Abstract
Viticulture is globally an important economic activity, and grapevine microbiomes hold a significant role in influencing yield and quality. Earlier studies showed that cultivar and agronomic management affect grapevine microbiome structure and, potentially, the quality of the end product. While microbial dynamics and ecology were established on some grapevine tissues, i.e., leaves and grapes, there is less knowledge deciphering microbiomes on other tissues, i.e., barks and buds. Moreover, although the impact on the microbiome of the so-called “vitivinicultural terroir” is well established, there are limited data considering microbiomes of genetically diverse cultivars within the same environment. Our study aims to explore microbiome diversity on bud and bark tissues of 37 different grapevine cultivars under the same environment and agronomic management. We targeted the V2-9 regions of the 16S rRNA gene of the microbiomes in bark and buds at the onset of new vegetation and bud expansion using Ion Torrent sequencing technology. Our results show that these tissues display high bacterial diversity regardless of cultivars’ use. Proteobacteria, Bacteroidetes, and Actinobacteria were the most prevalent among 11 detected phyla. The genotype of the cultivar seems to affect bacterial diversity and structure (p < 0.001) within the same environment. Our approach highlights the efficiency of high-throughput sequencing to unfold microbiomes of several grapevine parts that could be an important source of microbial inoculation and an important molecular fingerprint of the wine and grape end products.
1. Introduction
Viticulture is an economically important crop spanning an area of 7.5 mha with a total production exceeding 78 × 106 tons and a revenue of more than 30 billion Euros of wine [1].
Therefore understating the dynamics of product quality are of great economic importance [1]. Due to their abundance and activity, Vitis vinifera associated microbes are key in regulating growth, plant health, and grape and wine quality [2,3,4]. Even beneficial microbes in wine production, i.e., lactic acid bacteria, can cause undesirable changes in wine flavor under certain conditions, rendering the wine undrinkable. Similarly, several associated bacteria can cause some serious wine spoilage [5]. Despite the great progress in describing grapevine microbiomes and their implications on their growth, yield, and product quality, little is known about how different cultivars (as microbial hosts) can shape associated microbial assemblage.
A grapevine’s successful establishment depends mainly on the selection of the appropriate cultivar. The trunk bark, as a permanent part of the vine and microbial host, is of much importance not only as a potential source of microbial inoculums for leaves and grapes but also as an indicator for microbial diversity in vineyards [6,7]. Despite the importance of trunk bark in microbial dynamics, the majority of studies have focused on soil, leaves, berries, and wine associated microbial communities [4,8,9,10,11,12,13,14,15,16,17,18]. Current perceptions entail that trunk bark could be an additional source of grape berry pathogenic fungi, i.e., Uncinulanecator [19,20,21], Botrytis cinerea, Fusarium laterium, Penicillium spp., Phomopsis viticola [13,22]. Important pathogenic bacteria, Xylella fastidiosa, Agrobacterium tumefaciens, and Xylophilus ampelinus, belonging to Proteobacteria, were also detected in trunk bark, causing pierce’s, crown gall, and blight disease, respectively [6]. Very few studies have uncovered the presence of bacteria in grapevine buds, such as Xylophilus ampelinus spp. that have the ability to overwinter in the axillary buds [23]. However, although Pseudomonas syringae (Proteobacteria), which causes leaf spots, necrotic lesions, loss of inflorescences, etc., was found latent in the grapevine suckers/shoots [24], others proved its ability to overwinter not only in shoots but also in buds and other parts of many fruit crops [25].
Cultivar selection is an essential decision affecting product yield and quality; therefore, deciphering the abundance and diversity of vine-associated microbes requires systematic investigation. Previous research explored the abundance of specific pathogenic strains due to their economic importance in grape production and winemaking [13]. Fewer attempts have been made to describe the microbial diversity of V. vinifera, and those revealed the dominance of Acidobacteria, Actinobacteria, Bacteroidetes, Verrucomicrobia, and Chloroflexi microbial groups in Merlot, Dolcetto, and Sangiovese [6,7]. Some studies suggest that grape varieties themselves condition the microbial population of grapes, and therefore the microbial structure during fermentation [4,26]. Herein, we aim to decipher Vitis vinifera microbial community composition and diversity to assess the role of cultivar-genotype in shaping the microbial community. Thus, bark tissue and buds were collected to explore the cultivar’s bacterial community and its relationships with a wide selection of important national and international cultivars exploiting 16S rRNA gene sequencing on Vitis barks and buds samples. The aim of the study was to assess the microbial community of bark tissue and buds in a grapevine collection consisting of thirty-seven international and domestic cultivars.
2. Materials and Methods
2.1. Vine Cultivars and Sampling
A total of 37 grapevine cultivars (Table 1) were investigated in this study, collected from the Vine Cultivar Collection (VCC) curated by the Lab. of Viticulture, School of Agriculture, Aristotle University of Thessaloniki (AUTh) at Thermi, 570 01 Thessaloniki, Greece (N40.53829, E22.99633). The collection is established in a calcareous sandy loam soil. A semi-conventional system, with low-input agrochemicals is applied to the collection. Specifically, the collection had not received any chemical input the winter before the collection of samples.

Table 1.
List of cultivars analyzed. Fruit color is presented as B, black; R, red; W, white.
Samples were collected from fourteen (14) well-known cosmopolitan cultivars, as well as 24 domestic Greek cultivars presented in Table 1. One cane (40 cm long) was cut from 5 independent vines (trees) from each cultivar. The canes were pooled together and treated as a single sample. From each sample, buds were cut off, and bark peeled off the cane; 10 g from both organs were collected aseptically, equally mixed, and stored at −80 °C for further analyses.
