Identification of Bacterial and Fungal Communities in the Roots of Orchids and Surrounding Soil in Heavy Metal Contaminated Area of Mining Heaps
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sampling and DNA Extraction
2.3. Chemical Analysis
2.4. Polymerase Chain Reaction and Library Preparation for Next-Generation Sequencing
2.5. Data Analysis
3. Results and Discussion
Bacterial Communities
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Taylor, D.L.; Bruns, T.D. Independent, specialized invasion of ectomycorrhizal mutualism by two nonphotosynthetic orchids. Proc. Natl. Acad. Sci. USA 1997, 94, 4510–4515. [Google Scholar] [CrossRef] [PubMed]
- McCormick, M.K.; Whigham, D.F.; O’Neill, J.P. Mycorrhizal diversity in photosynthetic terrestrial orchids. New Phytol. 2004, 163, 425–438. [Google Scholar] [CrossRef]
- Gill, D.; Otte, D.; Endler, J.A. Fruiting failure, pollinator inefficiency and speciation in orchids, Speciation and Its Consequences. In Speciation and Its Consequences; Otte, D., Endler, J.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1989; pp. 458–481. [Google Scholar]
- Otero, J.T.; Flanagan, N.S. Orchid diversity—Beyond deception. Trends Ecol. Evol. 2006, 21, 64–65. [Google Scholar] [PubMed]
- Banásová, V. Vegetation of copper and antimony mine heaps. Biol. Práce 1976, 22, 1–109. [Google Scholar]
- Rasmussen, H.N. Terrestrial Orchids: From Seed to Mycotrophic Plant; Cambridge University Press: Cambridge, UK, 1995; p. 433. [Google Scholar]
- George, E.; Häussler, K.U.; Vetterlein, D.; Gorgus, E.; Marschner, H. Water and nutrient translocation by hyphae of Glomus mosseae. Can. J. Bot. 1992, 70, 2130–2137. [Google Scholar] [CrossRef]
- Jasper, D.A.; Abbott, L.K.; Robson, A.D. Hyphae of a vesicular-arbuscular mycorrhizal fungus maintain infectivity in dry soil, except when the soil is disturbed. New Phytol. 1989, 112, 101–107. [Google Scholar] [CrossRef]
- Thomas, R.S.; Franson, R.L.; Bethlenfalvay, G.J. Separation of arbuscular mycorrhizal fungus and root effect on soil aggregation. Soil Sci. Soc. Am. J. 1993, 57, 77–81. [Google Scholar] [CrossRef]
- Ashida, J. Adaptation of fungi to metal toxicants. Annu. Rev. Phytopathol. 1965, 3, 153–174. [Google Scholar] [CrossRef]
- Fourest, E.; Roux, J.C. Heavy Metal Biosorption by Fungal Mycelial By-Products Mechanisms and Influence of pH. Appl. Microbiol. Biotechnol. 1992, 37, 399–403. [Google Scholar] [CrossRef]
- Hamilton, W.A. Microbially influenced corrosion as a model system for the study of metal microbe interactions: A unifying electron transfer hypothesis. Biofouling 2003, 19, 65–76. [Google Scholar] [CrossRef]
- Remenár, M.; Harichová, J.; Zámocký, M.; Pangallo, D.; Szemes, T.; Budiš, J.; Šoltys, K.; Ferianc, P. Metagenomics of a nickel-resistant bacterial community in an anthropogenic nickel-contaminated soil in southwest Slovakia. Biologia 2017, 72, 971–981. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kraková, L.; Šoltys, K.; Otlewska, A.; Pietrzak, K.; Purkrtová, S.; Savická, D.; Puškárová, A.; Bučková, M.; Szemes, T.; Budiš, J.; et al. Comparison of methods for identification of microbial communities in book collections: Culture-dependent (sequencing and MALDI-TOF MS) and culture-independent (Illumina MiSeq). Int. Biodeterior. Biodegrad. 2018, 131, 51–59. [Google Scholar] [CrossRef]
- Böhmer, M.; Smol’ak, D.; Ženišová, K.; Čaplová, Z.; Pangallo, D.; Puškárová, A.; Bučková, M.; Cabicarová, T.; Budiš, J.; Šoltýs, K.; et al. Comparison of the Microbial Diversity During Two Different Wine Fermentation Processes. Appl. Sci. 2020, 367, fnaa150. [Google Scholar] [CrossRef]
- Hong, C.; Si, Y.; Xing, Y.; Li, Y. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 2015, 22, 10788–10799. [Google Scholar] [CrossRef]
- Xiao, E.Z.; Krumins, V.; Tang, S.; Xiao, T.F.; Ning, Z.P.; Lan, X.L.; Sun, W.M. Correlating microbial community profiles with geochemical conditions in a watershed heavily contaminated by an antimony tailing pond. Environ. Pollut. 2016, 215, 141–153. [Google Scholar] [CrossRef]
- Račko, M.; Ozdín, D.; Kučerová, G.; Jurkovič, L.; Vaculík, M. Occurrence and uptake of heavy metals by selected terrestrial orchids in extreme conditions of initial soils on previous mining sites. Biologia 2020, in press. [Google Scholar] [CrossRef]
- Ďuďa, R.; Ozdín, D. Minerály Slovenska; Granit: Praha, Czech Republic, 2012; 480p. [Google Scholar]
- Števko, M.; Ozdín, D. Supergene native silver and acanthite from the Jasenie-Soviansko base metals deposit, Nízke Tatry Mts. (Slovak Republic). Bull. Mineral. Petrolog. Odd. Nár. Muz. 2012, 20, 47–51. [Google Scholar]
- Grecula, P. (Ed.) Mineral Deposits of the Slovak Ore Mountains; Geocomplex: Bratislava, Slovakia, 1995; Volume 1, 834p. [Google Scholar]
- Cao, L.; Qiu, Z.; Dai, X.; Tan, H.; Lin, Y.; Zhou, S. Isolation of Endophytic Actinomycetes From Roots and Leaves of Banana (Musa Acuminata) Plants and Their Activities Against Fusarium oxysporumf. sp. cubense. World J. Microbiol. Biotechnol. 2004, 20, 501–504. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–321. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 20 November 2015).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Budiš, J.; Krampl, W.; Kucharík, M.; Hekel, R.; Lichvár, M.; Smol’ak, D.; Böhmer, M.; Baláž, A.; Ďuriš, F.; Gazdarica, J.; et al. SnakeLines: Integrated set of computational pipelines for paired-end sequencing reads. 2019. Available online: https://github.com/jbudis/snakelines/ (accessed on 18 October 2019).
