Detecting Diabetic Retinopathy Using Embedded Computer Vision
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Training and Testing Dataset
| while IFS = ’,’ read -r f1 f2 |
| do |
| imageEye = $(echo $f2 | cut -d ’_’ -f 2) |
| if [$imageEye == ’left’ -a $f2 == ’1’ ] |
| then |
| echo ’done left’ |
| else |
| echo ’done right’ |
| fi |
| done < “file_name” |
2.2. Convolution Neural Network
2.3. Comparison of Various Tools
2.4. Nvidia Jetson TX2 and Nvidia Digits
2.5. Training and Testing Procedure
3. Results and Discussion
3.1. Data Preprocessing
3.2. Hyperparameters Optimization
3.3. Accuracy Rate for Inference Model
3.3.1. Caffe Model
3.3.2. Keras Model
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mayo Clinic. Diabetic Retinopathy. Available online: https://www.mayoclinic.org/diseases-conditions/diabetic-retinopathy/symptoms-causes/syc-20371611 (accessed on 20 June 2020).
- National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, Centers for Diabetes Control and Prevention; Department of Health and Human Services: Atlanta, GA, USA, 2020.
- Timmons, J. Diabetes: Facts, Statistics, and You. Available online: https://www.healthline.com/health/diabetes/facts-statistics-infographic (accessed on 20 June 2020).
- Cheloni, R.; Gandolfi, S.A.; Signorelli, C.; Odone, A. Global Prevalence of Diabetic Retinopathy: Protocol for a Systematic Review and Meta-Analysis. BMJ Open 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Wong, T.Y.; Sabanayagam, C. Epidemiology of Diabetic Retinopathy, Diabetic Macular Edema and Related Vision Loss. Eye Vis. 2015, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Genentech. Retinal Diseases Fact Sheet. Genentech: Breakthrough Science. One Moment, One Day, One Person at a Time. Available online: https://www.gene.com/stories/retinal-diseases-fact-sheet (accessed on 20 June 2020).
- Mayo Clinic. Diabetic Retinopathy: Overview. Available online: https://g.co/kgs/WTnvDF (accessed on 20 June 2020).
- Mayo Clinic. Diabetic Retinopathy: Diagnosis. Available online: https://www.mayoclinic.org/diseases-conditions/diabetic-retinopathy/diagnosis-treatment/drc-20371617 (accessed on 20 June 2020).
- Getting an Eye Exam Without Insurance: What to Expect (Costs and More), Nvision. Available online: https://www.nvisioncenters.com/insurance/eye-exam/ (accessed on 10 July 2020).
- How Much Does an Intravenous Fluorescein Angiography Cost Near Me? MDsave. Available online: https://www.mdsave.com/procedures/intravenous-fluorescein-angiography/d482fbcc (accessed on 10 July 2020).
- Diabetic Retinopathy Detection, Identify Signs of Diabetic Retinopathy in Eye Images, Kaggle. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection (accessed on 18 January 2020).
- Lam, C.; Yi, D.; Guo, M.; Lindsey, T. Automated Detection of Diabetic Retinopathy using Deep Learning. AMIA Joint Summits on Translational Science Proceedings, 18 May 2018. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5961805/ (accessed on 18 January 2020).
- Wu, Z.; Shi, G.; Chen, Y.; Shi, F.; Chen, X.; Coatrieux, G.; Yang, J.; Luo, L.; Li, S. Coarse-to-fine classification for diabetic retinopathy grading using convolutional neural network. Artif. Intell. Med. 2020, 108, 101936. [Google Scholar] [CrossRef] [PubMed]
- Gargeya, R.; Leng, T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA Innov. Healthcare Deliv. 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Alfian, G.; Syafrudin, M.; Fitriyani, N.L.; Anshari, M.; Stasa, P.; Svub, J.; Rhee, J. Deep Neural Network for Predicting Diabetic Retinopathy from Risk Factors. Mathematics 2020, 8, 1620. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Cheung, C.Y.-L.; Lim, G. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images from Multiethnic Populations with Diabetes. JAMA Orig. Investig. 2017, 318, 2211–2223. [Google Scholar] [CrossRef] [PubMed]
- Datta, N.S.; Banerjee, R.; Dutta, H.S.; Mukhopadhyay, S. Hardware Based Analysis on Automate Early Detection of Diabetic-Retinopathy. Proc. Technol. 2012, 4, 256–260. [Google Scholar] [CrossRef]
- Zalewski, S. Earlier Detection of Diabetic Retinopathy with Smartphone AI, Health News, Medical Breakthroughs & Research for Health Professionals, 29 April 2019. Available online: https://labblog.uofmhealth.org/health-tech/earlier-detection-of-diabetic-retinopathy-smartphone-ai (accessed on 18 January 2020).
- NVIDIA Jetson TX2. Available online: https://developer.nvidia.com/embedded/jetson-tx2-developer-kit (accessed on 18 January 2020).