2.2. DNA Extraction
Frozen plant samples were mixed vigorously (10 min at max speed) with 25 mL of TENP buffer. The supernatant (20 mL) was collected by centrifugation (3000× g; 5 min) in a new 50 mL tube. The supernatant was centrifuged at 14,500 rpm for 10 min and the pellet was collected and stored at −80 °C for DNA extraction [11]. DNA was extracted using a NucleoSpin® Plant II kit (Macherey-Nagel GmbH, Dylan, Germany) according to the manufacturer’s instructions. All DNA samples were kept at −20 °C for further use. Samples DNA quantity and quality were assessed by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc., Walsham, MA, USA) and visualized in 1.0% agarose gel electrophoresis.
2.3. High-Throughput Sequencing and Statistical Analysis
Samples were analyzed for bacterial taxonomy using the Ion 16S™ Metagenomics Kit according to the manufacturer’s instructions (#A26216, Thermo Fisher Scientific, Inc., Walsham, MA, USA). Briefly, the 16S rDNA metagenomic library consisted of pooled and tagged (Ion Xpress Barcode Adapters, #4471250, #4474009, #4474518, Thermo Fisher Scientific, Inc., Walsham, MA, USA) amplicons targeting V2–4–8 and V3–6, 7–9 sets; the combination of the two primer pools allows for sequence-based identification of broad range of bacteria within a mixed population [27]. Briefly, 3 ng of high-quality DNA was amplified using 2X Environmental Master Mix with the provided 10X 16S Primer Sets (Thermo Fisher Scientific, Inc., Walsham, MA, USA). PCR products were purified with AMPure® XP beads solution (#A63881, Beckman Coulter, Breia, CA, USA), quantified, pooled, and end-repaired using Ion Plus Fragment Library Kit (#4471252, Thermo Fisher Scientific, Inc., Walsham, MA, USA). Then, amplicons were adapter-ligated, nick repaired, quantified using an Ion Universal Library Quantitation Kit (#A26217, Thermo Fisher Scientific, Inc., Walsham, MA, USA), and the library was pooled and diluted to the required concentration. The library template was prepared using Ion 520™ & Ion 530™ Kit–OT2 (#A27751, Thermo Fisher Scientific, Inc., Walsham, MA, USA) and was loaded on a Ion 520™ Chip (#A27762, Thermo Fisher Scientific, Inc., Walsham, MA, USA). Sequencing was done at the Plant Genetics and Breeding Lab of AUTH on a GeneStudio S5 platform using Ion Torrent™ sequencing technology (Thermo Fisher Scientific, Inc., Walsham, MA, USA). Sequencing progress and raw sequences were processed through the Ion Torrent Suite (ver. 5.12.2, Thermo Fisher Scientific, Inc., Walsham, MA, USA), then BAM files were uploaded to the Ion Reporter Platform to obtain Ion 16S Metagenomics report through Ion 16S Metagenomics Workflow (Thermo Fisher Scientific, Inc., Walsham, MA, USA). Ion 16S Metagenomics Workflow uses a premium curated Applied Biosystems™ MicroSEQ™ ID 16S rRNA database and a curated Greengenes database, that are not user customizable (i.e., thresholds and cut-off values). Therefore, Fastq files (QC-, adaptor-, and barcode trimmed) were exported using the FileExporter plugin through Ion Torrent Suite and uploaded on the MG-RAST server [28] under the project number mgp91606. After MG-RAST QC (dynamic error removal (DRISEE), dynamic trimming (DYNAMICTRIM), denoising and normalization (FASTQ-MCF); see details on MG-RAST under Processing Information per submitted sample) taxonomy was assigned using Greengenes database (v.13_8) at 97% identity (e value 10−5). The resulting taxonomy matrix (OTUs Table; not shown) was used for all subsequent analysis. Descriptive diversity indices (α-diversity, rarefaction curves) and statistics (PCoA, Clustering based on Bray-Curtis and UPGMA, PERMANOVA, and ANOSIM) were calculated using PAST3 software [29]. Venn diagrams were carried out according to Heberle et al. [30].
3. Results
3.1. Sequence Analysis
High-throughput sequencing was performed using an Ion Torrent platform GENE STUDIO S5 at the Laboratory of Plant Genetics and Breeding, School of Agriculture, Aristotle University of Thessaloniki, Greece. Seven hypervariable regions (V2, V3, V4, V6, V7, V8, and V9) of the 16S rRNA gene were amplified and sequenced. Sequencing yielded 3,836,209 reads in total, with an average of 15,775 classified sequences per cultivar with an average read length of 234 bp. Accordingly, V3 provided the greatest sequencing depth that is the number of reads generated per sample (33% of total reads), followed by V6 and 7 (20%), V4 (18%), V8 (18%), V2 (10%), and, V9 (>0.1%) for profiling the bacterial diversity. The proportion of the unclassified reads ranged from the lowest of Limniona (60%) to the highest of Mavro (97%) with an average of 84% and were excluded from further analyses. Though the rarefaction curve was not parallel with the x-axis, the Good’s coverage of bacterial diversity reached 99.9%, with the majority of microbial diversity being captured (Figure S1).
A 463 bacterial distinct OTUs were detected excluding singletons (with a threshold of 97% similarity; Greengenesv. 13_8); identifying 11 phyla, 22 classes, 42 orders, 98 families, and 204 genera (Figure 1).
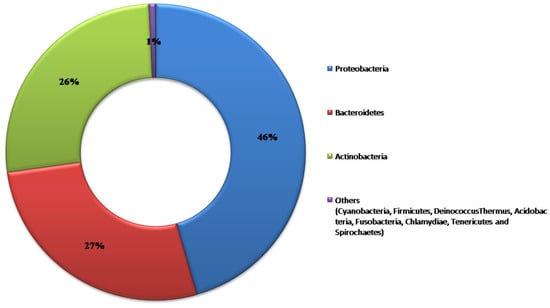
Figure 1.