- Köster, J.; Rahmann, S. Snakemake—S scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef]
- Turnau, K.; Gawroński, S.; Ryszka, P.; Zook, D. Mycorrhizal-based phytostabilization of Zn–Pb tailings: Lessons from the Trzebionka mining works (Southern Poland). In Bio-Geo Interactions in Metal-Contaminated Soils; Soil Biology; Varma, A., Kothe, E., Eds.; Springer: Berlin, Germany, 2012; Volume 31, pp. 327–348. [Google Scholar]
- Punz, W.; Sieghardt, H. The response of roots of herbaceous plant species to heavy metals. Environ. Exp. Bot. 1993, 33, 85–98. [Google Scholar] [CrossRef]
- Rufo, L.; de la Fuente, V. Successional dynamics of the climatophile vegetation of the mining territory of the Río Tinto basin (Huelva, Spain): Soil characteristics and implications for phytoremediation. Arid Land Res. Manag. 2010, 24, 301–327. [Google Scholar] [CrossRef]
- Dearnaley, J.D.W.; Martos, F.; Selosse, M.A. Orchid mycorrhizas: Molecular ecology, physiology, evolution and conservation aspects. In Fungal Associations, 2nd ed.; The Mycota (9); Hock, B., Ed.; Springer-Verlag: Berlin, Germany, 2012; pp. 207–230. [Google Scholar]
- Dearnaley, J.D.W. Further advances in orchid mycorrhizal research. Mycorrhiza 2007, 17, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Shefferson, R.P.; Kull, T.; Tali, K. Mycorrhizal interactions of orchids colonizing Estonian mine tailings hills. Am. J. Bot. 2008, 95, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Blagodatskaya, E.V.; Anderson, T.H. Interactive effects of pH and substrate quality on the fungal-to-bacterial ratio and QCO2 of microbial communities in forest soils. Soil Biol. Biochem. 1998, 30, 1269–1274. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Stępniewska, H.; Uzarowicz, Ł.; Błońska, E.; Kwasowski, W.; Słodczyk, Z.; Gałka, D.; Hebda, A. Fungal abundance and diversity as influenced by properties of Technosols developed from mine wastes containing iron sulphides: A case study from abandoned iron sulphide and uranium mine in Rudki, south-central Poland. Appl. Soil Ecol. 2020, 145, 103349. [Google Scholar] [CrossRef]
- Šimonovičová, A.; Kraková, L.; Pauditšová, E.; Pangallo, D. Occurrence and diversity of cultivable autochthonous microscopic fungi in substrates of old environmental loads from mining activities in Slovakia. Ecotoxicol. Environ. Saf. 2019, 172, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Jurkiewicz, A.; Turnau, K.; Mesjasz-Przybyłowicz, J.; Przybyłowicz, W.; Godzik, B. Heavy metal localisation in mycorrhizas of Epipactis atrorubens (Hoffm.) Besser (Orchidaceae) from zinc mine tailings. Protoplasma 2001, 218, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Šimonovičová, A.; Ferianc, P.; Vojtková, H.; Pangallo, D.; Hanajík, P.; Kraková, L.; Feketeová, Z.; Čerňanský, S.; Okenicová, L.; Žemberyová, M.; et al. Alkaline Technosol contaminated by former mining activity and its culturable autochthonous microbiota. Chemosphere 2017, 171, 89–96. [Google Scholar] [CrossRef]
- Timling, I.; Taylor, D.L. Peeking through a frosty window: Molecular insights into the ecology of Arctic soil fungi. Fungal Ecol. 2012, 5, 419–429. [Google Scholar] [CrossRef]
- Chávez, R.; Fierro, F.; García-Rico, R.O.; Vaca, I. Filamentous fungi from extreme environments as a promising source of novel bioactive secondary metabolites. Front. Microbiol. 2015, 6, 903. [Google Scholar] [CrossRef]
- Gupta, H.; Kumar, R.; Park, H.S.; Jeon, B.H. Photocatalytic efficiency of iron oxide nanoparticles for the degradation of priority pollutant anthracene. Geosyst. Eng. 2017, 20, 21–27. [Google Scholar] [CrossRef]
