Immunodeficient Rabbit Models: History, Current Status and Future Perspectives
Abstract
1. The Pre-Gene-Editing Age Where Mice Dominated
- Nude mice (Foxn1-/-). Forkhead box protein N1 (Foxn1) encodes a transcription factor required for both hair follicle and thymic development. KO of Foxn1 leads to a hairless phenotype and a failure of thymus development;
- SCID (severe combined immunodeficiency) mice (Prkdc-/-) or Rag deficient (Rag1/2-/-) mice. Prkdc encodes a polypeptide that is essential for the repair of double-strand breaks (DSBs) in the DNA; the breaks occur during somatic recombination of T cell receptor (TCR) and immunoglobulin (Ig) genes. The loss of Prkdc leads to T and B cell deficiency. Rag1 and Rag2 cluster closely in the genome and are critical elements for somatic recombination of TCR and Ig genes. The absence of Rag1 or 2 also results in both T and B cell deficiency.
- Il2rg deficiency on either the Prkdc or Rag deficient background. IL2rg mediates IL2, IL4, IL7, IL9, and IL15 high affinity signaling, which promotes the development of a myriad of immune cells.
- Additionally, the NOD/ShiLtJ strain exhibits deficiency in innate immune response, specifically, NK cells and antigen-presenting cells (i.e., macrophages). Coupling the previously described Prkdc or Rag mutation along with the Il2rg deficiency on a NOD background results in multigenic immunodeficiency, and a severely immunodeficient mouse model. Examples include NOD-Prkdcscid IL2rgTm1Wjl (NSG) and NOD-Rag1/2scid IL2rgTm1Wjl (NRG) lines.
2. The Gap between Mice and Humans
3. Gene Editing Tools and the Rise of Non-Murine Immunodeficient Animal Models
4. The Birth of Immunodeficient Rabbits
5. Where the ID Bunnies (Could) Rule
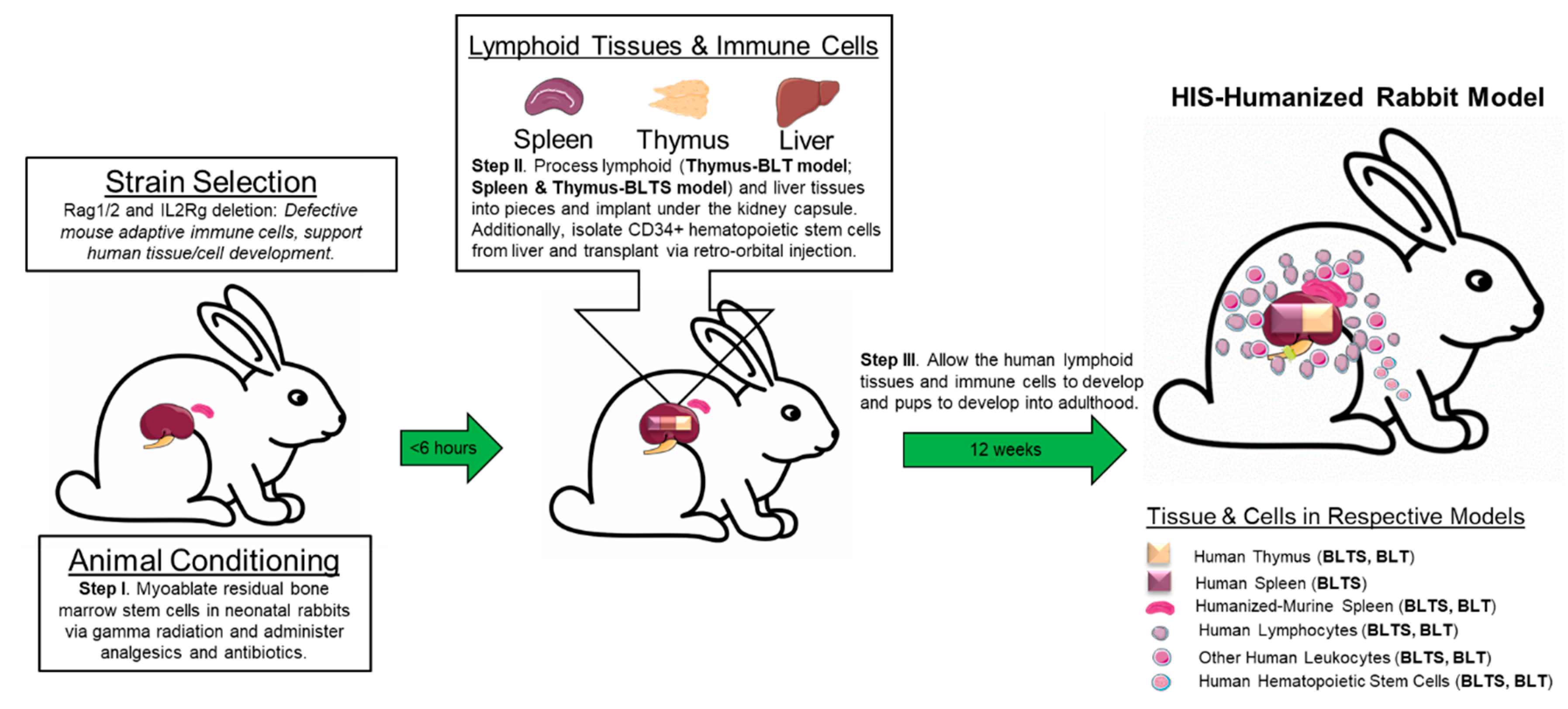
5.1. Humanized Rabbits for Infectious Diseases
5.2. CDX and PDX
5.3. Testing of Stem Cell-Derived Blood Vessels
5.4. Preclinical Development of Gene Editing Therapy for SCID
5.5. Hepatocytes Regeneration
Author Contributions
Funding
Conflicts of Interest
References
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nat. Cell Biol. 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed]
- Gossler, A.; Doetschman, T.; Korn, R.; Serfling, E.; Kemler, R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc. Natl. Acad. Sci. USA 1986, 83, 9065–9069. [Google Scholar] [CrossRef] [PubMed]
- Robertson, E.J.; Bradley, A.; Kuehn, M.; Evans, M.J. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nat. Cell Biol. 1986, 323, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, T.F. Use of rodents as models of human diseases. J. Pharm. BioAllied Sci. 2014, 6, 2–9. [Google Scholar] [CrossRef]
- Fischer, A.; Notarangelo, L.D.; Neven, B.; Cavazzana, M.; Puck, J.M. Severe combined immunodeficiencies and related disorders. Nat. Rev. Dis. Prim. 2015, 1, 15061. [Google Scholar] [CrossRef]
- Blundell, M.P.; Kinnon, C. Animal Models of Human Primary Immunodeficiency Diseases. eLS 2012. [Google Scholar] [CrossRef]
- Shultz, L.D.; Ishikawa, F.; Greiner, D.L. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007, 7, 118–130. [Google Scholar] [CrossRef]
- Cibelli, J.; Emborg, M.E.; Prockop, D.J.; Roberts, M.; Schatten, G.; Rao, M.; Harding, J.; Mirochnitchenko, O. Strategies for Improving Animal Models for Regenerative Medicine. Cell Stem Cell 2013, 12, 271–274. [Google Scholar] [CrossRef]
