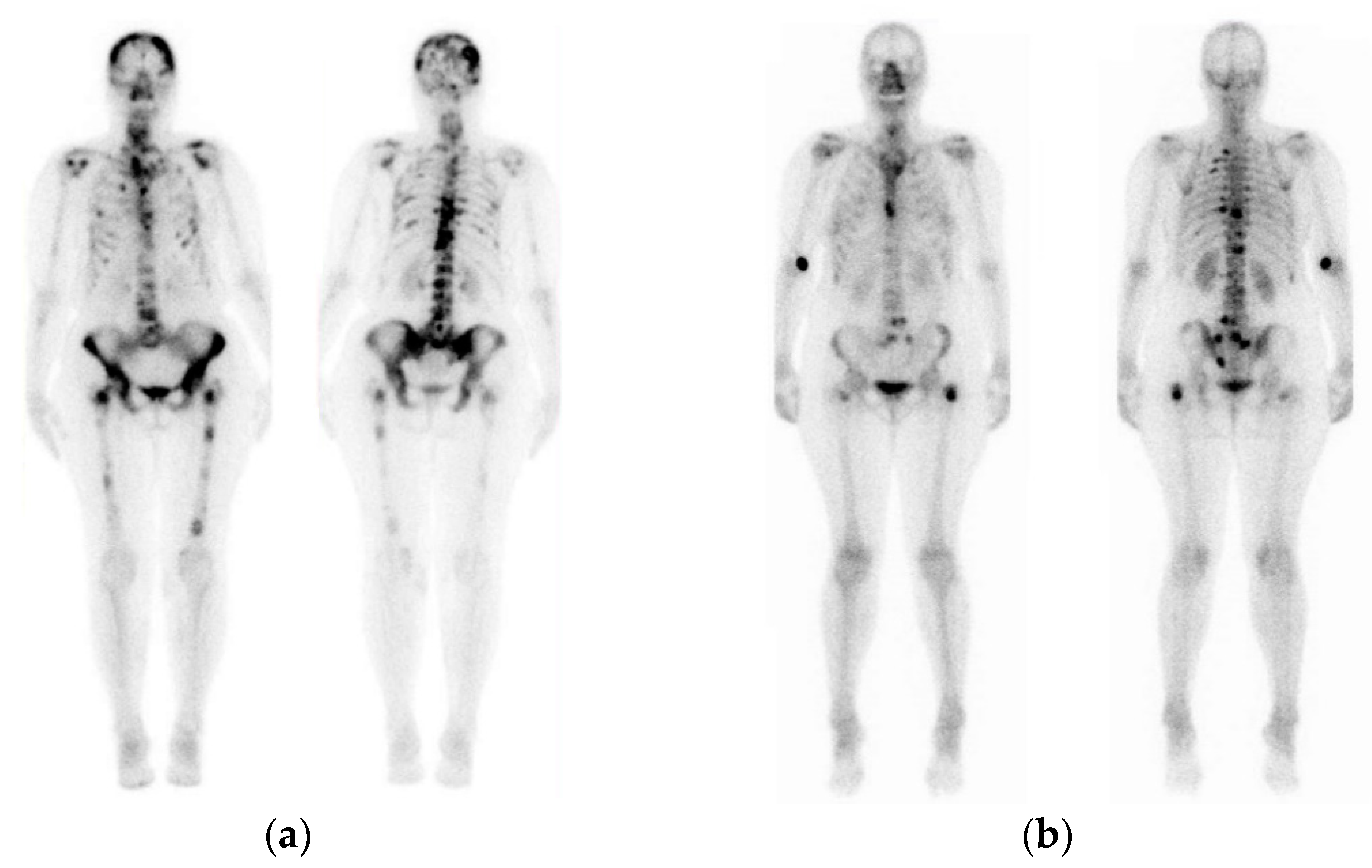
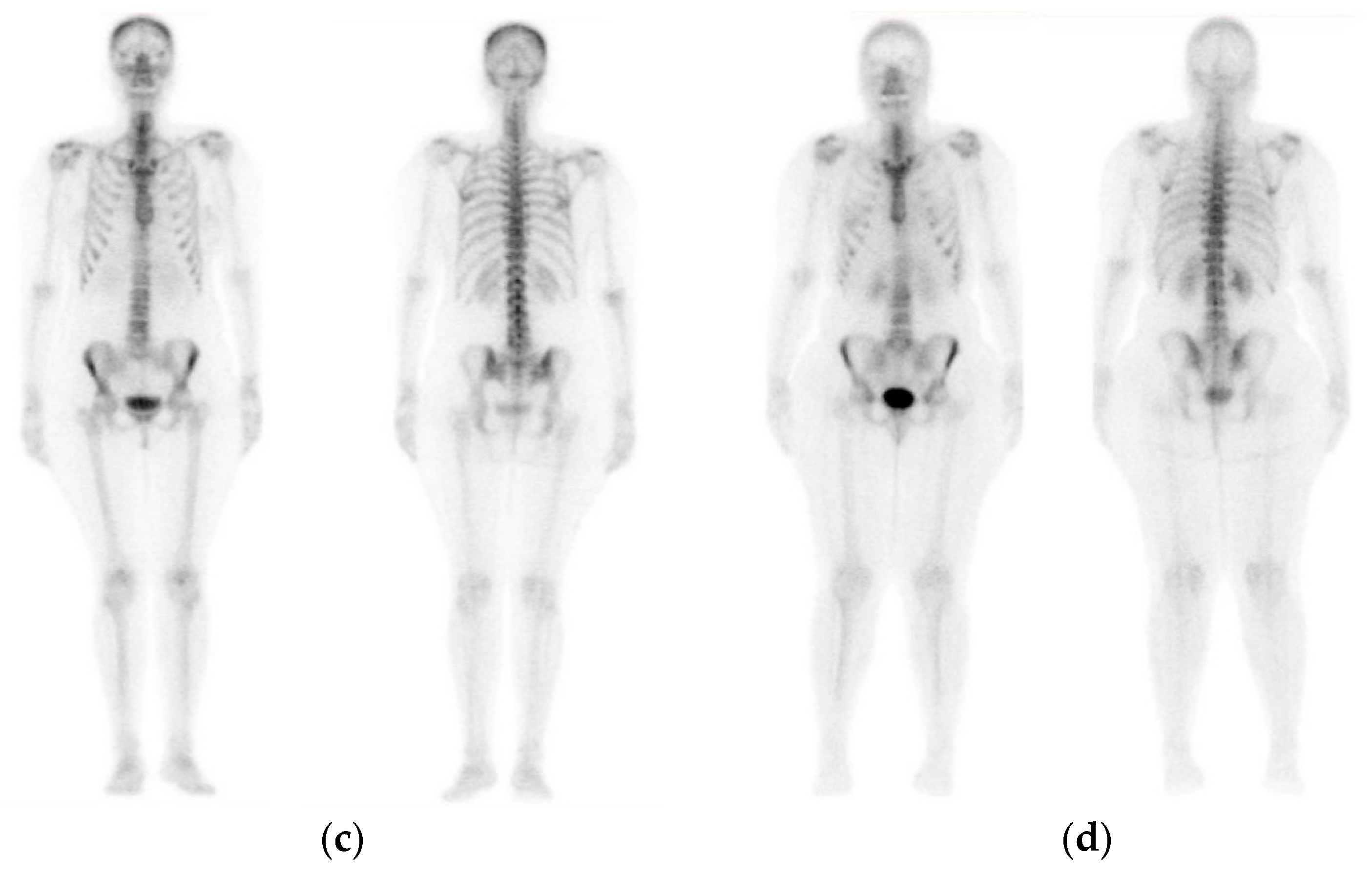





| Type of BC | Application Type | Modality | Reference | Data Available |
|---|---|---|---|---|
| Primary | classification | Histopathological images | Kumar K. et al. [51] | NO |
| Primary | detection | Mammograms | Swiderski, B. et al. [55] | YES |
| Primary | classification | Histopathological | Spanhol, F.A. et al. [53] | NO |
| Primary | mass detection | Mammograms | Wichakam, I. et al. [54] | NO |
| Primary | mass detection | Mammograms | Suzuki, S. et al. [52] | NO |
| Primary | segmentation | Mammograms | Kallenberg, M. et al. [56] | NO |
| Primary | mass detection | Mammograms | Fenton J.J. et al. [58] | NO |
| Primary | mass detection and classification | Mammograms | Cheng H. et al. [41] | NO |
| Primary | classification | Mammograms | Chougrad, H et al. [57] | NO |
| Metastatic in lymph nodes | prediction | Ultrasound images | Zhou Li-Q. et al. [59] | NO |
| Metastatic in lymph nodes | detection and classification | Histopathological | Steiner D.F. et al. [60] | NO |
| Application Type | Modality | Reference | Data Available |
|---|---|---|---|
| Metastatic prostate cancer classification | SPECT | [70] | NO |
| Metastatic prostate cancer classification | NaF PET/CT images | [71] | NO |
| Malignancy detection | FDG PET-CT | [72] | NO |
| Metastatic prostate cancer classification | FDG PET | [73] | NO |
| Patient’s sex prediction | FDG PET-CT | [74] | NO |
| predicts whether physician’s further diagnosis is required or not | FDG PET-CT | [75] | NO |
| Segmentation | SPECT | [76] | NO |
| Batch Size = 8 | Batch Size = 16 | Batch Size = 32 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | |
| Run 1 | 85.94 | 0.22 | 91.1 | 0.27 | 84.37 | 0.47 | 89.58 | 0.35 | 98.06 | 0.23 | 87.5 | 0.17 |