- Mohammadi, K.; Khalesro, S.; Sohrabi, Y.; Heidari, G. A review: Beneficial effects of the mycorrhizal fungi for plant growth. J. Appl. Environ. Biol. Sci. 2011, 1, 310–319. [Google Scholar]
- Baldrian, P. Interactions of heavy metals with white-rot fungi. Enzym. Microb. Technol. 2003, 32, 78–91. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, J.; Gao, C.; Guo, S. Mycena sp., a mycorrhizal fungus of the orchid Dendrobium officinale. Mycol. Prog. 2012, 11, 395–401. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, X.; Yang, H.; Sun, L. Bioremediation of heavy metal pollution utilizing composite microbial agent of Mucor circinelloides, Actinomucor sp. and Mortierella sp. J. Environ. Chem. Eng. 2017, 5, 3616–3621. [Google Scholar] [CrossRef]
- Niu, H.; Xu, X.S.; Wang, J.H.; Volesky, B. Removal of lead from aqueous solutions by Penicillium biomass. Biotechnol. Bioeng. 1993, 42, 785–787. [Google Scholar] [CrossRef] [PubMed]
- Chai, B.; Wu, Y.; Liu, P.; Liu, B.; Gao, M. Isolation and phosphate-solubilizing ability of a fungus, Penicillium sp. from soil of an alum mine. J. Basic Microbiol. 2011, 51, 5–14. [Google Scholar] [CrossRef]
- Mishra, S.; Chaudhury, G. Biosorption of Copper by Penicillium sp. Int. J. 1995, 14, 111–126. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, Minerals and Microbes: Geomicrobiology and Bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol. 2018, 18, 11. [Google Scholar] [CrossRef]
- Guo, H.; Nasir, M.; Lv, J.; Dai, Y.; Gao, J. Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotoxicol. Environ. Saf. 2017, 144, 300–306. [Google Scholar] [CrossRef]
- Marcin, C.; Marcin, G.; Justyna, M.P.; Katarzyna, K.; Maria, N. Diversity of microorganisms from forest soils differently polluted with heavy metals. Appl. Soil Ecol. 2013, 64, 7–14. [Google Scholar]
- Tipayno, S.C.; Truu, J.; Samaddar, S.; Truu, M.; Preem, J.K.; Oopkaup, K.; Espenberg, M.; Chatterjee, P.; Kang, Y.; Kim, K.; et al. The bacterial community structure and functional profile in the heavy metal contaminated paddy soils, surrounding a nonferrous smelter in South Korea. Ecol. Evol. 2018, 8, 6157–6168. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Y.; Chen, Q.; Li, Y.; Guo, D.; Nie, X.; Peng, X. Assessment of heavy metal pollution and the effect on bacterial community in acidic and neutral soils. Ecol. Indic. 2020, 117, 106626. [Google Scholar] [CrossRef]
- Brewer, T.E.; Handley, K.M.; Carini, P.; Gilbert, J.A.; Fierer, N. Genome reduction in an abundant and ubiquitous soil bacterium ‘Candidatus Udaeobacter copiosus’. Nat. Microbiol. 2016, 2, 16198. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Kube, M.; Bauer, M.; Teeling, H.; Lombardot, T.; Ludwig, W.; Gade, D.; Beck, A.; Borzym, K.; Heitmann, K.; et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 2003, 100, 8298–8303. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Yasouri, F.N. Plasmid mediated degradation of diazinon by three bacterial strains Pseudomonas sp., Flavobacterium sp. and Agrobacterium sp. Asian J. Chem. 2006, 18, 2437–2444. [Google Scholar]
- Lakshmi, C.V.; Kumar, M.; Khanna, S. Biotransformation of chlorpyrifos and bioremediation of contaminated soil. Int. Biodeter. Biodegr. 2008, 62, 204–209. [Google Scholar] [CrossRef]