- Diabetic Retinopathy. American Optometric Association. Available online: https://www.aoa.org/healthy-eyes/eye-and-vision-conditions/diabetic-retinopathy?sso=y (accessed on 18 January 2020).
- News, EyePACS, 9 November 2018. Available online: http://www.eyepacs.com/blog/news (accessed on 18 January 2020).
- Wolff, C. Using Otsu’s Methods to Generate Data for Training of Deep Learning Image Segmentation Models, SCE Developer, 17 May 2018. Available online: https://devblogs.microsoft.com/cse/2018/05/17/using-otsus-method-generate-data-training-deep-learning-image-segmentation-models/ (accessed on 10 June 2019).
- Omer, A.M.; Elfadil, M. Preprocessing of Digital Mammogram Image on Otsu’s Threshold. Am. Sci. Res. J. Eng. Technol. Sci. 2017, 37, 220–229. [Google Scholar]
- Raycad, Convolutional Neural Network (CNN), Medium. Available online: https://medium.com/@raycad.seedotech/convolutional-neural-network-cnn-8d1908c010ab (accessed on 10 June 2019).
- Amidi, A.; Amidi, S. Convolutional Neural Networks Cheat Sheet. Stanford University. Available online: https://stanford.edu/~shervine/teaching/cs-230/cheatsheet-convolutional-neural-networks (accessed on 15 June 2019).
- CS231n Convolutional Neural Networks for Visual Recognition, GitHub. Available online: https://cs231n.github.io/convolutional-networks/ (accessed on 16 June 2019).
- Saha, S. A Comprehensive Guide to Convolutional Neural Networks—The ELI5 Way, Medium, 15 December 2018. Available online: https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53 (accessed on 15 June 2019).
- Kim, S. A Beginner’s Guide to Convolutional Neural Networks (CNNs), Medium, 15 February 2019. Available online: https://towardsdatascience.com/a-beginners-guide-to-convolutional-neural-networks-cnns-14649dbddce8 (accessed on 15 June 2019).
- Leonel, J. Hyperparameters in Machine/Deep Learning, Medium, 7 April 2019. Available online: https://medium.com/@jorgesleonel/hyperparameters-in-machine-deep-learning-ca69ad10b981 (accessed on 16 June 2019).
- Geva, 7 Types of Activation Functions in Neural Networks: How to Choose? MissingLink.ai. Available online: https://missinglink.ai/guides/neural-network-concepts/7-types-neural-network-activation-functions-right/ (accessed on 16 June 2019).
- What is a Hidden Layer?—Definition from Techopedia, Techopedia.com. Available online: https://www.techopedia.com/definition/33264/hidden-layer-neural-networks (accessed on 16 June 2019).
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- TensorFlow. Available online: https://www.tensorflow.org/ (accessed on 16 June 2020).
- Franklin, D. NVIDIA Jetson TX2 Delivers Twice the Intelligence to the Edge, NVIDIA Developer Blog, 7 March 2017. Available online: https://devblogs.nvidia.com/jetson-tx2-delivers-twice-intelligence-edge/ (accessed on 22 September 2020).
- DIGITS Workflow. Available online: https://github.com/dusty-nv/jetson-inference/blob/master/docs/digits-workflow.md (accessed on 17 June 2020).
- NVIDIA Docker: GPU Server Application Deployment Made Easy. Available online: https://developer.nvidia.com/blog/nvidia-docker-gpu-server-application-deployment-made-easy/ (accessed on 17 June 2020).
- Cross-Validation: Evaluating Estimator Performance. Scikit-Learn. Available online: https://scikit-learn.org/stable/modules/cross_validation.html (accessed on 20 September 2020).
- Zhou, W.; Wu, H.; Wu, C.; Yu, X.; Yi, Y. Automatic Optic Disc Detection in Color Retina Images by Local Feature Spectrum Analysis. Comput. Math. Methods Med. 2018, 2018, 1942582. [Google Scholar] [CrossRef] [PubMed]
- Saalfeld, S. Ehcance Local Ontrast (CLAHE). ImageJ, 1 September 2010. Available online: https://imagej.net/Enhance_Local_Contrast_(CLAHE) (accessed on 1 October 2020).
