In Vitro and in Silico Analysis of miR-125a with rs12976445 Polymorphism in Breast Cancer Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Blood Sampling and Measurements
2.3. DNA Extraction, PCR, Restriction Analysis
2.4. Polymerase Chain Reaction, Sequencing, and Restriction Analysis
2.5. Preparing Sequence for in Silico Analysis
2.6. Predicting Protein Interactions
2.7. 2D Structure Modelling
2.8. Statistical Analysis
3. Results
3.1. In Vitro Analysis
3.2. In Silico Analysis of RNA Binding Proteins (RBPs)
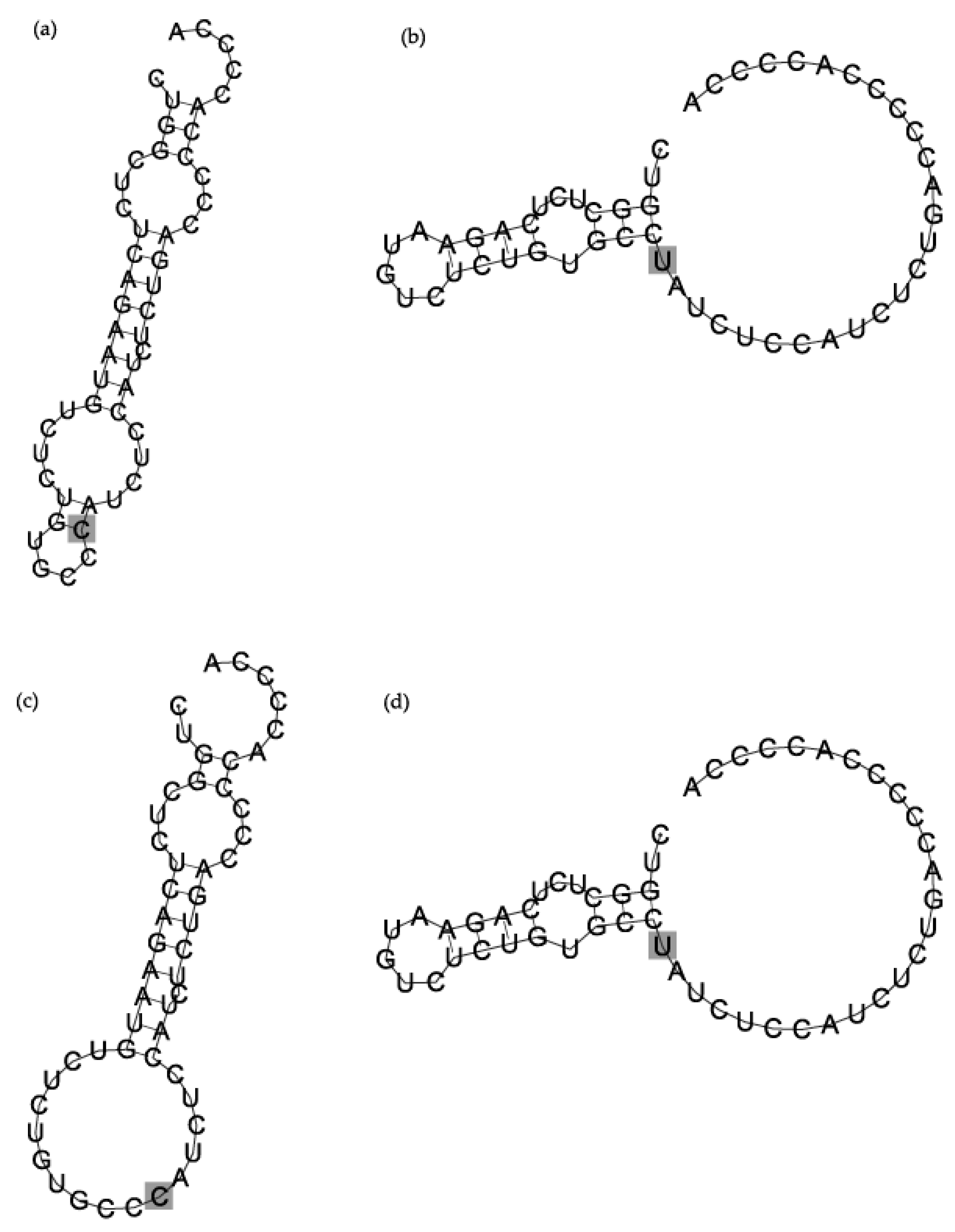
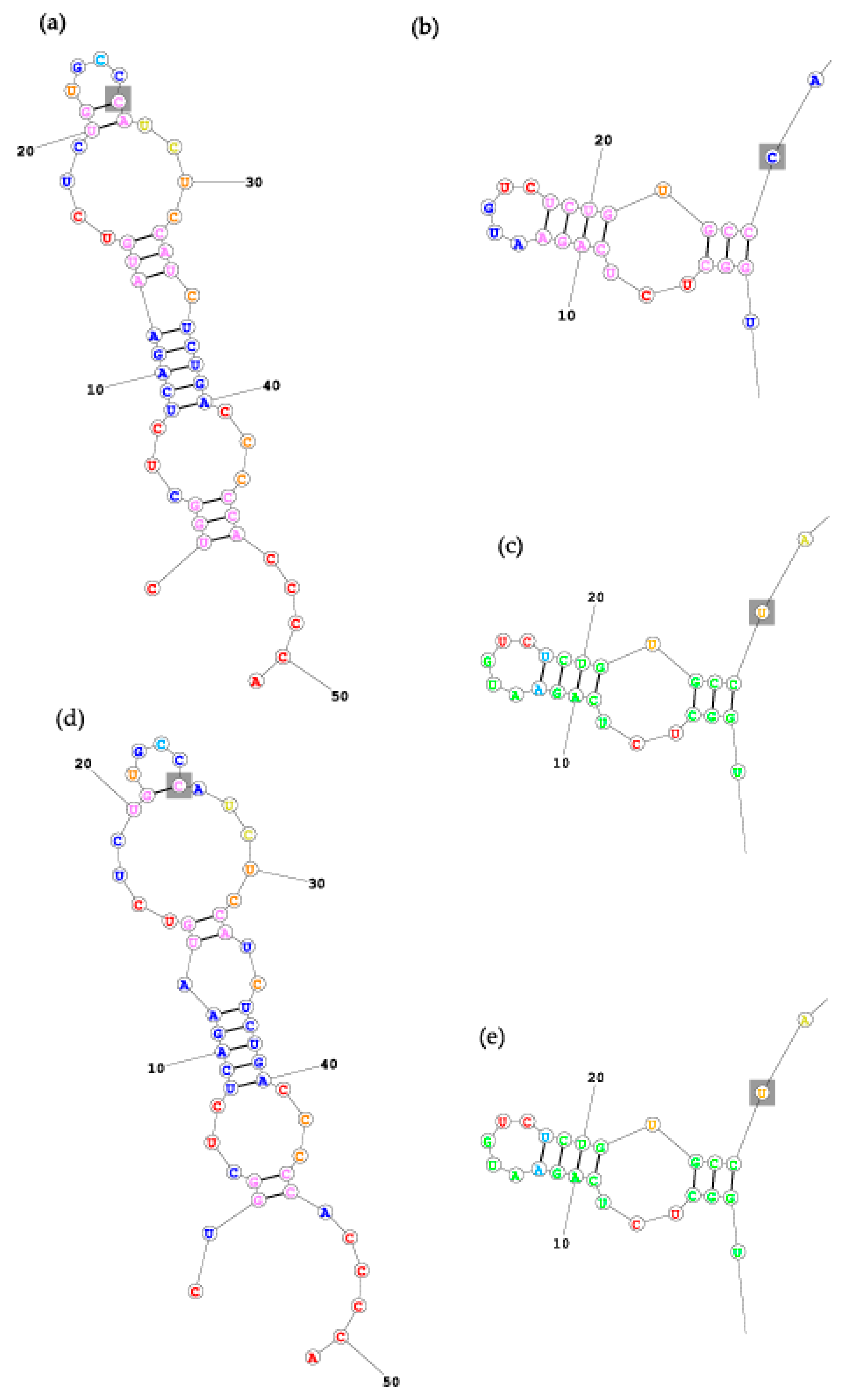
3.3. In Silico Modelling of pri-miR-125a Folding
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Keeney, M.G.; Couch, F.J.; Visscher, D.W.; Lindor, N.M. Non-BRCA familial breast cancer: Review of reported pathology and molecular findings. Pathology 2017, 49, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, P.; Read, G.H.; Weidhaas, J.B. Breast Cancer and miR-SNPs: The Importance of miR Germ-Line Genetics. Noncoding RNA 2019, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Mosallaei, M.; Simonian, M.; Esmaeilzadeh, E.; Bagheri, H.; Miraghajani, M.; Salehi, A.R.; Mehrzad, V.; Salehi, R. Single nucleotide polymorphism rs10889677 in miRNAs Let-7e and Let-7f binding site of IL23R gene is a strong colorectal cancer determinant: Report and meta-analysis. Cancer Genet. 2019, 239, 46–53. [Google Scholar] [CrossRef]
- Yan, Z.; Zhou, Z.; Li, C.; Yang, X.; Yang, L.; Dai, S.; Zhao, J.; Ni, H.; Shi, L.; Yao, Y. Polymorphisms in miRNA genes play roles in the initiation and development of cervical cancer. J. Cancer 2019, 10, 4747–4753. [Google Scholar] [CrossRef]
- Yin, Z.; Cui, Z.; Ren, Y.; Xia, L.; Wang, Q.; Zhang, Y.; He, Q.; Zhou, B. Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer 2016, 94, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Bahreini, F.; Ramezani, S.; Shahangian, S.S.; Salehi, Z.; Mashayekhi, F. miR-559 polymorphism rs58450758 is linked to breast cancer. Br. J. Biomed. Sci. 2019, 77, 29–34. [Google Scholar] [CrossRef]
- Linhares, J.J.; Azevedo, M., Jr.; Siufi, A.A.; de Carvalho, C.V.; WolgienMdel, C.; Noronha, E.C.; Bonetti, T.C.; da Silva, I.D. Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med. Genet. 2012, 13, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Huo, Q.; Wang, X.; Chen, X.; Long, L.; Jiang, L.; Ma, T.; Yang, Q. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. Gene 2013, 529, 125–130. [Google Scholar] [CrossRef]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef]