- Sun, W.; Dong, Y.; Gao, P.; Fu, M.; Ta, K.; Li, J. Microbial communities inhabiting oil-contaminated soils from two major oilfields in Northern China: Implications for active petroleum-degrading capacity. J. Microbiol. 2015, 53, 371–378. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Kim, J. Sphingomonas naphthae sp. nov., isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 4621–4627. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Q.; Liu, C.; Yu, J. Effect of different enrichment strategies on microbial community structure in petroleum-contaminated marine sediment in Dalian, China. Mar. Pollut. Bull. 2017, 117, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, C.; Li, H.; Bao, M.; Sun, P. Regulation of different electron acceptors on petroleum hydrocarbon biotransformation to final products in activated sludge biosystems. Bioprocess Biosyst. Eng. 2019, 42, 643–655. [Google Scholar] [CrossRef]
- Feng, X.; Liu, Z.; Jia, X.; Lu, W. Distribution of Bacterial Communities in Petroleum-Contaminated Soils from the Dagang Oilfield, China. Trans. Tianjin Univ. 2020, 26, 22–32. [Google Scholar] [CrossRef]
- Elshahed, M.S.; Youssef, N.H.; Luo, Q.; Najar, F.Z.; Roe, B.A.; Sisk, T.M.; Bühring, S.I.; Hinrichs, K.U.; Krumholz, L.R. Phylogenetic and Metabolic Diversity of Planctomycetes from Anaerobic, Sulfide- and Sulfur-Rich Zodletone Spring, Oklahoma. Appl. Environ. Microbiol. 2007, 73, 4707–4716. [Google Scholar] [CrossRef] [PubMed]
- Arp, D.; Sayavedra-Soto, L.; Hommes, N. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 2002, 178, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Coombs, J.T.; Franco, C.M.M. Isolation and identification of actinobacteria isolated from surface-sterilized wheat roots. Appl. Environ. Microbiol. 2003, 69, 5603–5608. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, D.K.; Kim, J. Flavobacterium olei sp. nov., a novel psychrotolerant bacterium isolated from oil-contaminated soil Free. Int. J. Syst. Evol. Microbiol. 2017, 67, 2211–2218. [Google Scholar] [CrossRef] [PubMed]

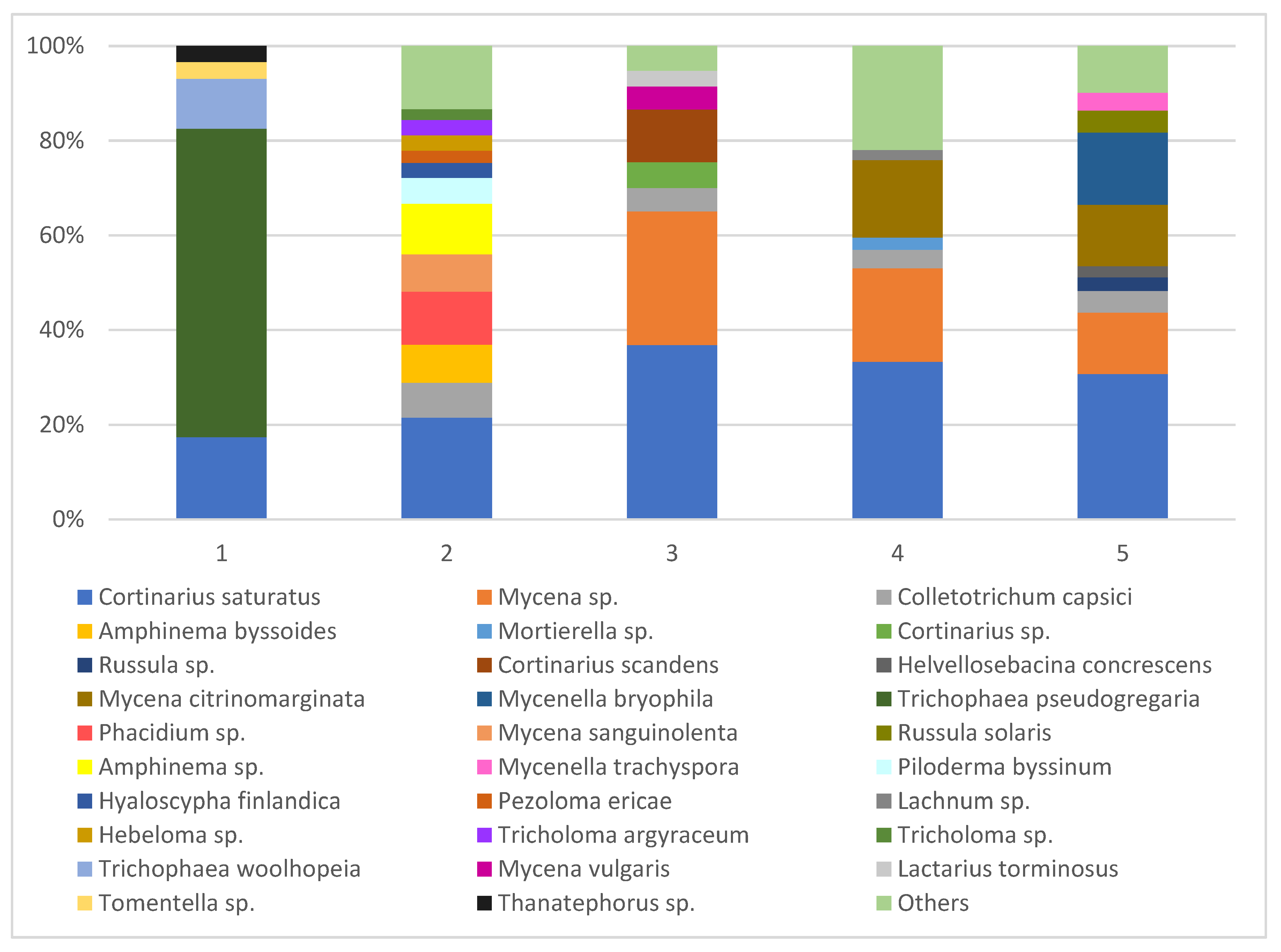

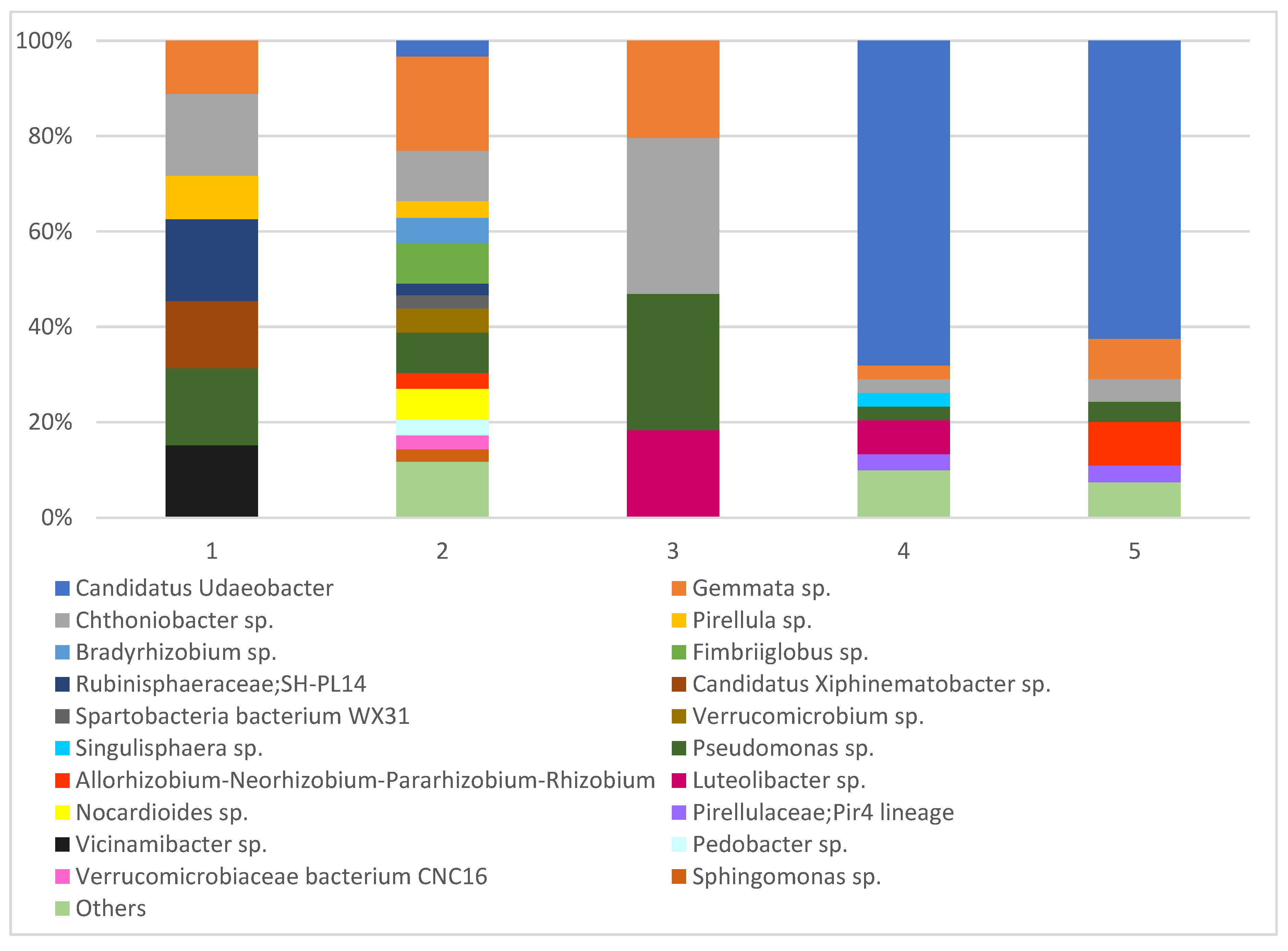
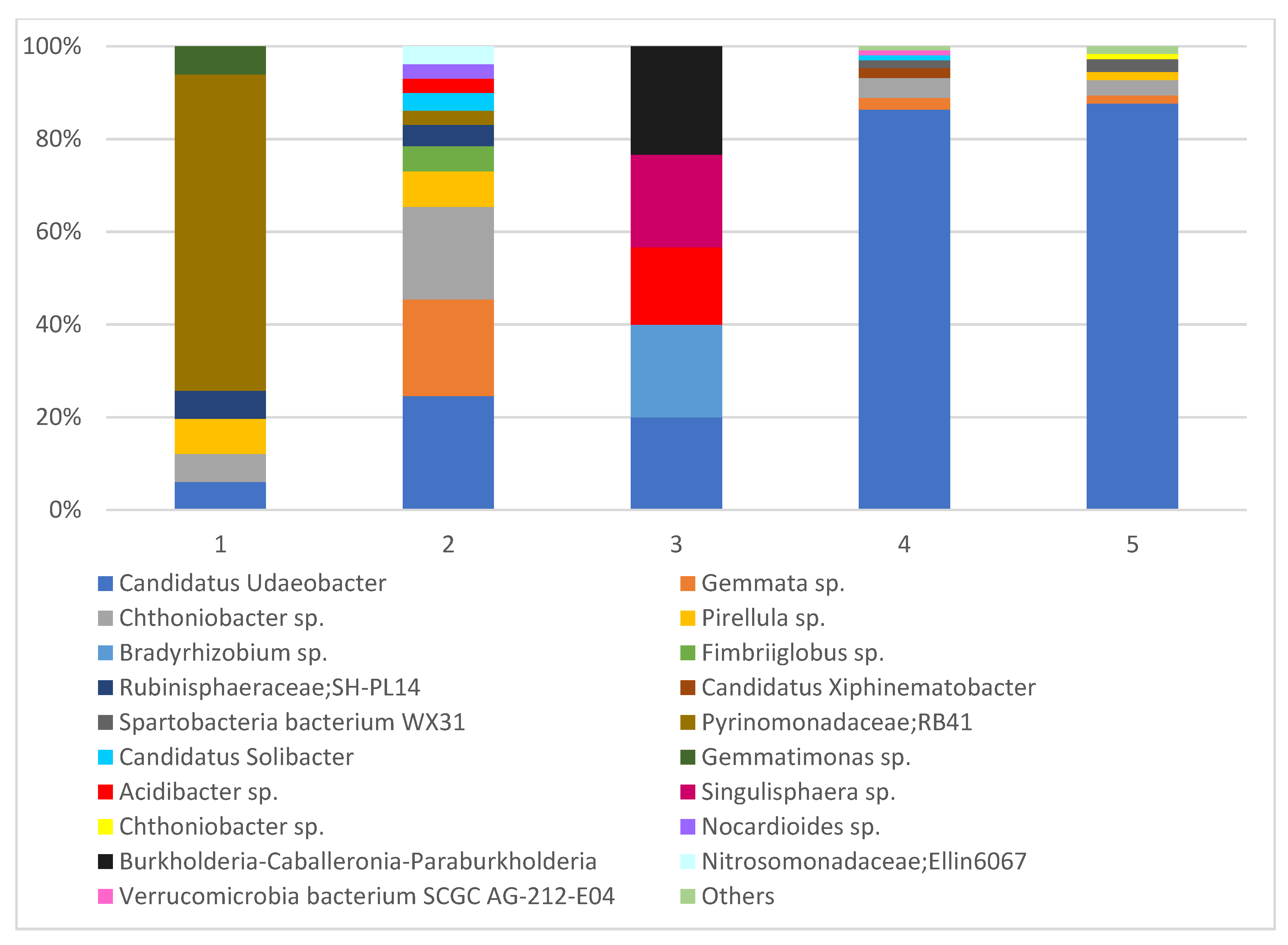
| The Most Abundant Strains of Fungi Detected in the Roots of Orchids and Surrounding Soils | 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Epipactis atrorubens | Epipactis atrorubens | Platanthera bifolia | Epipactis pontica | Cephalanthera longifolia | ||||||
| Ni-Co Mining Heap Dobšiná | Pb-Zn Mining Heap Jasenie | Fe Mining Heap Sirk | ||||||||
| pH 7.73 | pH 7.36 | pH 7.81 | pH 4.98 | pH 6.58 | ||||||
| Roots | Soil | Roots | Soil | Roots | Soil | Roots | Soil | Roots | Soil | |
| Ascomycota | ||||||||||
| Alatospora acuminata | − | − | − | + | − | − | − | − | − | − |
| Alatospora sp. | − | − | − | + | − | − | − | − | − | − |
| Arthrobotrys conoides | − | + | − | − | − | − | − | − | − | − |
| Arthrobotrys sp. | − | + | − | − | − | − | − | − | − | − |
| Aureobasidium pullulans | − | + | − | − | − | − | − | − | − | − |
| Bradymyces alpinus | − | + | − | − | − | − | − | − | − | − |
| Cadophora sp. | − | + | − | − | − | − | − | − | − | − |
| Capronia sp. | − | − | − | − | − | + | − | − | − | − |
| Cenococcum geophilum | − | − | − | − | − | − | − | − | + | + |
| Cenococcum sp. | − | − | − | − | − | − | + | + | + | + |
| Cistella sp. | − | − | − | + | − | − | − | − | − | − |
| Cladophialophora minutissima | − | − | − | + | − | − | − | − | − | − |
| Cladosporium sp. | − | + | − | − | − | − | − | − | − | − |
| Colletotrichum capsici | − | + | + | + | + | + | + | + | + | + |
| Dothidea eucalypti | − | − | − | + | − | − | − | − | − | − |
| Exophiala equina | − | − | − | + | − | − | + | + | − | + |
| Exophiala radicis | − | − | − | + | − | − | + | + | − | + |
| Exophiala sp. | − | − | + | + | − | − | + | + | − | + |
