Screening Patients with Early Stage Parkinson’s Disease Using a Machine Learning Technique: Measuring the Amount of Iron in the Basal Ganglia
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. MR Data Acquisition
2.3. QSM Reconstruction
2.4. ROI Selection
2.5. Classification Models
2.6. Feature Selection
2.7. Performance Assessment
3. Results
3.1. QSM and F-Scores for Patients with Early PD and HCs
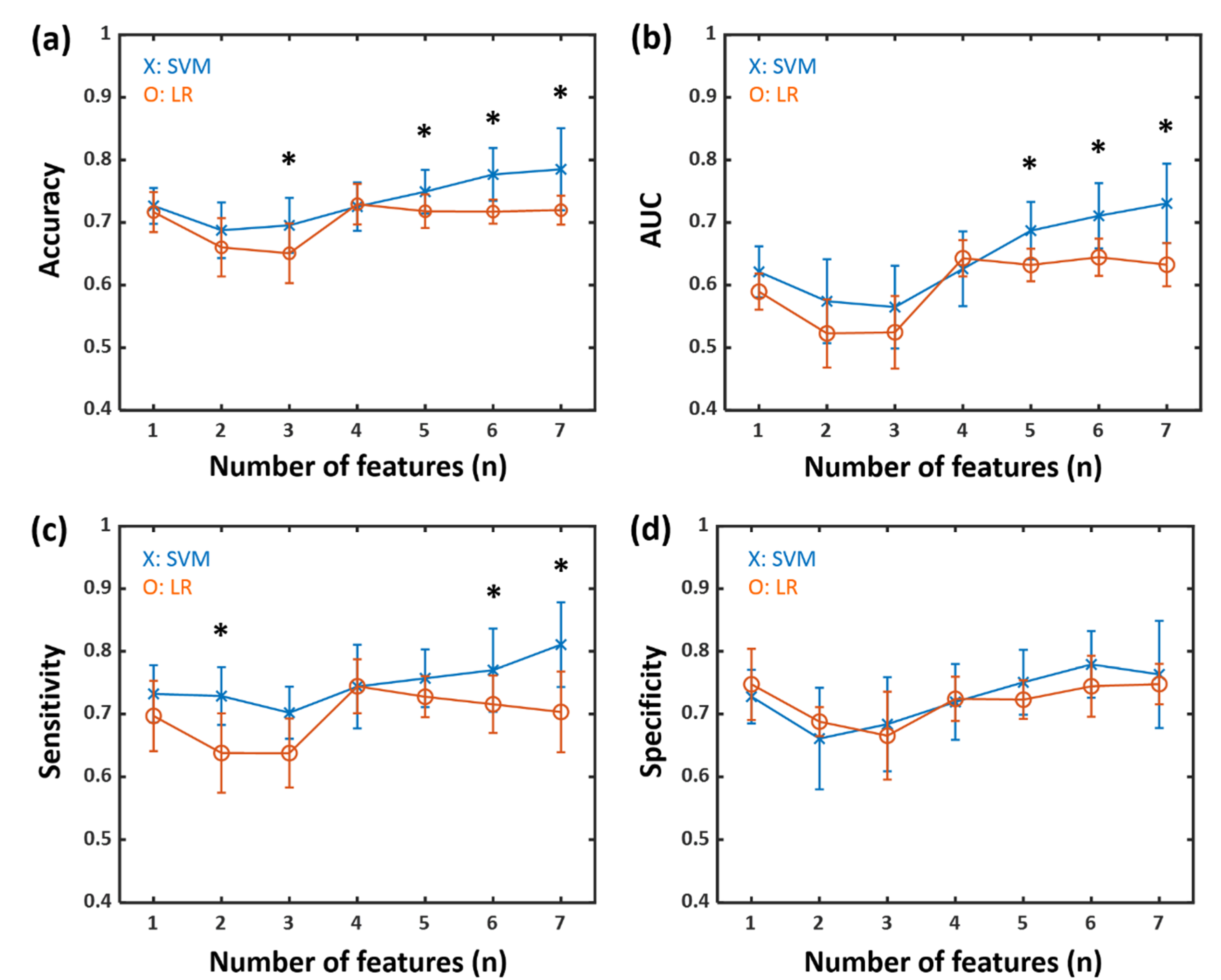
3.2. Classification Results for Patients with Early PD and HCs
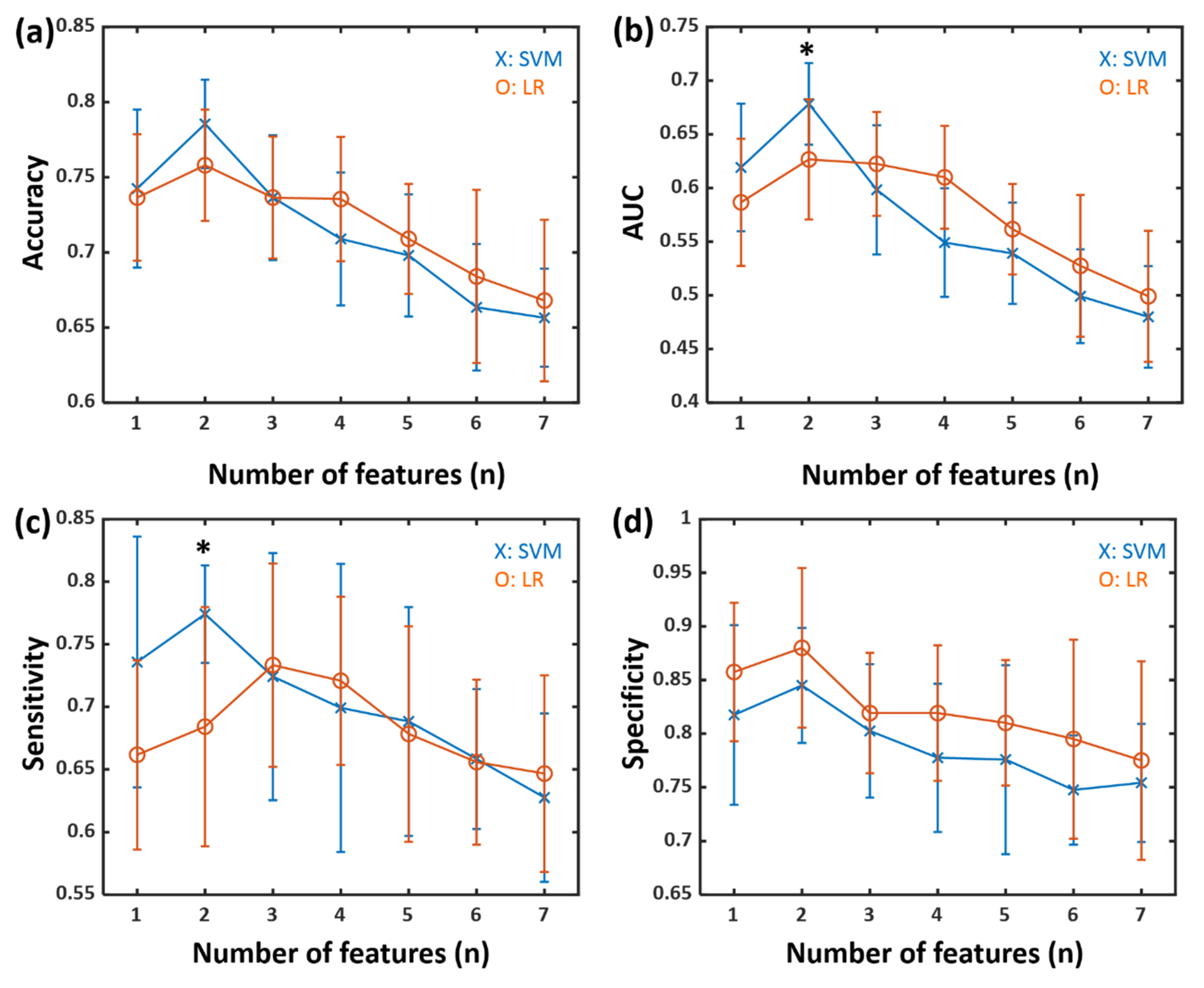
3.3. Classification Results for High and Low NMSS Score Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murman, D.L. Early treatment of Parkinson’s disease: Opportunities for managed care. Am. J. Manag. Care 2012, 18, S183–S188. [Google Scholar] [PubMed]
- De Lau, L.M.; Breteler, M.M. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006, 5, 525–535. [Google Scholar] [CrossRef]
- Chaudhuri, K.R.; Schapira, A.H.V. Non-motor symptoms of Parkinson’s disease: Dopaminergic pathophysiology and treatment. Lancet Neurol. 2009, 8, 464–474. [Google Scholar] [CrossRef]
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, K.R.; Healy, D.G.; Schapira, A.H.V. Non-motor symptoms of Parkinson’s disease: Diagnosis and management. Lancet Neurol. 2006, 5, 235–245. [Google Scholar] [CrossRef]
- Guan, X.; Xuan, M.; Gu, Q.; Huang, P.; Liu, C.; Wang, N.; Xu, X.; Luo, W.; Zhang, M. Regionally progressive accumulation of iron in Parkinson’s disease as measured by quantitative susceptibility mapping. NMR Biomed. 2017, 30, e3489. [Google Scholar] [CrossRef]