- Cavazzana-Calvo, M.; Hacein-Bey, S.; de Saint Basile, G.; Gross, F.; Yvon, E.; Nusbaum, P.; Selz, F.; Hue, C.; Certain, S.; Casanova, J.L.; et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 2000, 288, 669–672. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; Le Deist, F.; Wulffraat, N.; McIntyre, E.; Radford, I.; Villeval, J.-L.; Fraser, C.C.; Cavazzana-Calvo, M.; et al. A Serious Adverse Event after Successful Gene Therapy for X-Linked Severe Combined Immunodeficiency. N. Engl. J. Med. 2003, 348, 255–256. [Google Scholar] [CrossRef] [PubMed]
- Howe, S.J.; Mansour, M.R.; Schwarzwaelder, K.; Bartholomae, C.; Hubank, M.; Kempski, H.; Brugman, M.H.; Pike-Overzet, K.; Chatters, S.J.; De Ridder, D.; et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Investig. 2008, 118, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 2008, 118, 3132–3142. [Google Scholar] [CrossRef]
- Geurts, A.M.; Cost, G.J.; Freyvert, Y.; Zeitler, B.; Miller, J.C.; Choi, V.M.; Jenkins, S.S.; Wood, A.; Cui, X.; Meng, X.; et al. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science 2009, 325, 433. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2010, 29, 143–148. [Google Scholar] [CrossRef]
- Hockemeyer, D.; Wang, H.; Kiani, S.; Lai, C.S.; Gao, Q.; Cassady, J.P.; Cost, G.J.; Zhang, L.; Santiago, Y.; Miller, J.C.; et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011, 29, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Moscou, M.J.; Bogdanove, A.J. A Simple Cipher Governs DNA Recognition by TAL Effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; Dicarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Zhao, J.; Lai, L.; Ji, W.; Zhou, Q. Genome editing in large animals: Current status and future prospects. Natl. Sci. Rev. 2019, 6, 402–420. [Google Scholar] [CrossRef]
- Wiler, R.; Leber, R.; Moore, B.B.; VanDyk, L.F.; Perryman, L.E.; Meek, K. Equine severe combined immunodeficiency: A defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc. Natl. Acad. Sci. USA 1995, 92, 11485–11489. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.K.; Perryman, L.E.; Meek, K. A kinase-negative mutation of DNA-PK(CS) in equine SCID results in defective coding and signal joint formation. J. Immunol. 1997, 158, 3565–3569. [Google Scholar] [PubMed]
- Henthorn, P.S.; Somberg, R.L.; Fimiani, V.M.; Puck, J.M.; Patterson, D.F.; Felsburg, P.J. IL-2Rγ Gene Microdeletion Demonstrates That Canine X-Linked Severe Combined Immunodeficiency Is a Homologue of the Human Disease. Genomics 1994, 23, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Somberg, R.L.; Pullen, R.P.; Casal, M.L.; Patterson, D.F.; Felsburg, P.J.; Henthorn, P.S. A single nucleotide insertion in the canine interleukin-2 receptor gamma chain results in X-linked severe combined immunodeficiency disease. Veter. Immunol. Immunopathol. 1995, 47, 203–213. [Google Scholar] [CrossRef]
- Meek, K.; Kienker, L.; Dallas, C.; Wang, W.; Dark, M.J.; Venta, P.J.; Huie, M.L.; Hirschhorn, R.; Bell, T. SCID in Jack Russell Terriers: A New Animal Model of DNA-PKcs Deficiency. J. Immunol. 2001, 167, 2142–2150. [Google Scholar] [CrossRef]
- Ding, Q.; Bramble, L.; Yuzbasiyan-Gurkan, V.; Bell, T.; Meek, K. DNA-PKcs mutations in dogs and horses: Allele frequency and association with neoplasia. Gene 2002, 283, 263–269. [Google Scholar] [CrossRef]
- Mashimo, T.; Takizawa, A.; Voigt, B.; Yoshimi, K.; Hiai, H.; Kuramoto, T.; Serikawa, T. Generation of Knockout Rats with X-Linked Severe Combined Immunodeficiency (X-SCID) Using Zinc-Finger Nucleases. PLoS ONE 2010, 5, e8870. [Google Scholar] [CrossRef]