The abundance of bacteria among all grapevines based on the classified reads at the phylum level.
3.2. Microbial Diversity among Cultivars
Among the 11 phyla present in 37 cultivars, Proteobacteria, Bacteroidetes, and Actinobacteria were the most dominant, comprising 99% of the total OTUs (Figure 2). The proportion of the aforementioned phyla varied among the cultivars, ranging from 94% for Mavro to 100% for Karabraimis, with an average of 99% (Figure 2).
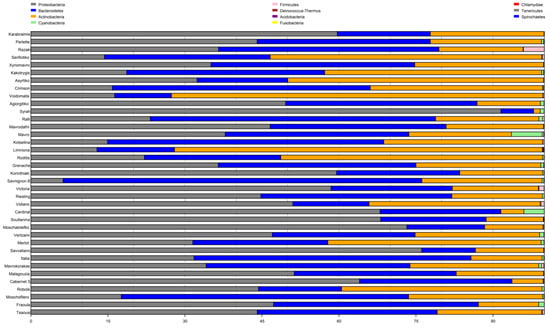
Figure 2.
The relative abundance of the 11 bacterial phyla of each grapevine based on the classified reads.
Microbial diversity indices varied among cultivars. Richness (Chao1) scores ranged among the cultivars from 183 to 544 (Table 2). Specifically, Cabernet Sauvignon held the lowest and Korinthiaki the highest Chao1 score. Regarding OTUs, the highest value was obtained by Limniona (266 and 164), both including and excluding singletons, respectively. The lowest values were shown by Cabernet Sauvignon with singletons (88), and by Grenache (37) excluding singletons (Table 2). Shannon diversity ranged from 1.2 (for Syrah) to 3.1 (for Mavrokorakas). Simpson’s evenness index ranged from 0.45 to 0.92 for Savvatiano and Mavrokorakas, respectively.

Table 2.
The richness and diversity indices of bacterial communities among the studied grapevines. The highest values in each column are in bold and lowest in italics.
3.3. Cultivar Genotype May Influence Bacterial Community Structure
Different Vitis vinifera cultivars planted in the same field exhibited distinctly different microbial composition. Noteworthy, PCoA (principal coordinate analysis) revealed one distantly apart cluster composed of Syrah, Cardinal, Soultanina, and Vertzami (Figure 3) diverged by 67% from other cultivars based on Bray–Curtis dissimilarities calculated for the composition of the bacterial communities at the genus level.
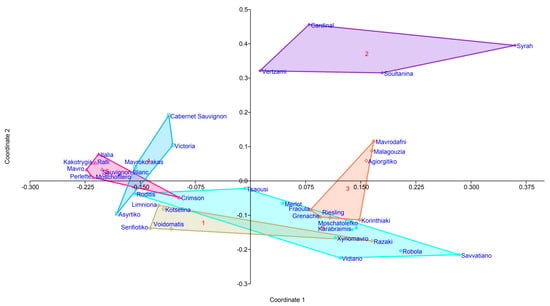
Figure 3.
PCoA constructed using Bray–Curtis at the genus level for 37 grapevines.
The first two PCoA axes (1 and 2) explain 35% of the total variation (Figure 3). Six groups were separated by hierarchical clustering (Figure 4). Significant differences in the microbial community structure were confirmed by PERMANOVA (p < 0.001 and F = 6.617) and ANOSIM (p < 0.001 and R = 0.6964) statistical analyses. Based on Venn diagram analyses (data not shown), 26 of the 37 cultivars showed the presence of cultivar-specific bacterial genera. The abundance of cultivar-specific genera ranged from 1–13 (Table 3), forming six distinct groups of cultivars. These six groups comprise 64 bacterial genera; interestingly, each cultivar had a unique bacterial fingerprint. While the remaining 11 cultivars from one group share 140 bacterial genera (Table 3).
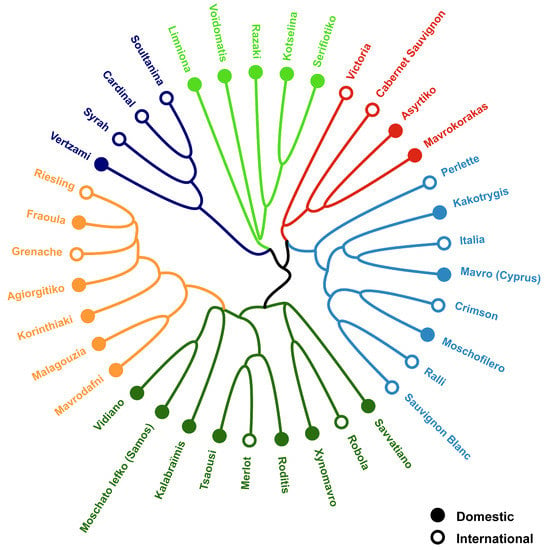
Figure 4.
Hierarchical cluster constructed using Bray–Curtis at the genus level for 37 grapevines.

Table 3.
The number of unique genera detected for the studied grapevines.
Of the total 204 bacterial genera identified, the following eleven: Curtobacterium, Fischerella, Frigoribacterium, Hymenobacter, Kineococcus, Massilia, Pantoea, Pseudomonas, Pedobacter, Rathayibacter, and Sphingomonas were present in all cultivars, except the Riesling. In the latter, the Pantoea genus was absent. The above-mentioned 11 genera are considered as a core microbiome. The relative abundance of this core microbiome ranged from 68–99%, with Limniona harboring the lowest and Robola the highest, respectively (Figure 5).