- Sofic, E.; Riederer, P.; Heinsen, H.; Beckmann, H.; Reynolds, G.; Hebenstreit, G.; Youdim, M. Increased iron (III) and total iron content in post mortem substantia nigra of parkinsonian brain. J. Neural. Transm. 1988, 74, 199–205. [Google Scholar] [CrossRef]
- Zecca, L.; Youdim, M.B.; Riederer, P.; Connor, J.R.; Crichton, R.R. Iron, brain ageing and neurodegenerative disorders. Nat. Rev. Neurosci. 2004, 5, 863–873. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, T. Quantitative susceptibility mapping (QSM): Decoding MRI data for a tissue magnetic biomarker. Magn. Reson Med. 2015, 73, 82–101. [Google Scholar] [CrossRef]
- Barbosa, J.H.O.; Santos, A.C.; Tumas, V.; Liu, M.; Zheng, W.; Haacke, E.M.; Salmon, C.E.G. Quantifying brain iron deposition in patients with Parkinson’s disease using quantitative susceptibility mapping, R2 and R2. Magn. Reson Imaging 2015, 33, 559–565. [Google Scholar] [CrossRef]
- Haller, S.; Badoud, S.; Nguyen, D.; Garibotto, V.; Lovblad, K.; Burkhard, P. Individual detection of patients with Parkinson disease using support vector machine analysis of diffusion tensor imaging data: Initial results. AJNR Am. J. Neuroradiol. 2012, 33, 2123–2128. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.; Badoud, S.; Nguyen, D.; Barnaure, I.; Montandon, M.; Lovblad, K.; Burkhard, P. Differentiation between Parkinson disease and other forms of Parkinsonism using support vector machine analysis of susceptibility-weighted imaging (SWI): Initial results. Eur. Radiol. 2013, 23, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Becker, G.; Müller, A.; Braune, S.; Büttner, T.; Benecke, R.; Greulich, W.; Klein, W.; Mark, G.; Rieke, J.; Thümler, R. Early diagnosis of Parkinson’s disease. J. Neurol. 2002, 249, iii40–iii48. [Google Scholar] [CrossRef]