- Mashimo, T.; Takizawa, A.; Kobayashi, J.; Kunihiro, Y.; Yoshimi, K.; Ishida, S.; Tanabe, K.; Yanagi, A.; Tachibana, A.; Hirose, J.; et al. Generation and Characterization of Severe Combined Immunodeficiency Rats. Cell Rep. 2012, 2, 685–694. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, J.; He, J.; Liu, J.; Wang, H.; Liu, Y.; Jiang, T.; Zhang, Q.; Fu, X.; Xu, Y. An Immune System-Modified Rat Model for Human Stem Cell Transplantation Research. Stem Cell Rep. 2018, 11, 514–521. [Google Scholar] [CrossRef]
- He, D.; Zhang, J.; Wu, W.; Yi, N.; He, W.; Lu, P.; Li, B.; Yang, N.; Wang, D.; Xue, Z.; et al. A novel immunodeficient rat model supports human lung cancer xenografts. FASEB J. 2018, 33, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Ménoret, S.; Ouisse, L.-H.; Tesson, L.; Delbos, F.; Garnier, D.; Remy, S.; Usal, C.; Concordet, J.-P.; Giovannangeli, C.; Chenouard, V.; et al. Generation of Immunodeficient Rats with Rag1 and Il2rg Gene Deletions and Human Tissue Grafting Models. Transplantation 2018, 102, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Iwamoto, M.; Saito, Y.; Fuchimoto, D.; Sembon, S.; Suzuki, M.; Mikawa, S.; Hashimoto, M.; Aoki, Y.; Najima, Y.; et al. Il2rg Gene-Targeted Severe Combined Immunodeficiency Pigs. Cell Stem Cell 2012, 10, 753–758. [Google Scholar] [CrossRef]
- Watanabe, M.; Nakano, K.; Matsunari, H.; Matsuda, T.; Maehara, M.; Kanai, T.; Kobayashi, M.; Matsumura, Y.; Sakai, R.; Kuramoto, M.; et al. Generation of Interleukin-2 Receptor Gamma Gene Knockout Pigs from Somatic Cells Genetically Modified by Zinc Finger Nuclease-Encoding mRNA. PLoS ONE 2013, 8, e76478. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Shibata, H.; Nakano, K.; Abe, T.; Uosaki, H.; Ohnuki, T.; Hishikawa, S.; Kunita, S.; Watanabe, M.; Nureki, O.; et al. Production and rearing of germ-free X-SCID pigs. Exp. Anim. 2017, 67, 139–146. [Google Scholar] [CrossRef]
- Suzuki, S.; Iwamoto, M.; Hashimoto, M.; Suzuki, M.; Nakai, M.; Fuchimoto, D.; Sembon, S.; Eguchi-Ogawa, T.; Uenishi, H.; Onishi, A. Generation and characterization of RAG2 knockout pigs as animal model for severe combined immunodeficiency. Veter. Immunol. Immunopathol. 2016, 178, 37–49. [Google Scholar] [CrossRef]
- Boettcher, A.N.; Li, Y.; Ahrens, A.P.; Kiupel, M.; Byrne, K.A.; Loving, C.L.; Cino-Ozuna, A.G.; Wiarda, J.E.; Adur, M.; Schultz, B.; et al. Novel Engraftment and T Cell Differentiation of Human Hematopoietic Cells in ART−/−IL2RG−/Y SCID Pigs. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Choi, Y.-J.; Lee, K.; Park, W.-J.; Kwon, D.-N.; Park, C.; Do, J.T.; Song, H.; Cho, S.-K.; Park, K.-W.; Brown, A.N.; et al. Partial loss of interleukin 2 receptor gamma function in pigs provides mechanistic insights for the study of human immunodeficiency syndrome. Oncotarget 2016, 7, 50914–50926. [Google Scholar] [CrossRef]
- Kang, J.-T.; Cho, B.; Ryu, J.; Ray, C.; Lee, E.-J.; Yun, Y.-J.; Ahn, S.; Lee, J.; Ji, D.-Y.; Jue, N.; et al. Biallelic modification of IL2RG leads to severe combined immunodeficiency in pigs. Reprod. Biol. Endocrinol. 2016, 14, 1–9. [Google Scholar] [CrossRef][Green Version]
- Lei, S.; Ryu, J.; Wen, K.; Twitchell, E.; Bui, T.; Ramesh, A.; Weiss, M.; Li, G.; Samuel, H.; Clark-Deener, S.; et al. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci. Rep. 2016, 6, 25222. [Google Scholar] [CrossRef]