- Potenza, N.; Panella, M.; Castiello, F.; Mosca, N.; Amendola, E.; Russo, A. Molecular mechanisms governing microRNA-125a expression in human hepatocellular carcinoma cells. Sci. Rep. 2017, 7, 10712. [Google Scholar] [CrossRef]
- Li, G.; Ao, S.; Hou, J.; Lyu, G. Low expression of miR-125a-5p is associated with poor prognosis in patients with gastric cancer. Oncol. Lett. 2019, 18, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zeng, X.; Guo, H.; Wang, T.; Wei, L.; Zhang, Y.; Zhao, J.; Ma, X.; Zhang, N. MicroRNA-125a-5p modulates radioresistance in LTEP-a-2 non-small cell lung cancer cells by targeting SIRT7. Cancer Biomark 2020, 27, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Zhou, L.; Wu, S.; Shi, X.; Jiang, G.; Niu, S.; Ding, D. miR-125a-5p inhibits colorectal cancer cell epithelial-mesenchymal transition, invasion and migration by targeting TAZ. Onco Targets Ther. 2019, 12, 3481–3489. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhan, Y.; Xu, Z.; Li, Y.; Luo, A.; Ding, F.; Cao, X.; Chen, H.; Liu, Z. ZEB1 induced miR-99b/let-7e/miR-125a cluster promotes invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2017, 398, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, C.M.; Qi, L.; He, T.Z.; Shi-Guo, L.; Hao, C.J.; Cui, Y.; Zhang, N.; Xia, H.F.; Ma, X. Two common SNPs in pri-miR-125a alter the mature miRNA expression and associate with recurrent pregnancy loss in a Han-Chinese population. RNA Biol. 2011, 8, 861–872. [Google Scholar] [CrossRef]
- Morales, S.; De Mayo, T.; Gulppi, F.A.; Gonzalez-Hormazabal, P.; Carrasco, V.; Reyes, J.M.; Gomez, F.; Waugh, E.; Jara, L. Genetic Variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 Are Associated with Breast Cancer Susceptibility in a South American Population. Genes 2018, 9, 427. [Google Scholar] [CrossRef]
- Peterlongo, P.; Caleca, L.; Cattaneo, E.; Ravagnani, F.; Bianchi, T.; Galastri, L.; Bernard, L.; Ficarazzi, F.; Dall’olio, V.; Marme, F.; et al. The rs12975333 variant in the miR-125a and breast cancer risk in Germany, Italy, Australia and Spain. J. Med. Genet. 2011, 48, 703–704. [Google Scholar] [CrossRef]