| Run 2 | 79.69 | 0.65 | 75 | 1.1 | 84.37 | 0.45 | 79.17 | 0.54 | 92.18 | 0.18 | 84.37 | 0.28 |
| Run 3 | 87.5 | 0.3 | 98.21 | 0.1 | 87.5 | 0.58 | 85.42 | 0.41 | 82.81 | 0.28 | 81.25 | 0.39 |
| Run 4 | 89.1 | 0.49 | 92.86 | 0.38 | 92.18 | 0.21 | 85.42 | 0.33 | 85.93 | 0.34 | 90.62 | 0.49 |
| Run 5 | 92.19 | 0.34 | 82.15 | 0.6 | 93.75 | 0.21 | 91.7 | 0.27 | 98.43 | 0.008 | 90.62 | 0.18 |
| Average | 86.89 | 0.4 | 87.86 | 0.49 | 88.43 | 0.38 | 86.26 | 0.38 | 91.48 | 0.207 | 86.87 | 0.305 |
| Batch Size = 8 | Batch Size = 16 | Batch Size = 32 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | |
| Run 1 | 93.75 | 0.21 | 89.28 | 0.37 | 93.75 | 0.15 | 83.33 | 0.37 | 89.06 | 0.42 | 93.75 | 0.207 |
| Run 2 | 95.31 | 0.35 | 94.64 | 0.26 | 90.62 | 0.29 | 89.58 | 0.23 | 95.31 | 0.16 | 87.5 | 0.33 |
| Run 3 | 92.18 | 0.23 | 92.86 | 0.18 | 95.31 | 0.16 | 93.75 | 0.22 | 90.62 | 0.22 | 93.75 | 0.18 |
| Run 4 | 93.75 | 0.24 | 92.85 | 0.19 | 89.1 | 0.29 | 89.58 | 0.22 | 95.31 | 0.15 | 87.5 | 0.314 |
| Run 5 | 87.5 | 0.29 | 92.85 | 0.24 | 90.62 | 0.23 | 87.51 | 0.26 | 92.18 | 0.24 | 93.75 | 0.239 |
| Run 6 | 89.06 | 0.3 | 89.28 | 0.22 | 89.1 | 0.29 | 97.92 | 0.14 | 82.81 | 0.35 | 90.62 | 0.227 |
| Run 7 | 90.62 | 0.287 | 94.64 | 0.21 | 84.38 | 0.32 | 95.83 | 0.2 | 98.43 | 0.16 | 93.75 | 0.21 |
| Run 8 | 92.18 | 0.22 | 87.5 | 0.25 | 92.18 | 0.23 | 93.75 | 0.19 | 85.93 | 0.28 | 87.5 | 0.232 |
| Run 9 | 84.37 | 0.33 | 96.42 | 0.16 | 85.7 | 0.32 | 95.83 | 0.24 | 90.62 | 0.21 | 93.75 | 0.19 |
| Run 10 | 93.75 | 0.176 | 94.64 | 0.154 | 89.06 | 0.37 | 87.5 | 0.26 | 85.93 | 0.39 | 87.5 | 0.32 |
| Average | 91.25 | 0.263 | 92.49 | 0.224 | 89.98 | 0.27 | 91.46 | 0.23 | 90.62 | 0.26 | 90.94 | 0.25 |
| Dropout = 0.2 | Dropout = 0.4 | Dropout = 0.9 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Batch Size = 8 | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test |