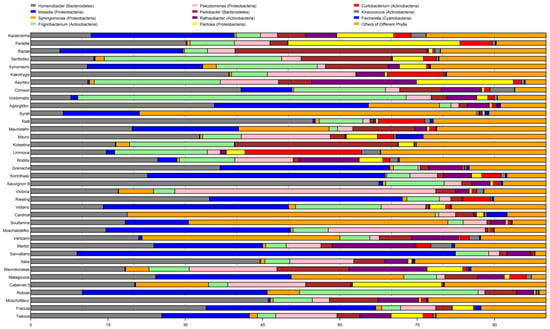
Figure 5.
The relative abundance of the top 11 bacterial genera among the 37 cultivars.
3.4. Assessment of Vine Bacteria of Economic Importance
The study revealed well-known bacteria with economic implications for viticulture and winemaking. Of all the studied cultivars, only seven cultivars presented such bacteria. Agrobacterium vitis and A. tumefaciens, which are responsible for crown and cane gall disease, were detected in Tsaousi and Riesling cultivars. Riesling cultivar harbored both A. vitis and A. tumefaciens, with the former presented in a 12-fold species abundance compared to Tsaousi, while Tsaousi contained only A. tumefaciens. Lactic acid bacteria (LAB), including Lactobacillus and Pediococcus, and acetic acid bacteria (AAC), such as Gluconobacter associated with wine production, were detected in low abundance. Specifically, Lactobacillus sp. was observed in Vidiano, Limniona, Voidomatis, and Kakotrygis, and Pediococcus spp. were only discovered in Mavro, while Gluconobacter was observed only in Serifiotiko cultivar.
4. Discussion
The aim of this study was to investigate the roles that Vitis vinifera cultivars might play, as microbial hosts, in shaping the differentiation of associated microbiomes. The hypothesis in the study was that if there is any influence of the genotype in the bacterial assemblage that overwinter on the vine canes, this should be evident as differences in the bacterial populations detected early in the spring on vines of different genotypes in the same vineyard, same soil and environment, and cultivated with the same agricultural practices. Here, we specifically focused on one-season-old canes’ barks and buds, which to our knowledge, no other study compared their associated microbiomes yet. High-throughput sequencing revealed rich microbiomes on these parts from several cultivars, supporting that trunk bark is an important microbial host (microbial terroir [7]). Interestingly, cane samples hosted several taxa that are typically identified in many studies across the world. On the other hand, a great portion of our sequencing data revealed unclassified microbial sequences. However, it is not uncommon for sequencing studies to result in high numbers of valid but unclassified reads. It could be due to experimental noise and richer microbiomes with some organisms less well described in the different databases [31]. Also, the differences in the microbiomes of the studied cultivars partly support the idea that vineyards consisting of different grape cultivars appear to harbor more diverse microbiomes. A possible selection pressure imposed on the microbiome by grapevine genotype might be one explanation, as indicated in other studies [4].
A multiple variable region 16S amplicon approach was applied to target seven hypervariable regions of the 16S rRNA gene in Vitis microbiomes, contrary to other previous studies focusing on a single region [4,6,7,11,12,16,26,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50]. The use of multiple variable regions of the bacterial 16S rRNA gene provides clear advantages compared to traditional single variable region approaches, particularly regarding detection specificity, and can better describe microbial diversity, i.e., higher resolution for lower-rank taxa [27,51]. A multiple variable region 16S amplicon approach could be incorporated in current workflows of Illumina [27] and Thermo Ion sequencing technologies (Ion 16S Metagenomics Kit, #A26216, Thermo Fisher Scientific Inc., USA) without any subsequent modifications after amplicon amplification and purification. Among the seven hypervariable regions of the 16S rRNA gene analyzed, the V3 region provided the greatest sequencing depth for profiling bacterial diversity, followed by V6-7 and V4. The later region (V4) also provided greater sequencing depth compared to the V5 region in another Vitis study [46]. Therefore, sequencing V3-4 regions, with high specificity primers excluding co-amplification of mitochondrial/chloroplast DNA, could be an effective alternative for future Vitis related studies.
Proteobacteria, Bacteroidetes, and Actinobacteria were the most abundant phyla. These phyla are considered as phyllosphere-associated generalists and have been detected in large numbers in several plant species [52,53] and soil samples as well [54,55]. Although their dominance detected in our study was in accordance with the findings of others [4,7,11,12,18,37,38,39,40,44,45,56,57,58,59,60,61], however, their niches were entirely neglected. The rest of the phyla that had a low presence were also detected in other studies in different amounts. For instance, the prevalence of Cyanobacteria was very low (0.5%) compared to 80% reported by [4], who examined phylosphere and carposphere, probably due to the different tissues examined. In addition, the detected phyla of Firmicutes, Chlamydiae, Acidobacteria, Verrucomicrobia, Chlorobi, Planctomycetes, Fusobacteria, Nitrospirae, and Spirochaetes were also reported in other Vitis studies [7,11,18].
Patterns of microbiome divergence among cultivars became evident, and the majority of Vitis microbiomes grouped close to the lower part of the plot (Figure 3). Interestingly, Vertzami, Cardinal, Soultanina, and Syrah cultivars formed a distinct group. Similar patterns of microbe divergence were also detected in other studies with different cultivars [4,62]. Moreover, the results of Venn diagrams revealed that Limniona hosted unique bacterial genera ranging from 40 to 72% of the total taxa when compared to Victoria and Grenache, respectively. Considering free-living microbes, it is well documented how geography and environment shape microbial community structure. In vineyard ecosystems it was recently demonstrated that microbial variability correlates with the geographic location of the cultivars, environmental characteristics and agronomic systems [63,64]. In a recent biogeographic investigation, geographic distance and environment appear to be important factors shaping Vitis associated Saccharomyces cerevisiae diversity [65]. In our study, geographic distance was not a factor controlling microbial diversity, because all cultivars were from the same vineyard. Interestingly, the genotype of the cultivars appeared to have a drastic impact on the bacterial community, influencing the fitness of certain V. vinifera associated microbes.