- Cai, T.; Li, J.; An, X.; Yan, N.; Li, D.; Jiang, Y.; Wang, W.; Shi, L.; Qin, Q.; Song, R.; et al. Polymorphisms in MIR499A and MIR125A gene are associated with autoimmune thyroid diseases. Mol. Cell. Endocrinol. 2017, 440, 106–115. [Google Scholar] [CrossRef]
- Jiao, L.; Zhang, J.; Dong, Y.; Duan, B.; Yu, H.; Sheng, H.; Huang, J.; Gao, H. Association between miR-125a rs12976445 and survival in breast cancer patients. Am. J. Transl. Res. 2014, 6, 869–875. [Google Scholar]
- Huang, X.; Zhang, T.; Li, G.; Guo, X.; Liu, X. Regulation of miR-125a expression by rs12976445 single-nucleotide polymorphism is associated with radiotherapy-induced pneumonitis in lung carcinoma patients. J. Cell. Biochem. 2019, 120, 4485–4493. [Google Scholar] [CrossRef]
- Quan, H.Y.; Yuan, T.; Hao, J.F. A microRNA125a variant, which affects its mature processing, increases the risk of radiationinduced pneumonitis in patients with nonsmallcell lung cancer. Mol. Med. Rep. 2018, 18, 4079–4086. [Google Scholar]
- Damodaran, M.; Paul, S.F.D.; Venkatesan, V. Genetic Polymorphisms in miR-146a, miR-196a2 and miR-125a Genes and its Association in Prostate Cancer. Pathol. Oncol. Res. 2018, 26, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Miskiewicz, J.; Tomczyk, K.; Mickiewicz, A.; Sarzynska, J.; Szachniuk, M. Bioinformatics Study of Structural Patterns in Plant MicroRNA Precursors. Biomed. Res. Int. 2017, 2017, 6783010. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Szachniuk, M. Discovering Structural Motifs in miRNA Precursors from the Viridiplantae Kingdom. Molecules 2018, 23, 1367. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Xu, K.; Cai, H.; Shen, Y.; Ni, Q.; Chen, Y.; Hu, S.; Li, J.; Wang, H.; Yu, L.; Huang, H.; et al. [Management of corona virus disease-19 (COVID-19): The Zhejiang experience]. Zhejiang Da Xue Xue Bao Yi Xue Ban 2020, 49, 147–157. [Google Scholar] [PubMed]
- Paz, I.; Kosti, I.; Ares, M., Jr.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef]
- Giudice, G.; Sanchez-Cabo, F.; Torroja, C.; Lara-Pezzi, E. ATtRACT-a database of RNA-binding proteins and associated motifs. Database (Oxford) 2016, 2016. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Sole, X.; Guino, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Guo, X.; Wu, Y.; Hartley, R.S. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009, 6, 575–583. [Google Scholar] [CrossRef]
