Enhancing Multi-tissue and Multi-scale Cell Nuclei Segmentation with Deep Metric Learning
Abstract
1. Introduction
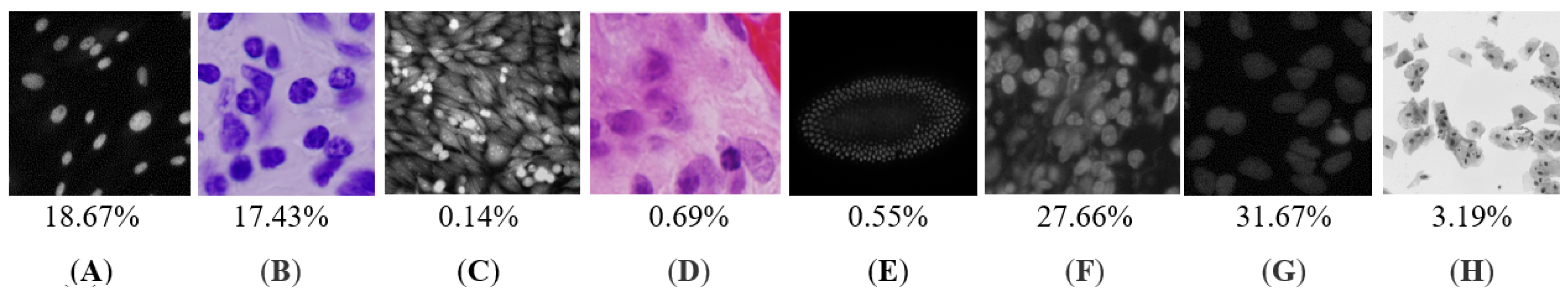
2. Materials and Methods
2.1. Deep Learning Approach
2.2. Deep Metric Embeddings
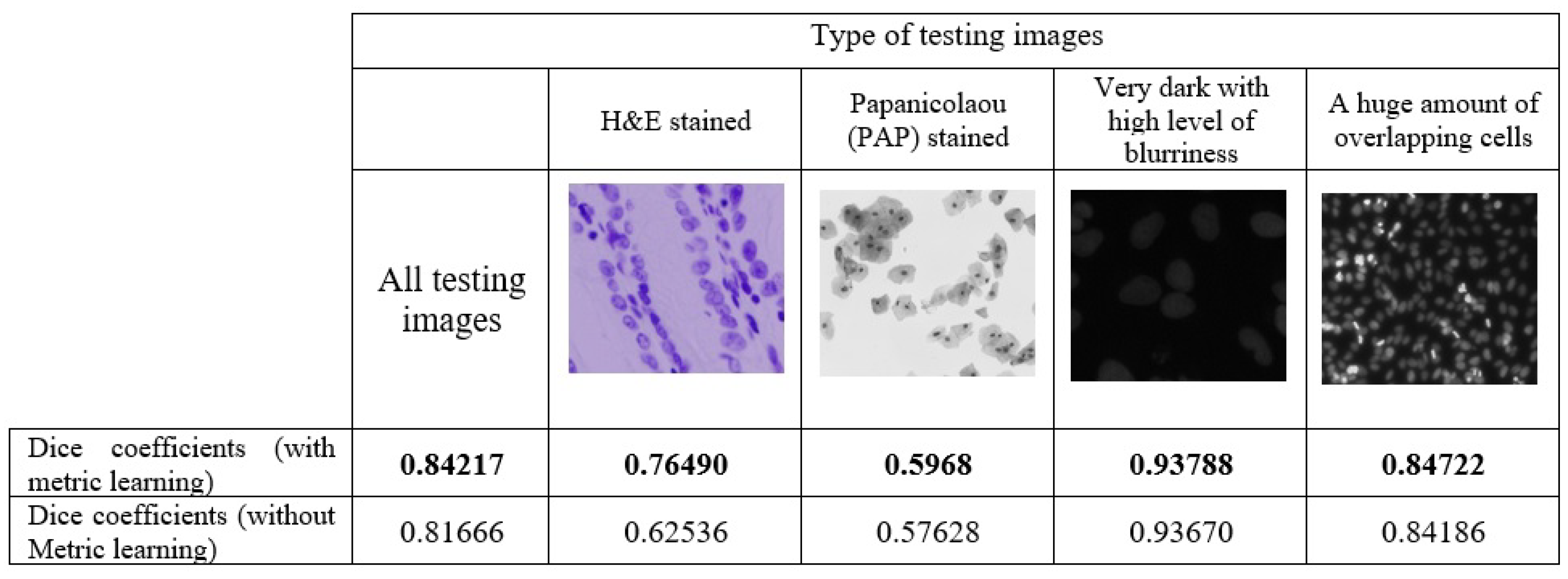
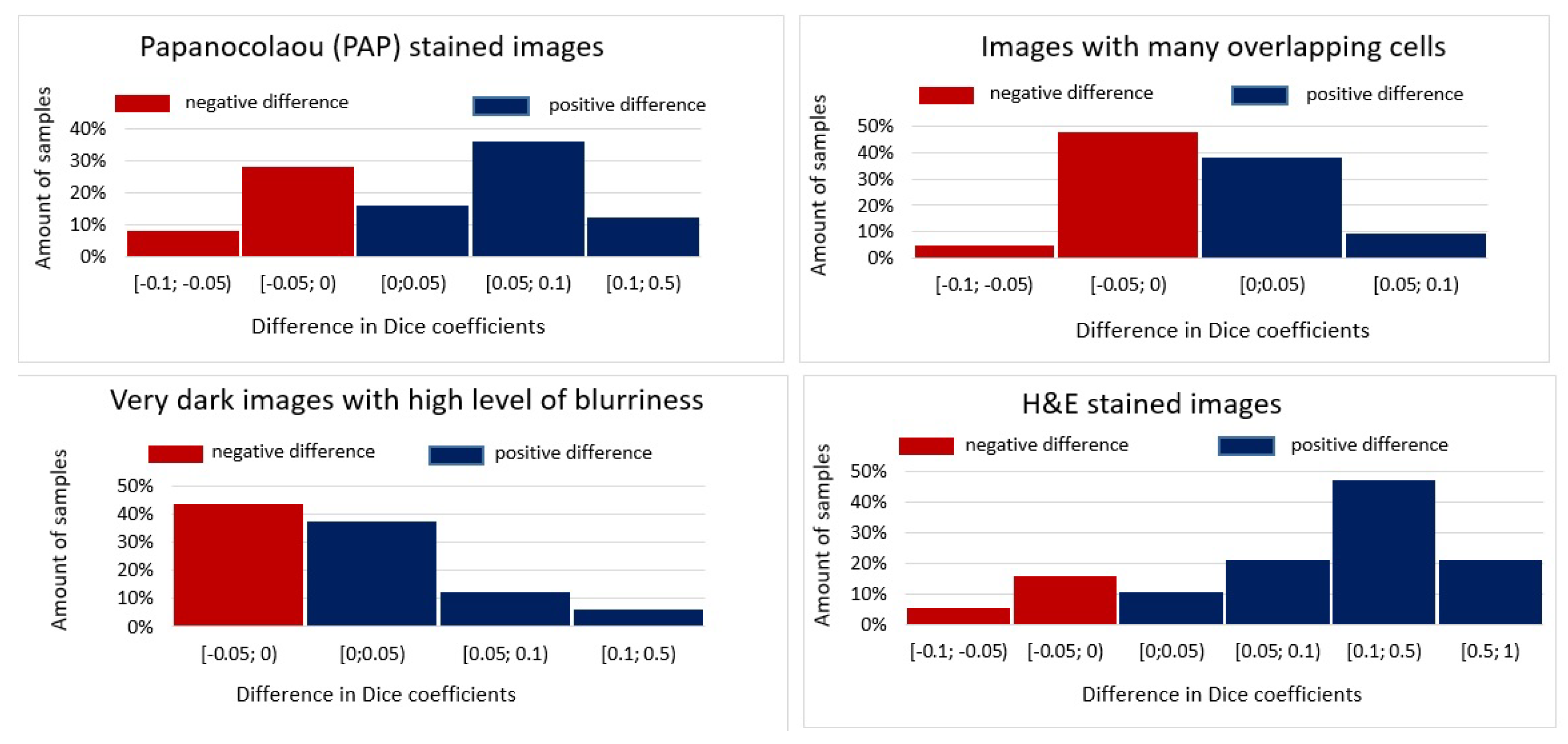
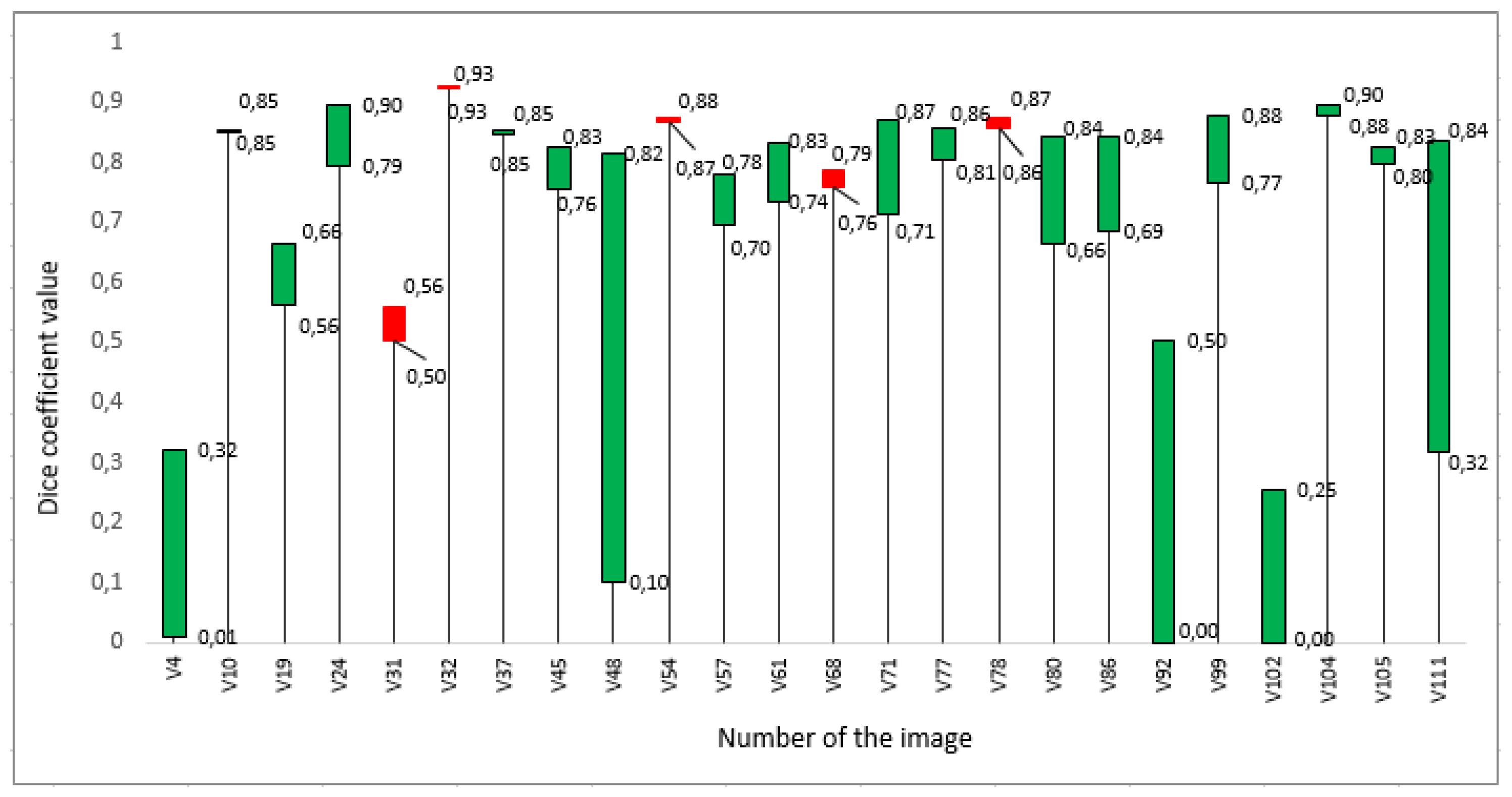
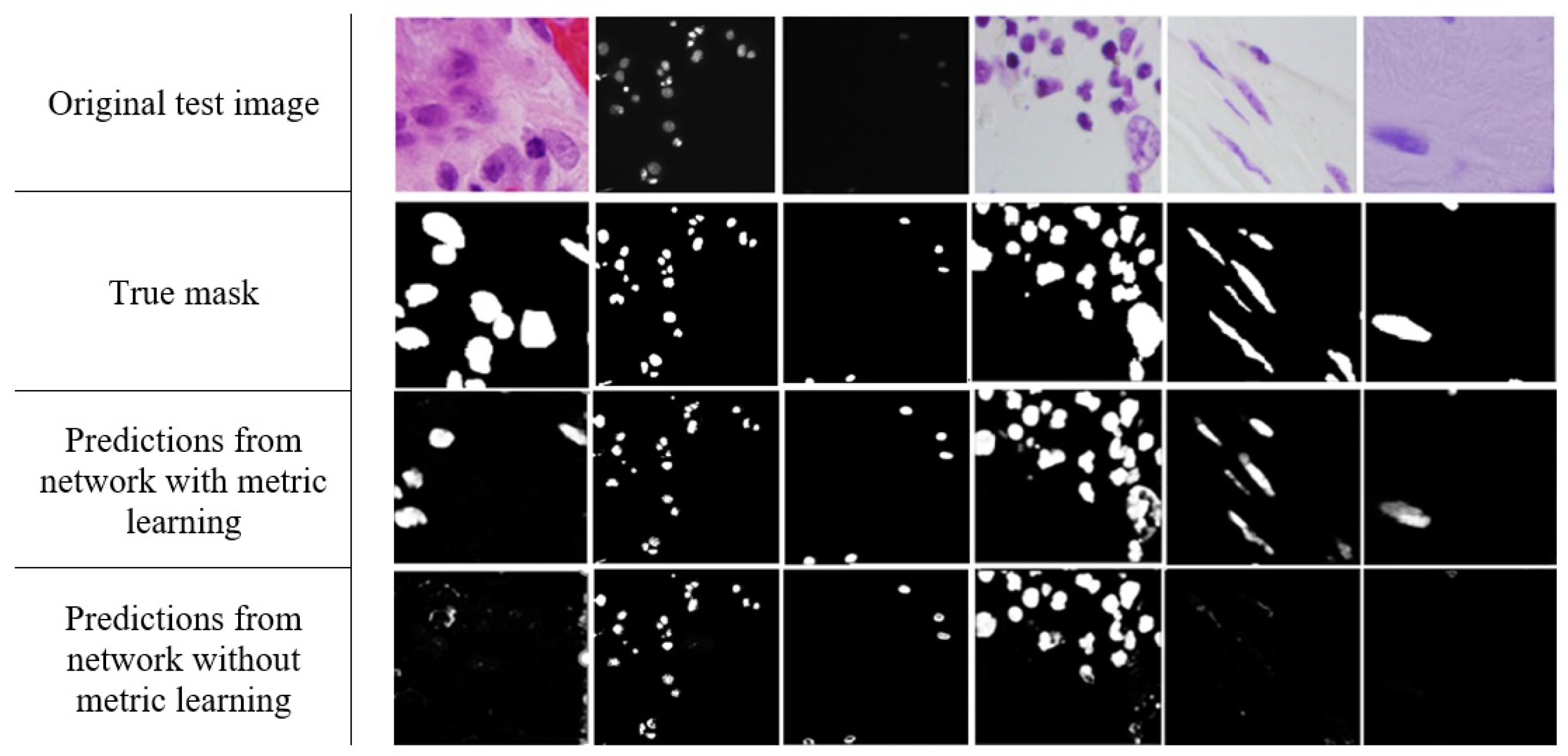
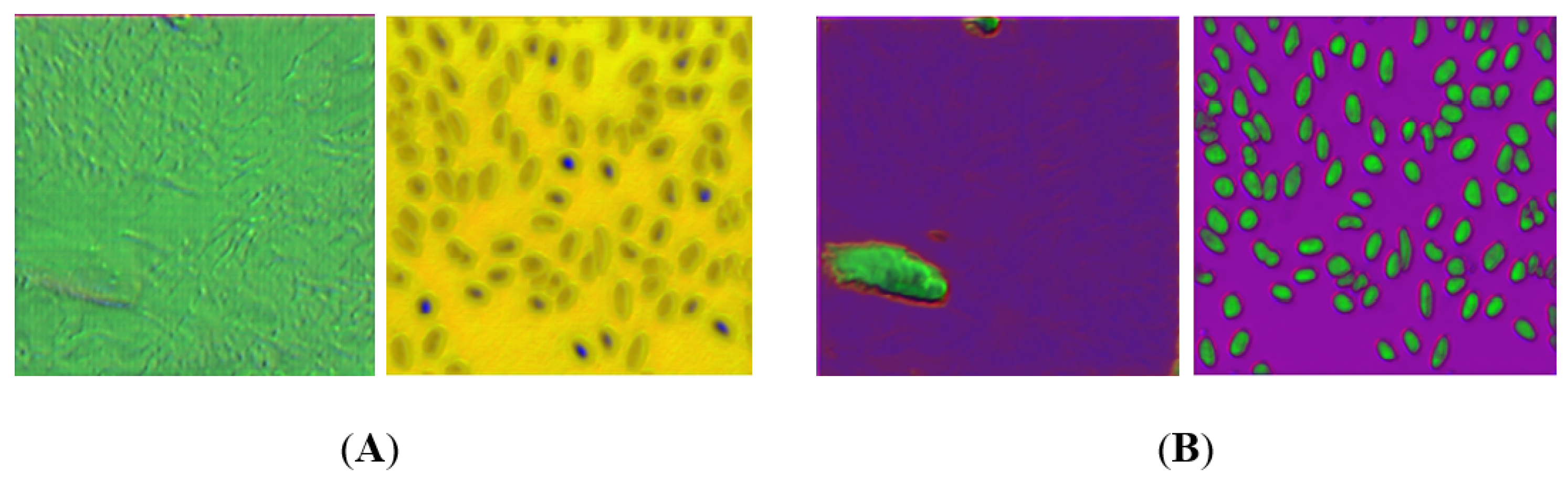
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AUC | Area under the ROC curve |
| CNN | Convolutional Neural Network |
| DenseNet | Dense Convolutional Network |