- Hughes, A.; Daniel, S.; Kilford, L.; Lees, A. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 1992, 55, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Goetz, C.G.; Tilley, B.C.; Shaftman, S.R.; Stebbins, G.T.; Fahn, S.; Martinez-Martin, P.; Poewe, W.; Sampaio, C.; Stern, M.B.; Dodel, R. Movement Disorder Society-sponsored revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS): Scale presentation and clinimetric testing results. Mov. Disord. 2008, 23, 2129–2170. [Google Scholar] [CrossRef]
- Koh, S.-B.; Kim, J.W.; Ma, H.-I.; Ahn, T.-B.; Cho, J.W.; Lee, P.H.; Chung, S.J.; Kim, J.-S.; Kwon, D.Y.; Baik, J.S. Validation of the Korean-version of the nonmotor symptoms scale for Parkinson’s disease. J. Clin. Neurol. 2012, 8, 276–283. [Google Scholar] [CrossRef]
- Shin, C.; Lee, S.; Lee, J.-Y.; Rhim, J.H.; Park, S.-W. Non-Motor Symptom Burdens Are Not Associated with Iron Accumulation in Early Parkinson’s Disease: A Quantitative Susceptibility Mapping Study. J. Korean Med. Sci. 2018, 33, e96. [Google Scholar] [CrossRef]
- Li, W.; Avram, A.V.; Wu, B.; Xiao, X.; Liu, C. Integrated Laplacian-based phase unwrapping and background phase removal for quantitative susceptibility mapping. NMR Biomed. 2014, 27, 219–227. [Google Scholar] [CrossRef]
- Li, W.; Wang, N.; Yu, F.; Han, H.; Cao, W.; Romero, R.; Tantiwongkosi, B.; Duong, T.Q.; Liu, C. A method for estimating and removing streaking artifacts in quantitative susceptibility mapping. Neuroimage 2015, 108, 111–122. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Lemon, S.C.; Roy, J.; Clark, M.A.; Friedmann, P.D.; Rakowski, W. Classification and regression tree analysis in public health: Methodological review and comparison with logistic regression. Ann. Behav. Med. 2003, 26, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Lemeshow, S.; Hosmer, D.W., Jr. A review of goodness of fit statistics for use in the development of logistic regression models. Am. J. Epidemiol 1982, 115, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Lin, C.-J. Combining SVMs with various feature selection strategies. In Feature Extraction. Studies in Fuzziness and Soft Computing; Guyon, I., Nikravesh, M., Gunn, S., Zadeh, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 207, pp. 315–324. [Google Scholar]
- Stone, M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. Ser. B 1974, 36, 111–133. [Google Scholar] [CrossRef]
- Tolosa, E.; Wenning, G.; Poewe, W. The diagnosis of Parkinson’s disease. Lancet Neurol. 2006, 5, 75–86. [Google Scholar] [CrossRef]
- Rajput, A.; Rozdilsky, B.; Rajput, A. Accuracy of clinical diagnosis in parkinsonism—A prospective study. Can. J. Neurol. Sci. 1991, 18, 275–278. [Google Scholar] [CrossRef]
- Ye, F.Q.; Allen, P.S.; Martin, W.W. Basal ganglia iron content in Parkinson’s disease measured with magnetic resonance. Mov. Disord. 1996, 11, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Langkammer, C.; Pirpamer, L.; Seiler, S.; Deistung, A.; Schweser, F.; Franthal, S.; Homayoon, N.; Katschnig-Winter, P.; Koegl-Wallner, M.; Pendl, T. Quantitative susceptibility mapping in Parkinson’s disease. PLoS ONE 2016, 11, e0162460. [Google Scholar] [CrossRef]