| Run 1 | 85.94 | 0.22 | 91.1 | 0.27 | 93.75 | 0.22 | 93.75 | 0.23 | 95.31 | 0.31 | 85.71 | 0.43 |
| Run 2 | 79.69 | 0.65 | 75 | 1.1 | 87.5 | 0.42 | 85.71 | 0.32 | 82.81 | 0.37 | 82.14 | 0.38 |
| Run 3 | 87.5 | 0.3 | 98.21 | 0.1 | 84.37 | 0.34 | 89.28 | 0.23 | 79.69 | 0.47 | 73.21 | 0.48 |
| Run 4 | 89.1 | 0.49 | 92.86 | 0.38 | 85.94 | 0.29 | 89.28 | 0.23 | 87.5 | 0.36 | 80.36 | 0.43 |
| Run 5 | 92.19 | 0.34 | 82.15 | 0.6 | 85.94 | 0.23 | 83.93 | 0.39 | 84.37 | 0.36 | 89.28 | 0.27 |
| Average | 86.89 | 0.4 | 87.86 | 0.49 | 87.52 | 0.32 | 87.68 | 0.3 | 85.94 | 0.37 | 82.14 | 0.4 |
| Dense Nodes = 16 | Dense Nodes = 32 | Dense Nodes = 64 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Batch size = 8 | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test | Acc. Val | Loss Val | Acc Test | Loss Test |
| Run 1 | 95.31 | 0.19 | 91.1 | 0.18 | 85.94 | 0.39 | 91.1 | 0.23 | 93.75 | 0.21 | 89.28 | 0.37 |
| Run 2 | 95.31 | 0.23 | 91.1 | 0.22 | 92.19 | 0.18 | 85.71 | 0.25 | 95.31 | 0.35 | 94.64 | 0.26 |
| Run 3 | 85.94 | 0.29 | 87.5 | 0.32 | 92.19 | 0.25 | 87.50 | 0.27 | 92.18 | 0.237 | 92.86 | 0.18 |
| Run 4 | 95.31 | 0.17 | 91.1 | 0.23 | 90.62 | 0.23 | 91.10 | 0.27 | 93.75 | 0.238 | 92.86 | 0.19 |
| Run 5 | 87.5 | 0.27 | 92.86 | 0.16 | 90.62 | 0.22 | 94.64 | 0.21 | 87.5 | 0.29 | 92.85 | 0.24 |
| Average | 91.87 | 0.23 | 90.73 | 0.22 | 90.31 | 0.25 | 90.01 | 0.25 | 92.50 | 0.27 | 92.50 | 0.25 |
| Runs | Precision | Recall | F1-Score | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|
| Run 1 | Malignant | 0.94 | 0.89 | 0.92 | 0.89 | 0.92 |
| Benign | 0.85 | 0.92 | 0.88 | 0.85 | 0.94 | |
| Run 2 | Malignant | 0.93 | 1.00 | 0.96 | 1.00 | 0.88 |
| Benign | 1.00 | 0.88 | 0.93 | 0.88 | 1.00 | |
| Run 3 | Malignant | 0.9 | 1.00 | 0.95 | 1.00 | 0.85 |
| Benign | 1.00 | 0.85 | 0.92 | 0.85 | 1.00 | |