The detected genera (Hymenobacter, Massilia, Frigoribacterium, Sphingomonas, Pseudomonas, Pedobacter, Pantoea, etc.) in the vineyard are known to have distinct beneficial and pathogenic roles. For instance, among Pseudomonas spp., Ps. putida had a presence of 11% among the other species of this genus and is known for its growth-promoting role [66,67]. Moreover, Ps. fluorescens (12%) and Ps. aeruginosa (0.02%) suppress different pathogens [68,69]. However, other Pseudomonas spp. such as Ps. viridiflava (15%) and Ps. syringae (0.4%) [68,69] are known for their pathogenicity. Only Massilia timonae was detected in this study, which is known for its plant-growth-promoting and protection in the rhizosphere and seed surface [70]. Massilia taxa have been reported in the phyllosphere of different plants, including lettuce and apple [71,72,73,74,75]. Moreover, the genus Sphingomonas generally acts as a plant-protective by suppressing disease symptoms and decreasing pathogen growth [53]. Researchers have demonstrated that the leaf bacterium Sphingomonas spp. can protect plants against the leaf-pathogenic Ps. syringae through substrate competition [74]. Also, Sphingomonas spp. are known for their plant growth-promoting through the production of plant growth-stimulating factors [75]. However, S. melonis was described as the causative agent of the brown-spot disease of yellow Spanish melons (Cucumis melo) [76] and was also detected with a presence of 97% among the other species of this genus. Furthermore, members of Pantoea act as biocontrol agents or plant-growth promoters [75], including Pantoea agglomerans (95%). This species works against different pathogens like Erwinia amylovora, causing fire blight disease of pear and apple fruit trees [77]. Some economically important bacterial genera such as Agrobacterium, Acetobacter, Gluconobacter, Pediococcus, Lactobacillus spp. were observed in trivial abundance. Although no observable effect is anticipated during this stage of the grapevine lifecycle, they might later be inoculants for the grapevine itself or its products, causing economic losses if the conditions are suitable. From our study, it is evident that cane tissues host both beneficial and pathogenic taxa, with important economic implications in viticulture. Thus, the dynamics of beneficial and pathogenic taxa through seasonal grapevine growth remain unclear. Future studies should investigate V. vinifera microbiomes at a greater temporal resolution to better understand the control and factors affecting the trade-off balance between beneficial and pathogenic taxa in vineyards.
Results of this study may be helpful to investigate the origin of the grapevine associated microbiomes in other tissues. Previous studies indicated that grapevine microbiomes on leaves and grapes originated primarily from the adjacent soil microbiome [6,7,37,39,47,78]. This could be due to the direct dissemination of soil particles or any other form of passive transportation, e.g., displacements of plant surface. Another concept of microbial inoculation considers direct inoculation of various vine parts through buds. Trunk bark was also considered as a permanent habitat and source of inoculation in a vineyard [6,7,79]. Hence, the buds, which are responsible for the annual growth cycle of grapevines [80], together with the barks of the one-season-old canes, could be the direct source of inoculants because they are in closer contact with leaves, inflorescences, and grapes than any other sources. The origin and mode of inoculation require further investigation and could potentially help us understand the importance of the local environment and cultivar selection in controlling microbial dynamics on V. vinifera cultivars.
5. Conclusions
Metagenomics mining revealed that a core microbiome is hosted by all studied V. vinifera cultivars conserved in the same environment. On the other hand, unique metagenomics fingerprints were unraveled for each cultivar, suggesting a role of cultivars’ genotype in shaping the associated microbial community. The vine microbiome’s complexity and diversity were highlighted, although a percentage of sequence data still remains unassigned. Thus, further microbial metagenomics research is vital for complete coverage of the microbial genome available in vineyards. The host-associated microbes require further investigation in greater temporal and spatial patterns to assess the importance of grapevine microbiomes to delineate the wines regional character and provide further tools for sustainable agriculture.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/23/8405/s1, Figure S1: Observed taxonomic units analysis (Rarefaction Curve) of bacterial species diversity in bark and buds of 37 vine varieties with singletons.