- He, N.; Ling, H.; Ding, B.; Huang, J.; Zhang, Y.; Zhang, Z.; Liu, C.; Chen, K.; Yan, F. Region-specific disturbed iron distribution in early idiopathic Parkinson’s disease measured by quantitative susceptibility mapping. Hum. Brain Mapp. 2015, 36, 4407–4420. [Google Scholar] [CrossRef]
- Salazar, D.A.; Vélez, J.I.; Salazar, J.C. Comparison between SVM and logistic regression: Which one is better to discriminate? Rev. Colomb Estadística 2012, 35, 223–237. [Google Scholar]
- Fernández-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D. Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]
- Long, D.; Wang, J.; Xuan, M.; Gu, Q.; Xu, X.; Kong, D.; Zhang, M. Automatic classification of early Parkinson’s disease with multi-modal MR imaging. PLoS ONE 2012, 7, e47714. [Google Scholar] [CrossRef] [PubMed]
- Marcus, G. Deep learning: A critical appraisal. arXiv 2018, arXiv:1801.00631. [Google Scholar]
- Milletari, F.; Ahmadi, S.-A.; Kroll, C.; Plate, A.; Rozanski, V.; Maiostre, J.; Levin, J.; Dietrich, O.; Ertl-Wagner, B.; Bötzel, K.; et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput. Vis. Image 2017, 164, 92–102. [Google Scholar] [CrossRef]
- Cobzas, D.; Sun, H.; Walsh, A.J.; Lebel, R.M.; Blevins, G.; Wilman, A.H. Subcortical gray matter segmentation and voxel-based analysis using transverse relaxation and quantitative susceptibility mapping with application to multiple sclerosis. J. Magn. Reson Imaging 2015, 42, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
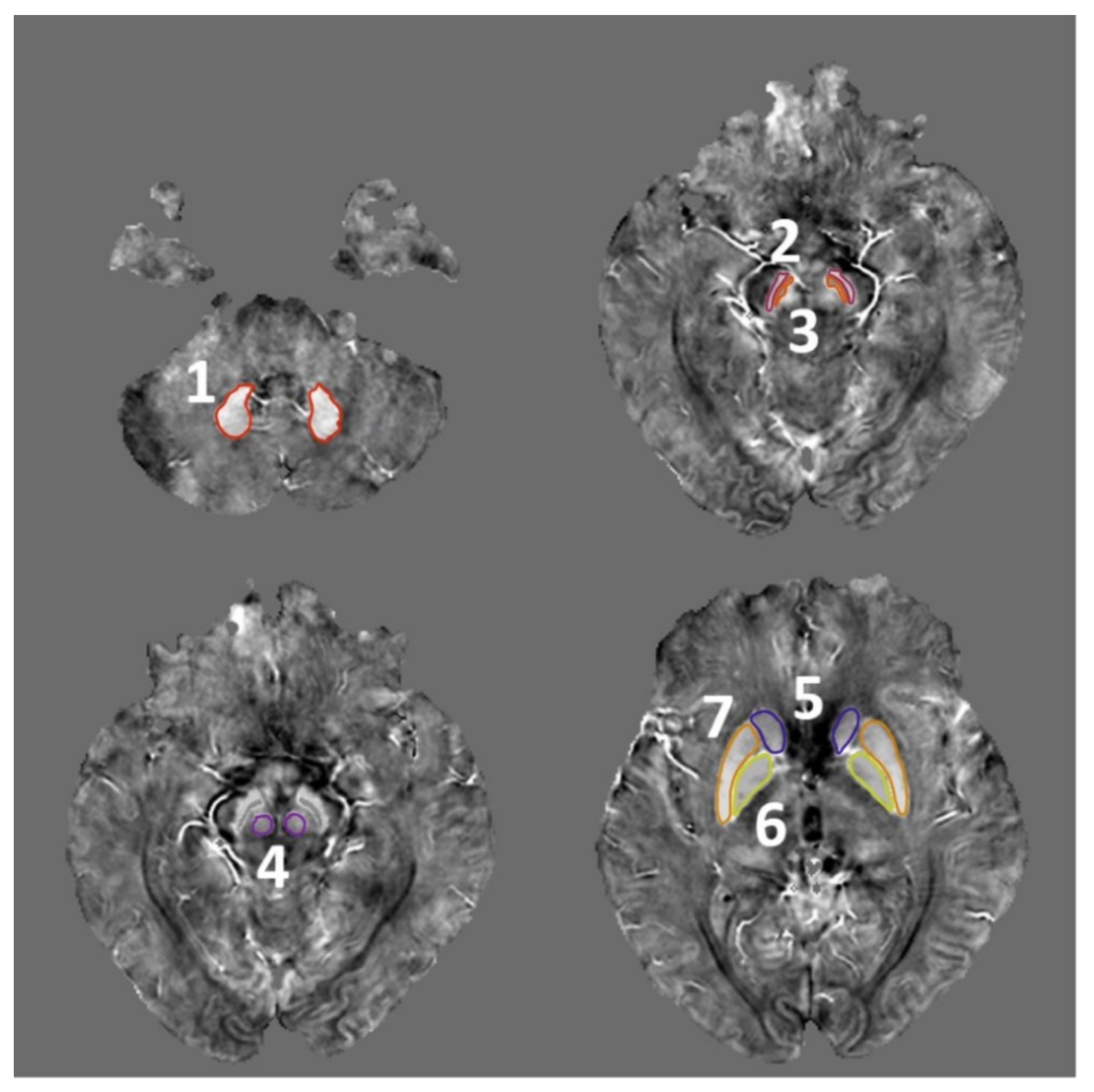
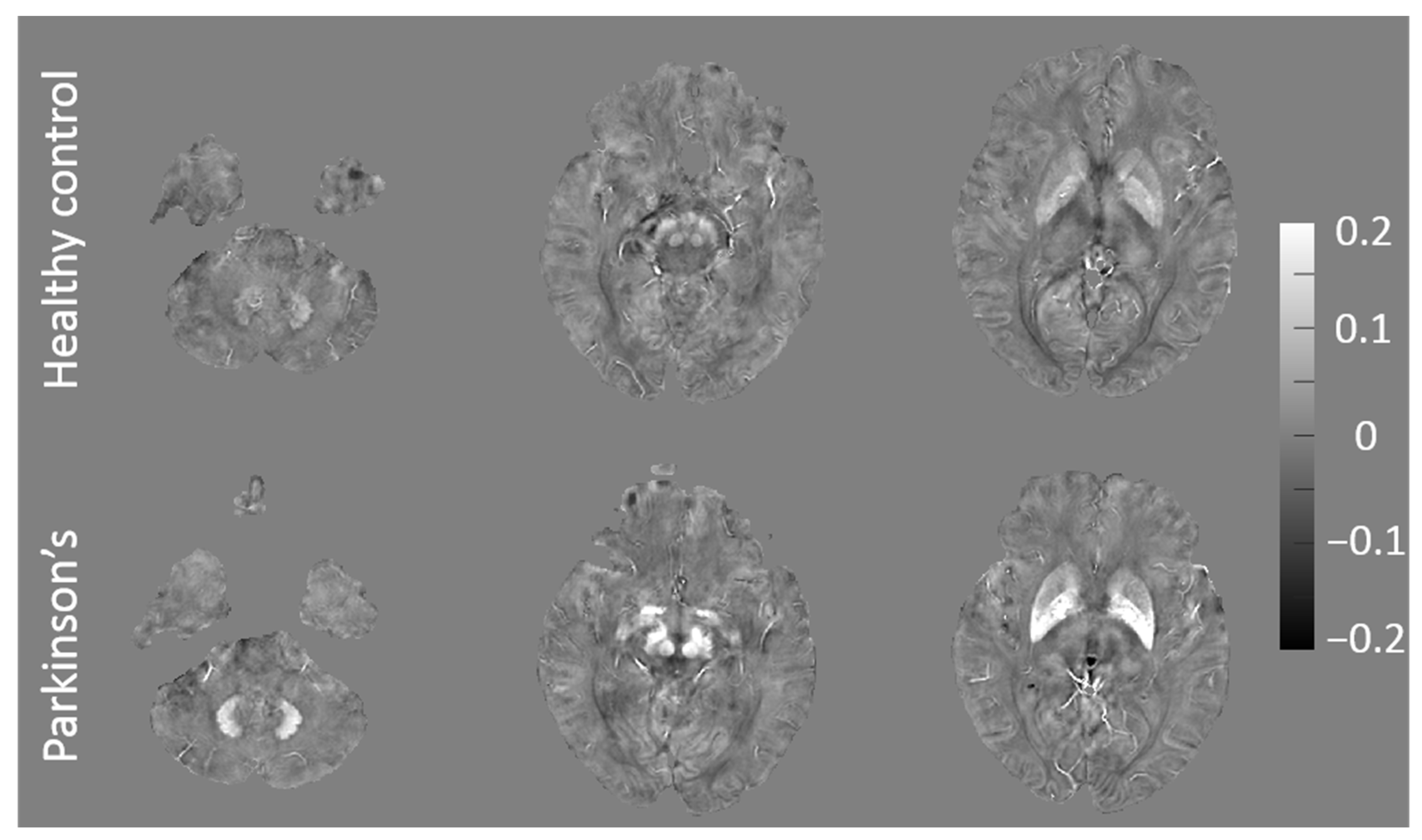
| Characteristic | Patients with Early PD a (n = 24) | HCs b (n = 27) | p-Value |