- Scott, G.K.; Goga, A.; Bhaumik, D.; Berger, C.E.; Sullivan, C.S.; Benz, C.C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 2007, 282, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Serpico, D.; Molino, L.; Di Cosimo, S. microRNAs in breast cancer development and treatment. Cancer Treat. Rev. 2014, 40, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Li, Z.; Zheng, H.; Ho, J.; Chan, M.T.V.; Wu, W.K.K. Neuro-oncological ventral antigen 1 (NOVA1): Implications in neurological diseases and cancers. Cell Prolif. 2017, 50, e12348. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, W.; Han, Q.; Wang, Y.; Li, C.; Zhang, P.; Xu, H. Post-translational modification control of RNA-binding protein hnRNPK function. Open Biol. 2019, 9, 180239. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.L.; Fu, X.; Huang, J.; Jia, T.T.; Zong, F.Y.; Mu, S.R.; Zhu, H.; Yan, Y.; Qiu, S.; Wu, Q.; et al. Genome-wide analysis of YB-1-RNA interactions reveals a novel role of YB-1 in miRNA processing in glioblastomamultiforme. Nucleic Acids Res. 2015, 43, 8516–8528. [Google Scholar] [CrossRef]
- He, H.; Xu, F.; Huang, W.; Luo, S.Y.; Lin, Y.T.; Zhang, G.H.; Du, Q.; Duan, R.H. miR-125a-5p expression is associated with the age of breast cancer patients. Genet. Mol. Res. 2015, 14, 17927–17933. [Google Scholar] [CrossRef]
- Li, W.; Duan, R.; Kooy, F.; Sherman, S.L.; Zhou, W.; Jin, P. Germline mutation of microRNA-125a is associated with breast cancer. J. Med. Genet. 2009, 46, 358–360. [Google Scholar] [CrossRef]
- Hu, Y.; Huo, Z.H.; Liu, C.M.; Liu, S.G.; Zhang, N.; Yin, K.L.; Qi, L.; Ma, X.; Xia, H.F. Functional study of one nucleotide mutation in pri-miR-125a coding region which related to recurrent pregnancy loss. PLoS ONE 2014, 9, e114781. [Google Scholar] [CrossRef]
- Li, C.; Lei, T. Rs12976445 Polymorphism is Associated with Risk of Diabetic Nephropathy Through Modulating Expression of MicroRNA-125 and Interleukin-6R. Med. Sci. Monit. 2015, 21, 3490–3497. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, B.; Hu, H.; Liu, F.; Tu, Y.; He, F. The association between differentially expressed micro RNAs in breast cancer cell lines and the micro RNA-205 gene polymorphism in breast cancer tissue. Oncol. Lett. 2018, 15, 2139–2146. [Google Scholar] [CrossRef]