| Geopora arenicola | − | + | − | − | − | − | − | − | − | − |
| Geopora sp. | − | + | − | − | − | − | − | − | − | − |
| Geoglossum fallax | − | − | − | − | − | − | + | + | − | − |
| Geomyces auratus | − | − | − | − | − | + | − | − | − | − |
| Gyoerffyella sp. | − | − | − | − | − | − | + | − | − | − |
| Herpotrichia sp. | − | − | − | − | − | − | − | − | + | − |
| Humaria hemisphaerica | − | − | − | − | − | − | + | − | − | − |
| Humaria sp. | − | − | − | − | − | − | + | − | − | − |
| Hyalodendriella_betulae | − | − | − | + | − | − | − | − | − | − |
| Hyalodendriella sp. | − | − | − | + | − | − | − | − | − | − |
| Hyaloscypha bicolor | − | − | − | − | − | + | − | + | − | − |
| Hyaloscypha finlandica | − | − | − | + | − | − | − | + | + | − |
| Hyaloscypha sp. | − | − | − | − | − | − | − | − | + | − |
| Infundichalara minuta | − | − | − | + | − | − | − | − | − | − |
| Lachnum sp. | − | − | − | − | − | − | + | − | − | − |
| Leohumicola sp. | − | − | − | + | − | − | − | − | − | − |
| Mollisia dextrinospora | − | − | − | + | − | − | − | − | − | − |
| Sporormiella intermedia | − | − | − | + | − | − | − | − | − | − |
| Paracladophialophora carceris | − | − | − | − | − | − | − | + | − | − |
| Paraphoma fimeti | − | + | − | − | − | − | − | − | − | − |
| Penicillium brunneoconidiatum | − | − | − | + | − | + | − | − | − | − |
| Penicillium spinulosum | − | − | − | + | − | + | − | − | − | − |
| Penicillium sp. | − | − | − | + | − | + | − | + | − | + |
| Peziza succosa | − | − | − | + | − | − | − | + | − | − |
| Pezoloma ericae | − | − | + | + | − | + | − | + | + | − |
| Phacidium sp. | − | − | + | − | − | − | − | − | − | − |
| Phialocephala fortinii | − | − | − | − | + | − | − | − | − | − |
| Phialocephala sp. | − | − | − | − | − | + | − | − | − | − |
| Plenodomus biglobosus | − | + | − | − | − | − | − | − | − | − |
| Pseudodictyosporium elegans | − | + | − | − | − | − | − | − | − | − |
| Pseudodictyosporium sp. | − | + | − | − | − | − | − | − | − | − |
| Sagenomella sp. | − | − | − | − | − | + | − | − | − | − |
| Talaromyces sp. | − | − | − | + | − | + | − | − | − | − |
| Tetracladium apiense | − | − | + | − | − | − | − | − | − | − |
| Tetracladium sp. | − | + | + | + | − | − | − | − | − | − |
| Tricharina gilva | − | + | − | − | − | − | − | − | − | − |
| Tricharina sp. | − | + | − | − | − | − | − | − | − | − |
| Trichocladium opacum | − | − | − | − | − | − | − | + | − | − |
| Trichophaea pseudogregaria | + | + | − | − | − | − | − | − | − | − |
| Trichophaea woolhopeia | + | − | + | + | − | − | − | − | − | − |
| Tuber rufum | − | − | − | − | − | − | − | − | − | + |
| Tuber sp. | − | − | − | − | − | − | − | − | − | + |
| Verrucaria muralis | − | + | − | − | − | − | − | − | − | − |
| Verrucaria ahtii | − | + | − | − | − | − | − | − | − | − |
| Basidiobolomycota | ||||||||||
| Basidiobolus magnus | − | − | − | + | − | − | − | − | − | − |
| Basidiobolus ranarum | − | − | − | + | − | − | − | − | − | − |
| Basidiomycota | ||||||||||
| Amphinema byssoides | − | − | + | + | − | − | − | − | − | − |
| Amphinema sp. | − | − | + | + | − | + | − | − | − | − |
| Apiotrichum dulcitum | − | − | − | − | − | − | − | + | − | − |
| Apiotrichum sp. | − | − | − | − | − | − | − | + | − | − |
| Coprinellus micaceus | − | − | − | + | − | − | − | − | − | − |
| Cortinarius casimiri | − | − | − | − | − | + | − | − | − | − |
| Cortinarius cyprinus | − | − | − | − | − | − | − | − | − | + |
| Cortinarius saturatus | + | + | + | + | + | + | + | + | + | + |
| Cortinarius scandens | − | − | − | − | + | + | − | − | − | − |