Author Contributions
M.A. performed experimental work, data analysis and prepared the original draft, G.G. performed sequencing and metagenomic analysis, wrote and revised the manuscript, P.V.M. contributed in conceptualization, funding acquisition, and revised the manuscript, A.N.P. contributed in conceptualization, experimental design, funding acquisition and revising the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors wish to thank N. Nikolaou for providing Vitis vinifera samples and access to the AUTH Vine collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- OIV. International Organisation of Vine and Wine 2019 Statistical Report on World Vitiviniculture; International Organisation of Vine and Wine Intergovernmental Organisation: Paris, France, 2019; Volume 23. [Google Scholar]
- Yang, C.H.; Crowley, D.E.; Borneman, J.; Keen, N.T. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 2001, 98, 3889–3894. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Ruppel, S. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol. 2014, 87, 2–17. [Google Scholar] [CrossRef]
- Singh, P.; Santoni, S.; This, P.; Péros, J.-P. Genotype-Environment Interaction Shapes the Microbial Assemblage in Grapevine’s Phyllosphere and Carposphere: An NGS Approach. Microorganisms 2018, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Renouf, V.; Claisse, O.; Lonvaud-Funel, A. Inventory and monitoring of wine microbial consortia. Appl. Microbiol. Biotechnol. 2007, 75, 149–164. [Google Scholar] [CrossRef]
- Martins, G.; Lauga, B.; Miot-Sertier, C.; Mercier, A.; Lonvaud, A.; Soulas, M.L.; Soulas, G.; Masneuf-Pomarède, I. Characterization of Epiphytic Bacterial Communities from Grapes, Leaves, Bark and Soil of Grapevine Plants Grown, and Their Relations. PLoS ONE 2013, 8, e073013. [Google Scholar] [CrossRef] [PubMed]
- Vitulo, N.; Lemos, W.J.F.; Calgaro, M.; Confalone, M.; Felis, G.E.; Zapparoli, G.; Nardi, T. Bark and grape microbiome of Vitis vinifera: Influence of geographic patterns and agronomic management on bacterial diversity. Front. Microbiol. 2019, 10, 3203. [Google Scholar] [CrossRef] [PubMed]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Grangeteau, C.; Roullier-Gall, C.; Rousseaux, S.; Gougeon, R.D.; Schmitt-Kopplin, P.; Alexandre, H.; Guilloux-Benatier, M. Wine microbiology is driven by vineyard and winery anthropogenic factors. Microb. Biotechnol. 2017, 10, 354–370. [Google Scholar] [CrossRef]
- Oliveira, M.; Arenas, M.; Lage, O.; Cunha, M.; Amorim, M.I. Epiphytic fungal community in Vitis vinifera of the Portuguese wine regions. Lett. Appl. Microbiol. 2018, 66, 93–102. [Google Scholar] [CrossRef]
- Wei, Y.J.; Wu, Y.; Yan, Y.Z.; Zou, W.; Xue, J.; Ma, W.R.; Wang, W.; Tian, G.; Wang, L.Y. High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS ONE 2018, 13, e0193097. [Google Scholar] [CrossRef]
- Gamalero, E.; Bona, E.; Novello, G.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Berta, G.; Lingua, G. Discovering the bacteriome of Vitis vinifera cv. Pinot Noir in a conventionally managed vineyard. Sci. Rep. 2020, 10, 6453. [Google Scholar] [CrossRef] [PubMed]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Setati, M.E.; Jacobson, D.; Andong, U.C.; Bauer, F. The Vineyard Yeast Microbiome, a Mixed Model Microbial Map. PLoS ONE 2012, 7, e52609. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.; Miot-Sertier, C.; Lauga, B.; Claisse, O.; Lonvaud-Funel, A.; Soulas, G.; Masneuf-Pomarède, I. Grape berry bacterial microbiota: Impact of the ripening process and the farming system. Int. J. Food Microbiol. 2012, 158, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Collins, T.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Fermentation Behavior Suggest Microbial Contribution to Regional. mBio 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- del Carmen Portillo, M.; Mas, A. Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT Food Sci. Technol. 2016, 72, 317–321. [Google Scholar] [CrossRef]
- Salvetti, E.; Campanaro, S.; Campedelli, I.; Fracchetti, F.; Gobbi, A.; Tornielli, G.B.; Torriani, S.; Felis, G.E. Whole-metagenome-sequencing-based community profiles of Vitis vinifera L. cv. Corvina berries withered in two post-harvest conditions. Front. Microbiol. 2016, 7, 937. [Google Scholar] [CrossRef]
- Cortesi, P.; Bisiach, M.; Ricciolini, M.; Gadoury, D.M. Cleistothecia of Uncinula necator—An additional source of inoculum in Italian vineyards. Plant Dis. 1997, 81, 922–926. [Google Scholar] [CrossRef]
- Grove, G.G. Perennation of Uncinula necator in vineyards of Eastern Washington. Plant Dis. 2004, 88, 242–247. [Google Scholar] [CrossRef]
- Behar, A.; Jurkevitch, E.; Yuval, B. Bringing back the fruit into fruit fly-bacteria interactions. Mol. Ecol. 2008, 17, 1375–1386. [Google Scholar] [CrossRef]
- Munkvold, G.P. Efficacy of Natural Epiphytes and Colonizers of Grapevine Pruning Wounds for Biological Control of Eutypa Dieback. Phytopathology 1993, 83, 624. [Google Scholar] [CrossRef]
- Komatsu, T.; Kondo, N. Winter habitat of Xylophilus ampelinus, the cause of bacterial blight of grapevine, in Japan. J. Gen. Plant Pathol. 2015, 81, 237–242. [Google Scholar] [CrossRef]
- Hall, S.J.; Dry, I.B.; Blanchard, C.L.; Whitelaw-Weckert, M.A. Phylogenetic relationships of Pseudomonas syringae pv. Syringae isolates associated with bacterial inflorescence rot in Grapevine. Plant Dis. 2016, 100, 607–616. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bultreys, A.; Kaluzna, M. Bacterial cankers caused by Pseudomonas syringae on stone fruit species with special emphasis on the pathovars syringae and morsprunorum race 1 and race 2. J. Plant Pathol. 2010, 92. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the diversity of grapevine microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef]