- Sato, K.; Oiwa, R.; Kumita, W.; Henry, R.; Sakuma, T.; Ito, R.; Nozu, R.; Inoue, T.; Katano, I.; Sato, K.; et al. Generation of a Nonhuman Primate Model of Severe Combined Immunodeficiency Using Highly Efficient Genome Editing. Cell Stem Cell 2016, 19, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Zschemisch, N.-H.; Glage, S.; Wedekind, D.; Weinstein, E.J.; Cui, X.; Dorsch, M.; Hedrich, H. Zinc-finger nuclease mediated disruption of Rag1 in the LEW/Ztm rat. BMC Immunol. 2012, 13, 60. [Google Scholar] [CrossRef] [PubMed]
- Ménoret, S.; Fontanière, S.; Jantz, D.; Tesson, L.; Thinard, R.; Rémy, S.; Usal, C.; Ouisse, L.; Fraichard, A.; Anegon, I. Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J. 2012, 27, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Sendai, Y.; Yamazaki, S.; Seki-Soma, M.; Hirose, K.; Watanabe, M.; Fukawa, K.; Nakauchi, H. Generation of Recombination Activating Gene-1-Deficient Neonatal Piglets: A Model of T and B Cell Deficient Severe Combined Immune Deficiency. PLoS ONE 2014, 9, e113833. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Guo, X.; Fan, N.; Song, J.; Zhao, B.; Ouyang, Z.; Liu, Z.; Zhao, Y.; Yan, Q.; Yi, X.; et al. RAG1/2 Knockout Pigs with Severe Combined Immunodeficiency. J. Immunol. 2014, 193, 1496–1503. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.-X.; Ying, B.; Li, R.; Tollefson, A.E.; Spencer, J.F.; Wold, W.S.M.; Song, S.-H.; Kong, I.; Toth, K.; Wang, Y.; et al. Characterization of an N-Terminal Non-Core Domain of RAG1 Gene Disrupted Syrian Hamster Model Generated by CRISPR Cas9. Viruses 2018, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fan, C.; Zhou, S.; Guo, Y.; Zuo, Q.; Ma, J.; Liu, S.; Wu, X.; Peng, Z.; Fan, T.; et al. Bioluminescent imaging of vaccinia virus infection in immunocompetent and immunodeficient rats as a model for human smallpox. Sci. Rep. 2015, 5, 11397. [Google Scholar] [CrossRef]
- Noto, F.K.; Steffey, V.A.; Tong, M.; Ravichandran, K.; Zhang, W.; Arey, A.; McClain, C.B.; Ostertag, E.; Mazhar, S.; Sangodkar, J.; et al. Sprague Dawley Rag2-Null Rats Created from Engineered Spermatogonial Stem Cells Are Immunodeficient and Permissive to Human Xenografts. Mol. Cancer Ther. 2018, 17, 2481–2489. [Google Scholar] [CrossRef]
- Lee, K.; Kwon, D.-N.; Ezashi, T.; Choi, Y.-J.; Park, C.; Ericsson, A.C.; Brown, A.N.; Samuel, M.S.; Park, K.-W.; Walters, E.M.; et al. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc. Natl. Acad. Sci. USA 2014, 111, 7260–7265. [Google Scholar] [CrossRef]
- Ménoret, S.; Iscache, A.-L.; Tesson, L.; Rémy, S.; Usal, C.; Osborn, M.J.; Cost, G.J.; Brüggemann, M.; Buelow, R.; Anegon, I. Characterization of immunoglobulin heavy chain knockout rats. Eur. J. Immunol. 2010, 40, 2932–2941. [Google Scholar] [CrossRef]
- Panzer, S.E.; Wilson, N.A.; Verhoven, B.M.; Xiang, D.; Rubinstein, C.D.; Redfield, R.R.; Zhong, W.; Reese, S.R. Complete B Cell Deficiency Reduces Allograft Inflammation and Intragraft Macrophages a Rat Kidney Transplant Model. Transplantation 2017, 102, 396–405. [Google Scholar] [CrossRef]
- Chen, F.; Wang, Y.; Yuan, Y.; Zhang, W.; Ren, Z.; Jin, Y.; Liu, X.; Xiong, Q.; Chen, Q.; Zhang, M.; et al. Generation of B Cell-Deficient Pigs by Highly Efficient CRISPR/Cas9-Mediated Gene Targeting. J. Genet. Genom. 2015, 42, 437–444. [Google Scholar] [CrossRef]
- Bell, T.G.; Butler, K.L.; Sill, H.B.; Stickle, J.E.; Ramos-Vara, J.A.; Dark, M.J. Autosomal Recessive Severe Combined Immunodeficiency of Jack Russell Terriers. J. Veter. Diagn. Investig. 2002, 14, 194–204. [Google Scholar] [CrossRef]