| Cortinarius subtilis | − | − | − | + | − | − | − | − | − | − |
| Cortinarius vernus | − | − | − | − | − | + | + | + | − | − |
| Cortinarius sp. | − | − | − | + | + | + | + | + | − | + |
| Cuphophyllus virgineus | − | − | − | − | − | − | + | + | − | − |
| Cuphophyllus sp. | − | − | − | − | − | − | − | + | − | − |
| Cutaneotrichosporon moniliiforme | − | − | − | − | − | − | − | − | + | − |
| Cutaneotrichosporon sp. | − | − | − | − | − | − | + | − | − | − |
| Filobasidium wieringae | − | + | − | − | − | − | − | − | − | − |
| Flagelloscypha minutissima | − | − | − | − | − | − | − | − | − | + |
| Ganoderma sp. | − | − | − | + | − | − | − | + | − | + |
| Hebeloma cylindrosporum | − | − | + | − | − | − | − | − | − | − |
| Hebeloma leucosarx | − | − | + | − | − | − | − | − | − | − |
| Hebeloma sp. | − | − | + | + | − | − | − | − | − | − |
| Helvellosebacina concrescens | − | − | − | − | − | − | − | − | + | + |
| Helvellosebacina sp. | − | − | − | − | − | − | + | + | − | + |
| Hymenogaster griseus | − | − | − | − | − | − | − | + | − | + |
| Hymenogaster rehsteineri | − | − | − | − | − | − | − | − | − | + |
| Hymenogaster sp. | − | − | − | − | − | − | − | − | − | + |
| Hypholoma fasciculare | − | − | − | + | − | − | − | − | − | − |
| Inocybe mixtilis | − | − | − | − | − | − | − | − | − | + |
| Inocybe sp. | − | + | − | − | − | − | − | − | − | − |
| Laccaria sp. | − | − | − | − | − | − | − | + | − | − |
| Lactarius circellatus | − | − | − | − | − | − | − | + | − | − |
| Lactarius torminosus | − | − | − | − | + | − | − | − | − | − |
| Lactarius sp. | − | − | − | − | + | − | − | − | − | − |
| Leccinum pseudoscabrum | − | − | − | − | − | − | + | + | − | − |
| Lycoperdon pyriforme | − | − | − | + | − | − | − | − | − | − |
| Lycoperdon sp. | − | − | − | + | − | − | − | − | − | − |
| Macrolepiota procera | − | − | − | − | − | − | − | + | − | − |
| Mallocybe sp. | − | + | − | − | − | − | − | − | − | − |
| Mycena citrinomarginata | − | − | − | − | − | − | + | + | + | − |
| Mycena leptocephala | − | − | − | − | − | − | + | + | − | − |
| Mycena olivaceomarginata | − | − | − | − | − | − | + | − | + | − |
| Mycena plumipes | − | − | − | − | − | − | + | + | − | − |
| Mycena pura | − | − | − | − | − | + | − | − | − | − |
| Mycena sanguinolenta | − | − | + | + | − | − | − | − | − | − |
| Mycena vulgaris | − | − | − | − | + | + | − | − | − | − |
| Mycena sp. | − | − | + | − | + | + | + | + | + | + |
| Mycenella bryophila | − | − | − | − | − | − | − | − | + | − |
| Mycenella trachyspora | − | − | − | − | − | − | − | − | + | − |
| Mycenella sp. | − | − | − | − | − | − | − | − | + | − |
| Odontia fibrosa | − | − | − | + | − | − | − | − | − | − |
| Odontia sp. | − | − | − | + | − | − | − | − | − | − |
| Phallus impudicus | − | − | − | + | − | − | − | + | − | + |
| Phallus ultraduplicatus | − | − | − | + | − | − | − | − | − | − |
| Phallus sp. | − | − | − | + | − | − | − | − | − | − |
| Piloderma byssinum | + | − | − | + | − | − | + | + | − | − |
| Psathyrella candolleana | − | − | − | − | − | − | − | − | − | + |
| Psathyrella sp. | − | − | − | − | − | − | − | − | − | + |
| Russula persicina | − | − | − | − | − | − | − | − | + | − |
| Russula solaris | − | − | − | − | − | − | + | + | + | + |
| Russula versicolor | − | − | − | − | − | − | + | − | − | − |
| Russula sp. | − | − | − | − | − | − | + | + | + | + |
| Saitozyma podzolica | − | − | − | + | − | − | + | + | − | + |
| Saitozyma sp. | − | − | − | − | − | − | − | + | − | + |
| Scleroderma bovista | − | − | − | − | − | − | − | − | − | + |