|---|---|---|---|
| Mean age, years (range) | 68.8 (56–79) | 65.3 (53–82) | 0.139 * |
| Sex (male:female) | 8:16 | 7:20 | 0.759 † |
| Mean disease duration, years (range) ‡ | 0.8 (0–3) | - | - |
| Mean NMSS c score (range) | 18 (4–47) | - | - |
| Mean Hoehn and Yahr stage (range) | 1.6 (1–2.5) | - | - |
| Mean MDS-UPDRS d (part III sum) (range) | 17.8 (4–40.5) | - | - |
| ROI c | Mean QSM k Value ± SD, ppm | F-score l | |
|---|---|---|---|
| Patients with Early PD (n = 24) | HCs (n = 27) | ||
| GP d | 0.142 ± 0.034 | 0.134 ± 0.038 | 14.634 |
| DN e | 0.101 ± 0.031 | 0.095 ± 0.026 | 11.461 |
| SNr f | 0.131 ± 0.044 | 0.115 ± 0.044 | 11.261 |
| SNc g | 0.130 ± 0.043 | 0.125 ± 0.041 | 9.349 |
| RN h | 0.102 ± 0.032 | 0.097 ± 0.034 | 9.251 |
| CN i | 0.070 ± 0.025 | 0.059 ± 0.021 | 7.497 |
| PUT j | 0.109 ± 0.049 | 0.094 ± 0.025 | 6.910 |
| The Number of Features | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Accuracy | ||||||||
| SVM a | 0.67 | 0.65 | 0.65 | 0.64 | 0.72 | 0.71 | 0.76 | |
| LR b | 0.58 | 0.55 | 0.59 | 0.70 | 0.69 | 0.70 | 0.72 | |
| AUC c | ||||||||
| SVM a | 0.58 | 0.45 | 0.40 | 0.48 | 0.62 | 0.63 | 0.70 | |
| LR b | 0.40 | 0.37 | 0.36 | 0.59 | 0.58 | 0.62 | 0.64 | |
| Sensitivity | ||||||||
| SVM a | 0.68 | 0.54 | 0.56 | 0.55 | 0.81 | 0.89 | 0.77 | |
| LR b | 0.49 | 0.49 | 0.52 | 0.69 | 0.62 | 0.64 | 0.72 | |
| Specificity | ||||||||
| SVM a | 0.70 | 0.76 | 0.72 | 0.75 | 0.67 | 0.60 | 0.84 | |
| LR b | 0.69 | 0.67 | 0.68 | 0.75 | 0.82 | 0.79 | 0.72 | |
| ROI d | Mean QSM Value ± SD, ppm | F-score | |
|---|---|---|---|
| High NMSS (n = 12) | Low NMSS (n = 12) | ||