- Bukin, Y.S.; Galachyants, Y.P.; Morozov, I.V.; Bukin, S.V.; Zakharenko, A.S.; Zemskaya, T.I. The effect of 16s rRNA region choice on bacterial community metabarcoding results. Sci. Data 2019, 6, 190007. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.; Ryan, P. PAST: Paleontological statistics software package for education education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16. [Google Scholar] [CrossRef]
- Siegwald, L.; Touzet, H.; Lemoine, Y.; Hot, D.; Audebert, C.; Caboche, S. Assessment of common and emerging bioinformatics pipelines for targeted metagenomics. PLoS ONE 2017, 12, e0169563. [Google Scholar] [CrossRef]
- Campisano, A.; Antonielli, L.; Pancher, M.; Yousaf, S.; Pindo, M.; Pertot, I. Bacterial endophytic communities in the grapevine depend on pest management. PLoS ONE 2014, 9, e112763. [Google Scholar] [CrossRef]
- Marzano, M.; Fosso, B.; Manzari, C.; Grieco, F.; Intranuovo, M.; Cozzi, G.; Mulè, G.; Scioscia, G.; Valiente, G.; Tullo, A.; et al. Complexity and dynamics of the winemaking bacterial communities in berries, musts, and wines from apulian grape cultivars through time and space. PLoS ONE 2016, 11, e0157383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, X.; Zhong, Q.; Huang, Z.; Bai, Z. Relations among epiphytic microbial communities from soil, leaves and grapes of the grapevine. Front. Life Sci. 2017, 10, 73–83. [Google Scholar] [CrossRef]
- Novello, G.; Gamalero, E.; Bona, E.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Lingua, G.; Berta, G. The rhizosphere bacterial microbiota of Vitis vinifera cv. Pinot Noir in an integrated pest management vineyard. Front. Microbiol. 2017, 8, 1528. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, V.; Sandionigi, A.; Bruni, I.; Bruno, A.; Lovicu, G.; Casiraghi, M.; Labra, M. Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS ONE 2017, 12, e0184615. [Google Scholar] [CrossRef]
- Mezzasalma, V.; Sandionigi, A.; Guzzetti, L.; Galimberti, A.; Grando, M.S.; Tardaguila, J.; Labra, M. Geographical and cultivar features differentiate grape microbiota in Northern Italy and Spain vineyards. Front. Microbiol. 2018, 9, 946. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.; Chen, D.; Howell, K. From the Vineyard to the Winery: How Microbial Ecology Drives Regional Distinctiveness of Wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef]
- Chou, M.Y.; Vanden Heuvel, J.; Bell, T.H.; Panke-Buisse, K.; Kao-Kniffin, J. Vineyard under-vine floor management alters soil microbial composition, while the fruit microbiome shows no corresponding shifts. Sci. Rep. 2018, 8, 11039. [Google Scholar] [CrossRef]
- Canfora, L.; Vendramin, E.; Felici, B.; Tarricone, L.; Florio, A.; Benedetti, A. Vineyard microbiome variations during different fertilisation practices revealed by 16s rRNA gene sequencing. Appl. Soil Ecol. 2018, 125, 71–80. [Google Scholar] [CrossRef]
- Gupta, V.V.S.R.; Bramley, R.G.V.; Greenfield, P.; Yu, J.; Herderich, M.J. Vineyard soil microbiome composition related to rotundone concentration in Australian cool climate “peppery” Shiraz grapes. Front. Microbiol. 2019, 10, 1607. [Google Scholar] [CrossRef]
- Campanaro, S.; Treu, L.; Vendramin, V.; Bovo, B.; Giacomini, A.; Corich, V. Metagenomic analysis of the microbial community in fermented grape marc reveals that Lactobacillus fabifermentans is one of the dominant species: Insights into its genome structure. Appl. Microbiol. Biotechnol. 2014, 98, 6015–6037. [Google Scholar] [CrossRef] [PubMed]
- Deyett, E.; Rolshausen, P.E. Endophytic microbial assemblage in grapevine. FEMS Microbiol. Ecol. 2020, 96, fiaa053. [Google Scholar] [CrossRef] [PubMed]
- Gobbi, A.; Kyrkou, I.; Filippi, E.; Ellegaard-Jensen, L.; Hansen, L.H. Seasonal epiphytic microbial dynamics on grapevine leaves under biocontrol and copper fungicide treatments. Sci. Rep. 2020, 10, 681. [Google Scholar] [CrossRef] [PubMed]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Wine fermentation microbiome: A landscape from different Portuguese wine appellations. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Joseph, C.M.L.; Allen, G.; Benson, A.K.; Mills, D.A. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS ONE 2012, 7, e36357. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Gilbert, J.A. Understanding grapevine-microbiome interactions: Implications for viticulture industry. Microb. Cell 2015, 2, 171–173. [Google Scholar] [CrossRef]
- Alonso, A.; De Celis, M.; Ruiz, J.; Vicente, J.; Navascués, E.; Acedo, A.; Ortiz-Álvarez, R.; Belda, I.; Santos, A.; Gómez-Flechoso, M.Á.; et al. Looking at the origin: Some insights into the general and fermentative microbiota of vineyard soils. Fermentation 2019, 5, 78. [Google Scholar] [CrossRef]
- Burns, K.N.; Bokulich, N.A.; Cantu, D.; Greenhut, R.F.; Kluepfel, D.A.; O’Geen, A.T.; Strauss, S.L.; Steenwerth, K.L. Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: Differentiation by vineyard management. Soil Biol. Biochem. 2016, 103, 337–348. [Google Scholar] [CrossRef]
- Hall, M.E.; O’Bryon, I.; Wilcox, W.F.; Osier, M.V.; Cadle-Davidson, L. The epiphytic microbiota of sour rot-affected grapes differs minimally from that of healthy grapes, indicating causal organisms are already present on healthy berries. PLoS ONE 2019, 14, e0211378. [Google Scholar] [CrossRef]