| Run 4 | Malignant | 1.00 | 0.88 | 0.93 | 0.88 | 1.00 |
| Benign | 0.88 | 1.00 | 0.94 | 1.00 | 0.88 | |
| Run 5 | Malignant | 1.00 | 0.88 | 0.94 | 0.88 | 1.00 |
| Benign | 0.88 | 1.00 | 0.94 | 1.00 | 0.88 | |
| Run 6 | Malignant | 0.87 | 0.93 | 0.9 | 0.93 | 0.88 |
| Benign | 0.94 | 0.88 | 0.91 | 0.88 | 0.93 | |
| Run 7 | Malignant | 0.97 | 0.93 | 0.95 | 0.93 | 0.97 |
| Benign | 0.94 | 0.97 | 0.95 | 0.97 | 0.93 | |
| Run 8 | Malignant | 0.82 | 0.96 | 0.89 | 0.96 | 0.82 |
| Benign | 0.97 | 0.82 | 0.89 | 0.82 | 0.96 | |
| Run 9 | Malignant | 0.97 | 0.94 | 0.96 | 0.94 | 0.96 |
| Benign | 0.93 | 0.96 | 0.95 | 0.96 | 0.94 | |
| Run 10 | Malignant | 0.94 | 0.97 | 0.96 | 0.97 | 0.93 |
| Benign | 0.96 | 0.93 | 0.94 | 0.93 | 0.97 | |
| Average | Malignant | 0.934 | 0.938 | 0.936 | 0.94 | 0.92 |
| Benign | 0.935 | 0.921 | 0.925 | 0.91 | 0.94 |
| Title 1 | Malignant | Benign |
|---|---|---|
| Malignant | 31 | 1 |
| Benign | 2 | 25 |
| Grayscale | CNN Architecture (2,4,8), Epochs = 200, Batch Size = 8, Dense = 16, Drop-Rate = 0.2, Pixel Size (150 × 150) | |||
|---|---|---|---|---|
| Runs | Accuracy (Validation) | Loss (Validation) | Accuracy (Testing) | Loss (Testing) |
| Run 1 | 85.94 | 0.31 | 85.71 | 0.42 |
| Run 2 | 92.19 | 0.28 | 80.36 | 0.45 |
| Run 3 | 78.12 | 0.52 | 75 | 0.51 |
| Run 4 | 87.5 | 0.26 | 78.57 | 0.61 |
| Run 5 | 84.37 | 0.33 | 89.28 | 0.3 |
| Average | 85.62 | 0.34 | 81.78 | 0.46 |
| Grayscale | Precision | Recall | F1-score | Sensitivity | Specificity |
| Malignant | 0.82 | 0.97 | 0.89 | 0.82 | 0.96 |
| Benign | 0.96 | 0.76 | 0.85 | 0.96 | 0.82 |
| Confusion matrix | Malignant | Benign | |||
| Malignant | 32 | 1 | |||
| Benign | 7 | 22 |
| Average | Best CNN | ResNet50 | VGG16 | MobileNet | DenseNet |
|---|---|---|---|---|---|
| Acc. Validation | 92.50 | 91.87 | 82.49 | 88.13 | 93.75 |
| Loss Validation | 0.27 | 0.24 | 0.37 | 0.25 | 0.15 |