| GP e | 0.136 ± 0.029 | 0.149 ± 0.037 | 17.772 |
| RN f | 0.106 ± 0.028 | 0.099 ± 0.035 | 10.544 |
| DN g | 0.102 ± 0.033 | 0.100 ± 0.031 | 10.426 |
| SNr h | 0.129 ± 0.043 | 0.133 ± 0.045 | 9.070 |
| SNc i | 0.127 ± 0.045 | 0.134 ± 0.042 | 9.063 |
| CN j | 0.074 ± 0.027 | 0.067 ± 0.023 | 7.846 |
| PUT k | 0.111 ± 0.042 | 0.107 ± 0.056 | 5.005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Oh, S.-H.; Park, S.-W.; Shin, C.; Kim, J.; Rhim, J.-H.; Lee, J.-Y.; Choi, J.-Y. Screening Patients with Early Stage Parkinson’s Disease Using a Machine Learning Technique: Measuring the Amount of Iron in the Basal Ganglia. Appl. Sci. 2020, 10, 8732. https://doi.org/10.3390/app10238732
Lee S, Oh S-H, Park S-W, Shin C, Kim J, Rhim J-H, Lee J-Y, Choi J-Y. Screening Patients with Early Stage Parkinson’s Disease Using a Machine Learning Technique: Measuring the Amount of Iron in the Basal Ganglia. Applied Sciences. 2020; 10(23):8732. https://doi.org/10.3390/app10238732
Chicago/Turabian StyleLee, Seon, Se-Hong Oh, Sun-Won Park, Chaewon Shin, Jeehun Kim, Jung-Hyo Rhim, Jee-Young Lee, and Joon-Yul Choi. 2020. "Screening Patients with Early Stage Parkinson’s Disease Using a Machine Learning Technique: Measuring the Amount of Iron in the Basal Ganglia" Applied Sciences 10, no. 23: 8732. https://doi.org/10.3390/app10238732
APA StyleLee, S., Oh, S.-H., Park, S.-W., Shin, C., Kim, J., Rhim, J.-H., Lee, J.-Y., & Choi, J.-Y. (2020). Screening Patients with Early Stage Parkinson’s Disease Using a Machine Learning Technique: Measuring the Amount of Iron in the Basal Ganglia. Applied Sciences, 10(23), 8732. https://doi.org/10.3390/app10238732