- Goto, T.; Hara, H.; Nakauchi, H.; Hochi, S.; Hirabayashi, M. Hypomorphic phenotype of Foxn1 gene-modified rats by CRISPR/Cas9 system. Transgenic Res. 2016, 25, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Ozuna, A.G.C.; Rowland, R.R.R.; Nietfeld, J.C.; Kerrigan, M.A.; Dekkers, J.C.M.; Wyatt, C. Preliminary Findings of a Previously Unrecognized Porcine Primary Immunodeficiency Disorder. Veter. Pathol. 2012, 50, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Waide, E.H.; Dekkers, J.C.M.; Ross, J.W.; Rowland, R.R.R.; Wyatt, C.R.; Ewen, C.L.; Evans, A.B.; Thekkoot, D.M.; Boddicker, N.J.; Serão, N.V.L.; et al. Not All SCID Pigs Are Created Equally: Two Independent Mutations in the Artemis Gene Cause SCID in Pigs. J. Immunol. 2015, 195, 3171–3179. [Google Scholar] [CrossRef]
- Jasper, P.J.; Rhee, K.-J.; Kalis, S.L.; Sethupathi, P.; Yam, P.-C.; Zhai, S.-K.; Knight, K. B lymphocyte deficiency in IgH-transgenic rabbits. Eur. J. Immunol. 2007, 37, 2290–2299. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, N.; Ruan, J.; Zhang, J.; Chen, Y.E.; Xu, J. Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Sci. Rep. 2017, 7, 12202. [Google Scholar] [CrossRef]
- Hashikawa, Y.; Hayashi, R.; Tajima, M.; Okubo, T.; Azuma, S.; Kuwamura, M.; Takai, N.; Osada, Y.; Kunihiro, Y.; Mashimo, T.; et al. Generation of knockout rabbits with X-linked severe combined immunodeficiency (X-SCID) using CRISPR/Cas9. Sci. Rep. 2020, 10, 9957. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhong, J.; Guo, X.; Chen, Y.; Zou, Q.; Huang, J.; Li, X.; Zhang, Q.; Jiang, Z.; Tang, C.; et al. Generation of RAG 1- and 2-deficient rabbits by embryo microinjection of TALENs. Cell Res. 2013, 23, 1059–1062. [Google Scholar] [CrossRef][Green Version]
- Flisikowska, T.; Thorey, I.S.; Offner, S.; Ros, F.; Lifke, V.; Zeitler, B.; Rottmann, O.; Vincent, A.; Zhang, L.; Jenkins, S.; et al. Efficient Immunoglobulin Gene Disruption and Targeted Replacement in Rabbit Using Zinc Finger Nucleases. PLoS ONE 2011, 6, e21045. [Google Scholar] [CrossRef] [PubMed]
- Esteves, P.J.; Abrantes, J.; Baldauf, H.-M.; Benmohamed, L.; Chen, Y.; Christensen, N.; González-Gallego, J.; Giacani, L.; Hu, J.; Kaplan, G.; et al. The wide utility of rabbits as models of human diseases. Exp. Mol. Med. 2018, 50, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Watanabe, T. Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol. Ther. 2003, 99, 261–282. [Google Scholar] [CrossRef]
- Peng, X. Transgenic Rabbit Models for Studying Human Cardiovascular Diseases. Comp. Med. 2012, 62, 472–479. [Google Scholar]
- Peng, X.; Knouse, J.A.; Hernon, K.M. Rabbit Models for Studying Human Infectious Diseases. Comp. Med. 2015, 65, 499–507. [Google Scholar]
- Fan, J.; Kitajima, S.; Watanabe, T.; Xu, J.; Zhang, J.; Liu, E.; Chen, Y.E. Rabbit models for the study of human atherosclerosis: From pathophysiological mechanisms to translational medicine. Pharmacol. Ther. 2015, 146, 104–119. [Google Scholar] [CrossRef]
- Pearce, J.M.S. Louis Pasteur and Rabies: A brief note. J. Neurol. Neurosurg. Psychiatry 2002, 73, 82. [Google Scholar] [CrossRef]
- Cambau, E.; Drancourt, M. Steps towards the discovery of Mycobacterium tuberculosis by Robert Koch, 1882. Clin. Microbiol. Infect. 2014, 20, 196–201. [Google Scholar] [CrossRef]
- Epstein, F.H.; Brown, M.S.; Kita, T.; Brown, M.S. Defective Lipoprotein Receptors and Atherosclerosis. N. Engl. J. Med. 1983, 309, 288–296. [Google Scholar] [CrossRef]
- Chang, M.C. Fertilization of Rabbit Ova in vitro. Nat. Cell Biol. 1959, 184, 466–467. [Google Scholar] [CrossRef]
- Biggers, J.D. Walter Heape, FRS: A pioneer in reproductive biology. Centenary of his embryo transfer experiments. Reproduction 1991, 93, 173–186. [Google Scholar] [CrossRef]