| Scleroderma sp. | − | − | − | − | − | − | − | − | − | + |
| Sebacina sp. | − | − | − | − | − | − | + | + | − | − |
| Solicoccozyma terricola | − | − | − | + | − | − | + | + | − | + |
| Solicoccozyma sp. | − | − | − | − | − | − | − | + | − | − |
| Tephrocybe rancida | − | − | − | − | − | − | − | + | − | − |
| Thanatephorus sp. | + | − | − | − | − | − | − | − | − | − |
| Thelephora atra | − | − | + | + | − | − | + | + | − | − |
| Thelephora caryophyllea | − | − | − | + | − | − | − | − | − | − |
| Tomentella badia | − | − | − | − | − | − | + | + | − | − |
| Tomentella fuscocinerea | − | − | + | − | − | − | − | − | − | − |
| Tomentella lilacinogrisea | − | − | − | + | − | − | − | − | − | − |
| Tomentella pilosa | − | − | − | − | − | − | + | + | − | + |
| Tomentella sp. | + | + | − | + | − | + | + | + | − | + |
| Tremella sp. | − | − | − | − | − | − | − | − | − | + |
| Tricholoma argyraceum | − | − | + | + | − | − | − | − | − | − |
| Tricholoma sp. | − | − | + | + | − | − | − | − | − | − |
| Tylospora sp. | − | + | + | − | − | − | − | − | − | − |
| Chytridiomycota | ||||||||||
| Operculomyces laminatus | − | − | − | + | − | − | − | − | − | − |
| Rhizophlyctis rosea | − | − | − | + | − | − | − | − | − | − |
| Rhizophydium sp. | − | + | − | − | − | − | − | − | − | − |
| Spizellomyces pseudodichotomus | − | + | − | − | − | − | − | − | − | − |
| Monoblepharomycota | ||||||||||
| Sanchytrium sp. | − | − | − | + | − | − | − | − | − | + |
| Mortierellomycota | ||||||||||
| Mortierella alpina | − | + | − | − | − | − | − | − | − | + |
| Mortierella amoeboidea | − | − | − | − | − | − | − | − | + | + |
| Mortierella beljakovae | − | − | − | − | − | − | − | − | − | + |
| Mortierella clonocystis | − | − | − | + | − | − | − | + | − | − |
| Mortierella gamsii | − | − | − | − | − | − | + | + | − | + |
| Mortierella globulifera | − | − | − | − | − | − | − | + | − | − |
| Mortierella humilis | − | − | − | + | − | + | − | + | − | − |
| Mortierella minutissima | − | − | − | + | − | − | + | + | − | − |
| Mortierella paraensis | − | − | − | − | − | − | − | + | − | − |
| Mortierella pseudozygospora | − | − | − | − | − | − | − | + | − | + |
| Mortierella sarnyensis | − | − | − | − | − | − | − | + | − | + |
| Mortierella zonata | − | − | − | − | − | − | − | + | − | + |
| Mortierella sp. | − | + | + | + | − | + | + | + | + | + |
| Mucoromycota | ||||||||||
| Absidia cylindrospora | − | − | − | − | − | − | − | − | − | + |
| Absidia sp. | − | − | − | − | − | − | − | + | − | − |
| Mucor hiemalis | − | − | − | − | − | − | − | + | − | − |
| Mucor sp. | − | − | − | + | − | − | − | − | − | − |
| Umbelopsis isabellina | − | − | − | − | − | + | − | − | − | − |
| Umbelopsis sp. | − | − | − | + | − | + | − | − | − | − |
| Zygorhynchus sp. | − | − | − | + | − | − | − | − | − | − |
| Ʃ 171 species | 6 | 29 | 21 | 62 | 9 | 25 | 36 | 57 | 21 | 44 |
| Faith’s phylogenetic diversity | 509 | 2828 | 989 | 3378 | 429 | 1141 | 1053 | 2368 | 906 | 2190 |
| The Most Abundant Strains of Bacteria Detected in the Roots of Orchids and Surrounding Soils | 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Epipactis atrorubens | Epipactis atrorubens | Platanthera bifolia | Epipactis pontica | Cephalanthera longifolia | ||||||
| Ni-Co Mining Heap Dobšiná | Pb-Zn Mining Heap Jasenie | Fe Mining Heap Sirk | ||||||||
| pH 7.73 | pH 7.36 | pH 7.81 | pH 4.98 | pH 6.58 | ||||||
| Roots | Soil | Roots | Soil | Roots | Soil | Roots | Soil | Roots | Soil | |
| Acidobacteria | ||||||||||
| Candidatus solibacter | − | − | + | + | − | + | − | + | − | + |
| Pyrinomonadaceae RB41 | − | + | − | − | − | − | − | − | − | − |