- Schriefer, A.E.; Cliften, P.F.; Hibberd, M.C.; Sawyer, C.; Brown-Kennerly, V.; Burcea, L.; Klotz, E.; Crosby, S.D.; Gordon, J.I.; Head, R.D. A multi-amplicon 16S rRNA sequencing and analysis method for improved taxonomic profiling of bacterial communities. J. Microbiol. Methods 2018, 154, 6–13. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Kim, M.J.; Jeon, C.W.; Cho, G.; Kim, D.R.; Kwack, Y.B.; Kwak, Y.S. Comparison of microbial community structure in kiwifruit pollens. Plant Pathol. J. 2018, 34, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.J.; Tech, J.J. Grapevine microbiomics: Bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16S rRNA amplicons. Acta Hortic. 2011, 905, 31–42. [Google Scholar] [CrossRef]
- Miura, T.; Sánchez, R.; Castañeda, L.E.; Godoy, K.; Barbosa, O. Is microbial terroir related to geographic distance between vineyards? Environ. Microbiol. Rep. 2017, 9, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Hendgen, M.; Hoppe, B.; Döring, J.; Friedel, M.; Kauer, R.; Frisch, M.; Dahl, A.; Kellner, H. Effects of different management regimes on microbial biodiversity in vineyard soils. Sci. Rep. 2018, 8, 9393. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.H.; du Toit, M.; Setati, M.E. The grapevine and wine microbiome: Insights from high-throughput amplicon sequencing. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Stefanini, I.; Cavalieri, D. Metagenomic approaches to investigate the contribution of the vineyard environment to the quality of wine fermentation: Potentials and difficulties. Front. Microbiol. 2018, 9, 991. [Google Scholar] [CrossRef]
- Kearsey, M. The principles of QTL analysis (a minimal mathematics approach). J. Exp. Bot. 1998, 49, 1619–1623. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Morrison-Whittle, P.; Lee, S.A.; Goddard, M.R. Fungal communities are differentially affected by conventional and biodynamic agricultural management approaches in vineyard ecosystems. Agric. Ecosyst. Environ. 2017, 246, 306–313. [Google Scholar] [CrossRef]
- Drumonde-Neves, J.; Franco-Duarte, R.; Vieira, E.; Mendes, I.; Lima, T.; Schuller, D.; Pais, C. Differentiation of Saccharomyces cerevisiae populations from vineyards of the Azores Archipelago: Geography vs. Ecology. Food Microbiol. 2018, 74, 151–162. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Obini, M.; Ugoji, E.O. Comparison of plant growth-promotion with Pseudomonas aeruginosa and Bacillus subtilis in three vegetables. Braz. J. Microbiol. 2008, 39, 423–426. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Sarris, P.F.; Trantas, E.A.; Mpalantinaki, E.; Ververidis, F.; Goumas, D.E. Pseudomonas viridiflava, a multi host plant pathogen with significant genetic variation at the molecular level. PLoS ONE 2012, 7, e036090. [Google Scholar] [CrossRef]
- Bartoli, C.; Berge, O.; Monteil, C.L.; Guilbaud, C.; Balestra, G.M.; Varvaro, L.; Jones, C.; Dangl, J.L.; Baltrus, D.A.; Sands, D.C.; et al. The Pseudomonas viridiflava phylogroups in the P.syringae species complex are characterized by genetic variability and phenotypic plasticity of pathogenicity-related traits. Environ. Microbiol. 2014, 16, 2301–2315. [Google Scholar] [CrossRef]
- Rozpdek, P.; Domka, A.; Turnau, K. Chapter 29 Mycorrhizal Fungi and Accompanying Microorganisms in Improving Phytoremediation Techniques. In The Fungal Community: Its Organization and Role in the Ecosystem; CRC Press: Boca Raton, FL, USA, 2017; ISBN 9781498706650. [Google Scholar]
- Bassas-Galia, M.; Nogales, B.; Arias, S.; Rohde, M.; Timmis, K.N.; Molinari, G. Plant original Massilia isolates producing polyhydroxybutyrate, including one exhibiting high yields from glycerol. J. Appl. Microbiol. 2012, 112, 443–454. [Google Scholar] [CrossRef]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef]
- Yashiro, E.; McManus, P.S. Effect of streptomycin treatment on bacterial community structure in the apple phyllosphere. PLoS ONE 2012, 7, e37131. [Google Scholar] [CrossRef] [PubMed]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Tao, J.; Liu, T.; Liu, Y.; Xiao, N.; Li, T.; Gu, Y.; Yin, H.; Meng, D. Responses of phyllosphere microbiota and plant health to application of two different biocontrol agents. AMB Express 2019, 9. [Google Scholar] [CrossRef]
- Buonaurio, R.; Stravato, V.M.; Kosako, Y.; Fujiwara, N.; Naka, T.; Kobayashi, K.; Cappelli, C.; Yabuuchi, E. Sphingomonas melonis sp. nov., a novel pathogen that causes brown spots on yellow Spanish melon fruits. Int. J. Syst. Evol. Microbiol. 2002, 52, 2081–2087. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.A.; Lee, D.H.; Kim, B.Y.; Heu, S. Draft genome sequence of Pantoea agglomerans R190, a producer of antibiotics against phytopathogens and foodborne pathogens. J. Biotechnol. 2014, 188, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, L.; Fan, X.; Jiang, J.; Zheng, X.-b.; Sun, H.; Chonghuai, L. Genome-wide assessment of population structure, linkage disequilibrium and resistant QTLs in Chinese wild grapevine. Sci. Hortic. 2017, 215, 59–64. [Google Scholar] [CrossRef]
- Morrison-Whittle, P.; Goddard, M.R. Quantifying the relative roles of selective and neutral processes in defining eukaryotic microbial communities. ISME J. 2015, 9, 2003–2011. [Google Scholar] [CrossRef]
- Keller, M. Phenology and Growth Cycle; Academic Press: Cambridge, MA, USA, 2015; ISBN 9780124199873. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).