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Alom, M.Z.; Sasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent Residual Convolution Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation. J. Med. Imaging 2019, 6. [Google Scholar] [CrossRef]
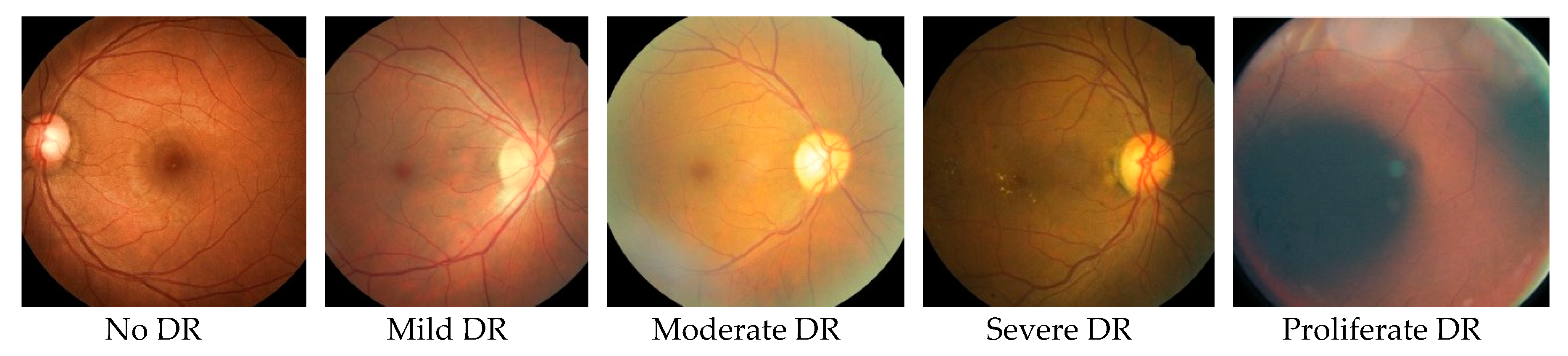
| Time Frame in Years | 0 | 3–5 | 5–10 | 10–15 | >15 |
|---|---|---|---|---|---|
| Stages of DR | Normal | Mild DR: Stage-1 non-proliferate | Moderate DR: Stage-2 non-proliferate | Severe DR: Stage-3 non-proliferate | Proliferate DR: Stage-4 proliferate |
| Changes in Retina | No retinopathy | A few small bulges in the blood vessels. | A few small bulges in the blood vessels. Spots of blood leakage. Deposits of cholesterol. | Larger spots of blood leakages. Irregular beading in veins. Growth of new blood vessels at the optic disc. Blockage of blood vessels. | Beading in veins. Growth of new blood vessels elsewhere in the retina. Clouding of vision. Complete vision loss. |
| DR Category/No. of Images | Training | Testing | ||
|---|---|---|---|---|
| Left Eye | Right Eye | Left Eye | Right Eye | |
| Normal (No DR) | 12,871 | 12,939 | 19,717 | 19,816 |
| Mild DR | 1212 | 1231 | 1905 | 1875 |
| Moderate DR | 2702 | 2590 | 3957 | 3904 |
| Severe DR | 425 | 448 | 613 | 601 |
| Proliferate DR | 353 | 355 | 596 | 610 |
| Solver Option | Sample Value |
|---|---|
| Training Epochs | 30 |
| Snapshot Interval (in epochs) | 1 |
| Validation Interval (in epochs) | 1 |
| Batch Size | 32 |
| Solver Option | Stochastic gradient descent (SGD) |
| Base Learning Rate | 0.01 |
| Resources | LMDB [NVIDIA Jetson TX2] | HDF5 [Google Engine] |
|---|---|---|
| RAM | 8 GB | 16 GB |
| CPU | 2 | 1 |
| Cores | 4 | 1 |
| Processing Time | 2 Hours 37 Minutes | 2 Hours 33 Minutes |
| Hyperparameters | Tested Value Being Used |
|---|---|
| Training Epochs | 100 |
| Random Seed | 100 |
| Batch Size | 32 |
| Solver Options | Gradient Descent |
| Base Learning Rate | 0.001 |
| Test Images | Classification | ||
|---|---|---|---|
| Type | No. of Images | Healthy | Unhealthy |
| No DR | 39,533 | 98% | 2% |
| Mild DR | 3780 | 97% | 3% |
| Moderate DR | 7861 | 90% | 10% |
| Severe DR | 1214 | 70% | 30% |
| Proliferate DR | 1206 | 60% | 40% |
| Test Images | Classification | ||
|---|---|---|---|
| Type | No. of Images | Healthy | Unhealthy |
| No DR | 39,533 | 97% | 3% |
| Mild DR | 3780 | 97% | 3% |
| Moderate DR | 7861 | 80% | 20% |
| Severe DR | 1214 | 59% | 41% |
| Proliferate DR | 1206 | 45% | 55% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vora, P.; Shrestha, S. Detecting Diabetic Retinopathy Using Embedded Computer Vision. Appl. Sci. 2020, 10, 7274. https://doi.org/10.3390/app10207274
Vora P, Shrestha S. Detecting Diabetic Retinopathy Using Embedded Computer Vision. Applied Sciences. 2020; 10(20):7274. https://doi.org/10.3390/app10207274
Chicago/Turabian StyleVora, Parshva, and Sudhir Shrestha. 2020. "Detecting Diabetic Retinopathy Using Embedded Computer Vision" Applied Sciences 10, no. 20: 7274. https://doi.org/10.3390/app10207274
APA StyleVora, P., & Shrestha, S. (2020). Detecting Diabetic Retinopathy Using Embedded Computer Vision. Applied Sciences, 10(20), 7274. https://doi.org/10.3390/app10207274