- Keppetipola, N.; Sharma, S.; Li, Q.; Black, D.L. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Placzek, W.J. PTBP1 enhances miR-101-guided AGO2 targeting to MCL1 and promotes miR-101-induced apoptosis. Cell Death Dis. 2018, 9, 552. [Google Scholar] [CrossRef] [PubMed]
- Storchel, P.H.; Thummler, J.; Siegel, G.; Aksoy-Aksel, A.; Zampa, F.; Sumer, S.; Schratt, G. A large-scale functional screen identifies Nova1 and Ncoa3 as regulators of neuronal miRNA function. EMBO J. 2015, 34, 2237–2254. [Google Scholar] [CrossRef] [PubMed]

| SNP Allele Frequencies (n = 304) | ||||||
| All subjects | Ca | Co | ||||
| Allele | Count | Proportion | Count | Proportion | Count | Proportion |
| T | 414 | 0.68 | 241 | 0.69 | 173 | 0.67 |
| C | 194 | 0.32 | 109 | 0.31 | 85 | 0.33 |
| SNP Genotype Frequencies (n = 304) | ||||||
| All subjects | Ca | Co | ||||
| Genotype | Count | Proportion | Count | Proportion | Count | Proportion |
| C/C | 24 | 0.08 | 14 | 0.08 | 10 | 0.08 |
| T/C | 146 | 0.48 | 81 | 0.46 | 65 | 0.5 |
| T/T | 134 | 0.44 | 80 | 0.46 | 54 | 0.42 |
| SNP Exact Test for Hardy–Weinberg Equilibrium (n = 304) | ||||||
| TT | TC | CC | T | C | p-value | |
| All subjects | 134 | 146 | 24 | 414 | 194 | 0.086 |
| Ca | 80 | 81 | 14 | 241 | 109 | 0.38 |
| Co | 54 | 65 | 10 | 173 | 85 | 0.16 |
| SNP Association with Response STATUS (n = 304, Crude Analysis) | ||||||
| Model | Genotype | Ca | Co | OR (95% CI) | p-value | |
| Codominant | T/T | 80 (45.7%) | 54 (41.9%) | 1.00 | 0.77 | |
| C/T | 81 (46.3%) | 65 (50.4%) | 1.19 (0.74–1.91) | |||
| C/C | 14 (8%) | 10 (7.8%) | 1.06 (0.44–2.56) | |||
| Dominant | T/T | 80 (45.7%) | 54 (41.9%) | 1.00 | 0.5 | |
| C/T+ C/C | 95 (54.3%) | 75 (58.1%) | 1.17 (0.74–1.85) | |||
| Recessive | T/T+ C/T | 161 (92%) | 119 (92.2%) | 1.00 | 0.94 | |
| C/C | 14 (8%) | 10 (7.8%) | 0.97 (0.41–2.25) | |||
| Over-dominant | T/T+ C/C | 94 (53.7%) | 64 (49.6%) | 1.00 | 0.48 | |
| C/T | 81 (46.3%) | 65 (50.4%) | 1.18 (0.75–1.86) | |||
| Log-additive | n/a | n/a | n/a | 1.10 (0.76–1.58) | 0.62 | |
| Receptor | Case | Variant | Group Status | p-Value | |
|---|---|---|---|---|---|
| P | N | ||||
| HER2 | Genotype | TT | 14 | 30 | 0.0606 |
| CT | 7 | 48 | |||
| CC | 1 | 6 | |||