- Song, J.; Zhang, J.; Xu, J.; Garcia-Barrio, M.; Chen, Y.E.; Yang, D. Genome engineering technologies in rabbits. J. Biomed. Res. 2020, 34, 1–13. [Google Scholar] [CrossRef]
- Mage, R.G.; Esteves, P.J.; Rader, C. Rabbit models of human diseases for diagnostics and therapeutics development. Dev. Comp. Immunol. 2019, 92, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, A.; Neves, F.; De Matos, A.L.; Abrantes, J.; Van Der Loo, W.; Mage, R.; Esteves, P.J. An overview of the lagomorph immune system and its genetic diversity. Immunogenetics 2015, 68, 83–107. [Google Scholar] [CrossRef]
- Pinheiro, A.; Lanning, D.; Alves, P.C.; Mage, R.G.; Knight, K.L.; Van Der Loo, W.; Esteves, P.J. Molecular bases of genetic diversity and evolution of the immunoglobulin heavy chain variable region (IGHV) gene locus in leporids. Immunogenetics 2011, 63, 397–408. [Google Scholar] [CrossRef]
- Katzourakis, A.; Tristem, M.; Pybus, O.G.; Gifford, R.J. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. USA 2007, 104, 6261–6265. [Google Scholar] [CrossRef]
- Alves, J.M.; Carneiro, M.; Cheng, J.Y.; De Matos, A.L.; Rahman, M.M.; Loog, L.; Campos, P.F.; Wales, N.; Eriksson, A.; Manica, A.; et al. Parallel adaptation of rabbit populations to myxoma virus. Science 2019, 363, 1319–1326. [Google Scholar] [CrossRef]
- Abrantes, J.; Van Der Loo, W.; Le Pendu, J.; Esteves, P.J. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): A review. Veter. Res. 2012, 43, 12. [Google Scholar] [CrossRef]
- Song, J.; Wang, G.; Hoenerhoff, M.J.; Ruan, J.; Yang, D.; Zhang, J.; Yang, J.; Lester, P.A.; Sigler, R.; Bradley, M.; et al. Bacterial and Pneumocystis Infections in the Lungs of Gene-Knockout Rabbits with Severe Combined Immunodeficiency. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Ma, Y.; Chen, Y.E.; Zhang, J.; Lin, T.-A.; Chen, C.-H.; Lin, W.-W.; Roach, M.; Ju, J.-C.; Yang, L.; et al. Recombinant Rabbit Leukemia Inhibitory Factor and Rabbit Embryonic Fibroblasts Support the Derivation and Maintenance of Rabbit Embryonic Stem Cells. Cell. Reprogramming 2012, 14, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Xu, J.; Gao, S.; Sung, L.Y.; Stone, D.; Joyner, M.; Zhang, J.; Chaubal, S.; Tian, X.; Chen, Y.E.; et al. 31 full-term and live rabbit clones produced by somatic cell nuclear transfer. Reprod. Fertil. Dev. 2006, 18, 124. [Google Scholar] [CrossRef]
- Lai, L.; Prather, R.S. Creating genetically modified pigs by using nuclear transfer. Reprod. Biol. Endocrinol. 2003, 1, 82. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ramsoondar, J.J.; Machaty, Z.; Costa, C.; Williams, B.L.; Fodor, W.L.; Bondioli, K.; Duffy, D.M. Production of α1,3-Galactosyltransferase-Knockout Cloned Pigs Expressing Human α1,2-Fucosylosyltransferase1. Biol. Reprod. 2003, 69, 437–445. [Google Scholar] [CrossRef]
- Lai, L.; Kwangwook, P.; Cheong, H.-T.; Kühholzer, B.; Samuel, M.; Bonk, A.; Im, G.-S.; Rieke, A.; Day, B.N.; Murphy, C.N.; et al. Transgenic pig expressing the enhanced green fluorescent protein produced by nuclear transfer using colchicine-treated fibroblasts as donor cells. Mol. Reprod. Dev. 2002, 62, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Schnieke, A.E.; Kind, A.J.; Ritchie, W.A.; Mycock, K.; Scott, A.R.; Ritchie, M.; Wilmut, I.; Colman, A.; Campbell, K.H.S. Human Factor IX Transgenic Sheep Produced by Transfer of Nuclei from Transfected Fetal Fibroblasts. Science 1997, 278, 2130–2133. [Google Scholar] [CrossRef] [PubMed]