| Acc. Testing | 92.50 | 91.87 | 83.75 | 85.36 | 95 |
| Loss Testing | 0.25 | 0.19 | 0.34 | 0.35 | 0.176 |
| Best CNN | ResNet50 | VGG16 | MobileNet | DenseNet | |
|---|---|---|---|---|---|
| Accuracy | 92.50 | 91.87 | 83.75 | 85.36 | 95 |
| Precision | 0.93 | 0.84 | 0.85 | 0.78 | 0.92 |
| Recall | 0.94 | 1 | 0.9 | 0.99 | 0.99 |
| F1-score | 0.94 | 0.91 | 0.86 | 0.87 | 0.95 |
| Sensitivity | 0.94 | 1 | 0.89 | 0.99 | 0.99 |
| Specificity | 0.92 | 0.78 | 0.75 | 0.66 | 0.9 |
| Best CNN | ResNet50 | VGG16 | MobileNet | DenseNet | |
|---|---|---|---|---|---|
| Accuracy | 92.50 | 91.87 | 83.75 | 85.36 | 95 |
| Precision | 0.94 | 1 | 0.87 | 0.99 | 0.98 |
| Recall | 0.92 | 0.78 | 0.75 | 0.66 | 0.9 |
| F1-score | 0.93 | 0.87 | 0.77 | 0.79 | 0.94 |
| Sensitivity | 0.91 | 0.78 | 0.75 | 0.66 | 0.91 |
| Specificity | 0.94 | 1 | 0.89 | 0.99 | 0.99 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Feleki, A. A Deep-Learning Approach for Diagnosis of Metastatic Breast Cancer in Bones from Whole-Body Scans. Appl. Sci. 2020, 10, 997. https://doi.org/10.3390/app10030997
Papandrianos N, Papageorgiou E, Anagnostis A, Feleki A. A Deep-Learning Approach for Diagnosis of Metastatic Breast Cancer in Bones from Whole-Body Scans. Applied Sciences. 2020; 10(3):997. https://doi.org/10.3390/app10030997
Chicago/Turabian StylePapandrianos, Nikolaos, Elpiniki Papageorgiou, Athanasios Anagnostis, and Anna Feleki. 2020. "A Deep-Learning Approach for Diagnosis of Metastatic Breast Cancer in Bones from Whole-Body Scans" Applied Sciences 10, no. 3: 997. https://doi.org/10.3390/app10030997
APA StylePapandrianos, N., Papageorgiou, E., Anagnostis, A., & Feleki, A. (2020). A Deep-Learning Approach for Diagnosis of Metastatic Breast Cancer in Bones from Whole-Body Scans. Applied Sciences, 10(3), 997. https://doi.org/10.3390/app10030997