| TT+CT | 21 | 78 | 0.2609 | ||
| CT+CC | 8 | 54 | |||
| Alleles frequency | T | 35 | 108 | 0.0814 | |
| C | 9 | 60 | |||
| ER | Genotype | TT | 36 | 8 | 0.2001 |
| CT | 37 | 17 | |||
| CC | 6 | 1 | |||
| TT+CT | 73 | 25 | 0.2891 | ||
| CT+CC | 33 | 18 | |||
| Alleles frequency | T | 109 | 33 | 0.5702 | |
| C | 49 | 19 | |||
| PR | Genotype | TT | 30 | 14 | 0.4922 |
| CT | 35 | 20 | |||
| CC | 6 | 1 | |||
| TT+CT | 65 | 34 | 1.0000 | ||
| CT+CC | 41 | 21 | |||
| Alleles frequency | T | 95 | 48 | 0.9297 | |
| C | 47 | 22 | |||
| Variant | Protein | Mode | Motif Containing Variant | ||
|---|---|---|---|---|---|
| Low | Medium | High | |||
| C | HNRNPK | ✓ | ✓ | CCAUCUC | |
| NOVA1 | ✓ | ✓ | ✓ | CCAU | |
| PCBP1 | ✓ | CAUCUCC | |||
| PTBP1 | ✓ | ✓ | ✓ | CCAUCU | |
| SRSF3 | ✓ | ✓ | ✓ | CCCAUCU | |
| SRSF5 | ✓ | CAUCUCC | |||
| U | BRUNOL4 | ✓ | UGUGCCU | ||
| BRUNOL5 | ✓ | UGUGCCU | |||
| PCBP1 | ✓ | CCUAUCU | |||
| PTBP1 | ✓ | ✓ | ✓ | CUAUCU | |
| SRSF5 | ✓ | CCUAUCU, UAUCUCC | |||
| Variant | Protein | Motif Containing Variant |
|---|---|---|
| C | NOVA1 | CCAU |
| YBX1 | CAUC | |
| U | PTBP1 | CUAU, CCUAU |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lehmann, T.P.; Miskiewicz, J.; Szostak, N.; Szachniuk, M.; Grodecka-Gazdecka, S.; Jagodziński, P.P. In Vitro and in Silico Analysis of miR-125a with rs12976445 Polymorphism in Breast Cancer Patients. Appl. Sci. 2020, 10, 7275. https://doi.org/10.3390/app10207275
Lehmann TP, Miskiewicz J, Szostak N, Szachniuk M, Grodecka-Gazdecka S, Jagodziński PP. In Vitro and in Silico Analysis of miR-125a with rs12976445 Polymorphism in Breast Cancer Patients. Applied Sciences. 2020; 10(20):7275. https://doi.org/10.3390/app10207275
Chicago/Turabian StyleLehmann, Tomasz P., Joanna Miskiewicz, Natalia Szostak, Marta Szachniuk, Sylwia Grodecka-Gazdecka, and Paweł P. Jagodziński. 2020. "In Vitro and in Silico Analysis of miR-125a with rs12976445 Polymorphism in Breast Cancer Patients" Applied Sciences 10, no. 20: 7275. https://doi.org/10.3390/app10207275
APA StyleLehmann, T. P., Miskiewicz, J., Szostak, N., Szachniuk, M., Grodecka-Gazdecka, S., & Jagodziński, P. P. (2020). In Vitro and in Silico Analysis of miR-125a with rs12976445 Polymorphism in Breast Cancer Patients. Applied Sciences, 10(20), 7275. https://doi.org/10.3390/app10207275