- McCreath, K.J.; Howcroft, J.; Campbell, K.H.S.; Colman, A.D.; Schnieke, A.; Kind, A. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nat. Cell Biol. 2000, 405, 1066–1069. [Google Scholar] [CrossRef]
- Cibelli, J.B.; Stice, S.L.; Golueke, P.J.; Kane, J.J.; Jerry, J.; Blackwell, C.; De León, F.A.P.; Robl, J.M. Cloned Transgenic Calves Produced from Nonquiescent Fetal Fibroblasts. Science 1998, 280, 1256–1258. [Google Scholar] [CrossRef]
- Yan, Q.; Zhang, Q.; Yang, H.; Zou, Q.; Tang, C.; Fan, N.; Lai, L. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen. 2014, 3, 12. [Google Scholar] [CrossRef][Green Version]
- Ballou, S.M.; Song, J.; Xu, J.; Lester, P.A. Husbandry Efforts for Housing and Maintaining Immunodeficient Transgenic Rabbits. Lab. Anim. Sci. Prof. 2019, 7, 58–59. Available online: https://www.aalas.org/articles/2019/09/01/husbandry-efforts-for-housing-and-maintaining-immunodeficient--transgenic-rabbits (accessed on 19 October 2020).
- Mocé, E.; Vicente, J.S. Rabbit sperm cryopreservation: A review. Anim. Reprod. Sci. 2009, 110, 1–24. [Google Scholar] [CrossRef]
- Lin, T.; Chen, C.; Sung, L.-Y.; Carter, M.; Chen, Y.; Du, F.; Ju, J.; Xu, J. Open-pulled straw vitrification differentiates cryotolerance of in vitro cultured rabbit embryos at the eight-cell stage. Theriogenology 2011, 75, 760–768. [Google Scholar] [CrossRef]
- Yue, F.; The Mouse ENCODE Consortium; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Samantha, K.; Davis, C.; et al. A comparative encyclopedia of DNA elements in the mouse genome. Nat. Cell Biol. 2014, 515, 355–364. [Google Scholar] [CrossRef]
- Shultz, L.D.; Brehm, M.A.; Garcia-Martinez, J.V.; Greiner, D.L. Humanized mice for immune system investigation: Progress, promise and challenges. Nat. Rev. Immunol. 2012, 12, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Akkina, R.; Barber, D.L.; Bility, M.T.; Bissig, K.-D.; Burwitz, B.J.; Eichelberg, K.; Endsley, J.J.; Garcia, J.V.; Hafner, R.; Karakousis, P.C.; et al. Small Animal Models for Human Immunodeficiency Virus (HIV), Hepatitis B, and Tuberculosis: Proceedings of an NIAID Workshop. Curr. HIV Res. 2020, 18, 19–28. [Google Scholar] [CrossRef]
- Allen, T.M.; Brehm, M.A.; Bridges, S.; Ferguson, S.; Kumar, P.; Mirochnitchenko, O.; Palucka, K.; Pelanda, R.; Sanders-Beer, B.; Shultz, L.D.; et al. Humanized immune system mouse models: Progress, challenges and opportunities. Nat. Immunol. 2019, 20, 770–774. [Google Scholar] [CrossRef]
- Samal, J.; Kelly, S.; Na-Shatal, A.; Elhakiem, A.; Das, A.; Ding, M.; Sanyal, A.; Gupta, P.; Melody, K.; Roland, B.; et al. Human immunodeficiency virus infection induces lymphoid fibrosis in the BM-liver-thymus-spleen humanized mouse model. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef]
- Day, C.-P.; Merlino, G.; Van Dyke, T. Preclinical Mouse Cancer Models: A Maze of Opportunities and Challenges. Cell 2015, 163, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Thrasher, A.J.; Zhang, F. Gene therapy and genome editing for primary immunodeficiency diseases. Genes Dis. 2019, 7, 38–51. [Google Scholar] [CrossRef]
- Azuma, H.; Paulk, N.K.; Ranade, A.; Dorrell, C.; Al-Dhalimy, M.; Ellis, E.; Strom, S.; Kay, M.A.; Finegold, M.J.; Grompe, M. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat. Biotechnol. 2007, 25, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, K.; Prasolava, T.K.; Wang, J.C.Y.; Mortin-Toth, S.M.; Khalouei, S.; Gan, O.I.; Dick, J.E.; Danska, J.S. Polymorphism in Sirpa modulates engraftment of human hematopoietic stem cells. Nat. Immunol. 2007, 8, 1313–1323. [Google Scholar] [CrossRef]