| Vicinamibacter sp. | + | − | − | − | − | − | − | − | − | − |
| Actinobacteria | ||||||||||
| Nocardioides sp. | + | − | + | + | − | − | + | − | − | − |
| Bacteroidetes | ||||||||||
| Chryseobacterium sp. | − | − | − | − | − | − | + | − | − | − |
| Flavobacterium sp. | − | − | + | − | − | − | − | − | − | − |
| Pedobacter sp. | − | − | − | − | − | − | + | − | − | − |
| Gemmatimonadetes | ||||||||||
| Gemmatimonas sp. | − | + | − | − | − | − | − | − | − | − |
| Planctomycetes | ||||||||||
| Fimbriiglobus sp. | − | − | + | + | − | − | − | − | − | − |
| Gemmata sp. | + | − | + | + | + | − | + | + | + | − |
| Pirellulaceae Pir4 lineage | − | − | + | + | − | − | − | − | − | − |
| Pirellula sp. | + | + | + | + | − | − | − | − | − | + |
| Rubinisphaeraceae SH-PL14 | + | + | + | + | − | − | − | − | − | − |
| Singulisphaera sp. | − | − | − | − | + | + | + | − | + | − |
| Proteobacteria | ||||||||||
| Acidibacter sp. | − | − | − | − | − | + | − | − | − | − |
| Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium | − | − | + | − | + | − | + | − | − | − |
| Bradyrhizobium sp. | − | − | + | − | − | + | − | − | − | − |
| Burkholderia-Caballeronia-Paraburkholderia | − | − | − | − | − | + | + | − | − | − |
| Nitrosomonadaceae Ellin6067 | − | − | − | + | − | − | − | − | − | − |
| Sphingomonas sp. | − | − | + | − | − | − | − | − | − | − |
| Pseudomonas sp. | − | − | − | − | − | − | + | − | − | − |
| Tenericutes | ||||||||||
| Candidatus phytoplasma fragariae | − | − | − | − | − | − | − | − | + | − |
| Verrucomicrobia | ||||||||||
| Candidatus udaeobacter | − | + | + | + | − | + | + | + | + | + |
| Candidatus xiphinematobacter | − | − | + | − | − | − | − | + | − | − |
| Chthoniobacter sp. | + | + | + | + | + | − | + | + | + | + |
| Luteolibacter sp. | − | − | + | − | − | − | − | − | − | − |
| Spartobacteria bacterium WX31 | − | − | − | − | − | − | + | + | + | + |
| Verrucomicrobia bacterium SCGC AG-212-E04 | − | − | − | − | − | − | − | + | − | − |
| Verrucomicrobiaceae bacterium CNC16 | + | − | − | − | − | − | − | − | − | − |
| Verrucomicrobium sp. | + | − | + | − | − | − | − | − | − | − |
| Ʃ 30 species | 8 | 6 | 16 | 10 | 4 | 6 | 11 | 7 | 6 | 5 |
| Faith’s phylogenetic diversity | 21.2 | 25.4 | 23.5 | 26.8 | 19.3 | 41.7 | 16.8 | 19.9 | 17.3 | 26.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Böhmer, M.; Ozdín, D.; Račko, M.; Lichvár, M.; Budiš, J.; Szemes, T. Identification of Bacterial and Fungal Communities in the Roots of Orchids and Surrounding Soil in Heavy Metal Contaminated Area of Mining Heaps. Appl. Sci. 2020, 10, 7367. https://doi.org/10.3390/app10207367
Böhmer M, Ozdín D, Račko M, Lichvár M, Budiš J, Szemes T. Identification of Bacterial and Fungal Communities in the Roots of Orchids and Surrounding Soil in Heavy Metal Contaminated Area of Mining Heaps. Applied Sciences. 2020; 10(20):7367. https://doi.org/10.3390/app10207367
Chicago/Turabian StyleBöhmer, Miroslav, Daniel Ozdín, Matúš Račko, Michal Lichvár, Jaroslav Budiš, and Tomáš Szemes. 2020. "Identification of Bacterial and Fungal Communities in the Roots of Orchids and Surrounding Soil in Heavy Metal Contaminated Area of Mining Heaps" Applied Sciences 10, no. 20: 7367. https://doi.org/10.3390/app10207367
APA StyleBöhmer, M., Ozdín, D., Račko, M., Lichvár, M., Budiš, J., & Szemes, T. (2020). Identification of Bacterial and Fungal Communities in the Roots of Orchids and Surrounding Soil in Heavy Metal Contaminated Area of Mining Heaps. Applied Sciences, 10(20), 7367. https://doi.org/10.3390/app10207367