| Year | Pre 2009 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | B: breeding | Z: ZFN | T: TALEN | C: Cas9 | |||||||||
| G: gene targeting | |||||||||||||
| M: meganuclease | |||||||||||||
| IL2RG | |||||||||||||
| Dog | B [24,25] | ||||||||||||
| Rat | Z [28] | Z [29] | C [30] | C [31] | |||||||||
| T [32] | |||||||||||||
| Pig | G [33] | Z [34,35] | G [36] | C [37] | |||||||||
| T [38] | |||||||||||||
| C [39,40] | |||||||||||||
| NHP | Z/T [41] | ||||||||||||
| Rag1 | |||||||||||||
| Rat | Z [42] | M [43] | C [31] | ||||||||||
| Pig | G [44] | ||||||||||||
| T [45] | |||||||||||||
| Hamster | C [46] | ||||||||||||
| Rag2 | |||||||||||||
| Rat | T [47] | T [48] | C [31] | ||||||||||
| Pig | T [45,49] | G [36] | |||||||||||
| C [40] | |||||||||||||
| IgM | |||||||||||||
| Rat | Z [50] | C [51] | |||||||||||
| Pig | C [52] | ||||||||||||
| Prkdc | |||||||||||||
| Dog | B [26,27,53] | ||||||||||||
| Horse | B [22,23] | ||||||||||||
| Rat | Z [29] | C [30] | |||||||||||
| Foxn1 | |||||||||||||
| Rat | C [54] | ||||||||||||
| Artemis | |||||||||||||
| Pig | B [55] | B [56] |
| Year | Pre 2009 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Tg: transgenic | Z: ZFN | T: TALEN | C: Cas9 | |||||||||
| VDJ-Cmu | Tg [57] | ||||||||||||
| IL2RG | C [58] | C [59] | |||||||||||
| Rag1 | T [60] | C [58] | |||||||||||
| Rag2 | T [60] | C [58] | |||||||||||
| IgM | Z [61] | ||||||||||||
| Prkdc | C [58] | ||||||||||||
| Foxn1 | C [58] |
| 1. Transplantation of stem cell-derived blood vessels, and examine long term safety and efficacy. |
| 2. PDX/CDX for drug development and screening for cancer and other diseases. |
| 3. Humanized livers for pharmacokinetics and hepatotoxicity studies. |
| 4. Long term efficacy and safety evaluation of neuronal stem cell-based therapy. |
| 5. A bioreactor for production of human hepatocytes. |
| 6. A bioreactor for production of human T-cells for CAR-T cell expansion. |
| 7. A large animal model with the human immune system for the study of infectious diseases. |
| 8. A large animal model with the human immune system for antibody discovery. |
| 9. A large animal model with the human immune system for TCR discovery. |
| 10. Development and validation of imaging and diagnostic tools. |
| 11. Study graft-versus-host disease (GVHD) in a large animal model. |
| 12. Development of gene therapy and/or gene editing therapy for primary immunodeficient diseases. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Pallas, B.; Yang, D.; Zhang, J.; Agarwal, Y.; Chen, Y.E.; Bility, M.; Xu, J. Immunodeficient Rabbit Models: History, Current Status and Future Perspectives. Appl. Sci. 2020, 10, 7369. https://doi.org/10.3390/app10207369
Song J, Pallas B, Yang D, Zhang J, Agarwal Y, Chen YE, Bility M, Xu J. Immunodeficient Rabbit Models: History, Current Status and Future Perspectives. Applied Sciences. 2020; 10(20):7369. https://doi.org/10.3390/app10207369
Chicago/Turabian StyleSong, Jun, Brooke Pallas, Dongshan Yang, Jifeng Zhang, Yash Agarwal, Y. Eugene Chen, Moses Bility, and Jie Xu. 2020. "Immunodeficient Rabbit Models: History, Current Status and Future Perspectives" Applied Sciences 10, no. 20: 7369. https://doi.org/10.3390/app10207369
APA StyleSong, J., Pallas, B., Yang, D., Zhang, J., Agarwal, Y., Chen, Y. E., Bility, M., & Xu, J. (2020). Immunodeficient Rabbit Models: History, Current Status and Future Perspectives. Applied Sciences, 10(20), 7369. https://doi.org/10.3390/app10207369