| ResNet | Residual Neural Network |
| ROC | Receiver Operating Characteristics |
References
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Xing, F.; Yang, L. Robust Nucleus/Cell Detection and Segmentation in Digital Pathology and Microscopy Images: A Comprehensive Review. IEEE Rev. Biomed. Eng. 2016, 9, 234–263. [Google Scholar] [CrossRef]
- Mahmood, F.; Borders, D.; Chen, R.; McKay, G.N.; Salimian, K.J.; Baras, A.; Durr, N.J. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans. Med. Imaging 2018. [Google Scholar] [CrossRef]
- Aprupe, L.; Litjens, G.; Brinker, T.J.; van der Laak, J.; Grabe, N. Robust and accurate quantification of biomarkers of immune cells in lung cancer micro-environment using deep convolutional neural networks. PeerJ 2019, 7, e6335. [Google Scholar] [CrossRef] [PubMed]
- Höfener, H.; Homeyer, A.; Weiss, N.; Molin, J.; Lundström, C.F.; Hahn, H.K. Deep learning nuclei detection: A simple approach can deliver state-of-the-art results. Comput. Med. Imaging Graph. 2018, 70, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef] [PubMed]
- Vu, Q.D.; Graham, S.; Kurc, T.; To, M.N.N.; Shaban, M.; Qaiser, T.; Koohbanani, N.A.; Khurram, S.A.; Kalpathy-Cramer, J.; Zhao, T.; et al. Methods for Segmentation and Classification of Digital Microscopy Tissue Images. Front. Bioeng. Biotechnol. 2019, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Ray, N. Cell Detection with Deep Convolutional Neural Network and Compressed Sensing. arXiv 2017, arXiv:1708.03307. [Google Scholar]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for Nuclei Detection, Segmentation, and Classification in Digital Histopathology: A Review—Current Status and Future Potential. IEEE Rev. Biomed. Eng. 2014, 7, 97–114. [Google Scholar] [CrossRef]
- Salvi, M.; Molinari, F. Multi-tissue and multi-scale approach for nuclei segmentation in H&E stained images. Biomed. Eng. Online 2018, 17, 89. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Roth, J.; Goodman, A.; Becker, T.; Karhohs, K.W.; Broisin, M.; Csaba, M.; McQuin, C.; Singh, S.; Theis, F.; et al. Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images. BioRxiv 2019, 335216. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, F.; Xi, W.; Chen, H.; Gao, P.; He, C. Joint Multi-frame Detection and Segmentation for Multi-cell Tracking. arXiv 2019, arXiv:1906.10886. [Google Scholar]
- Hernandez, D.E.; Chen, S.W.; Hunter, E.E.; Steager, E.B.; Kumar, V. Cell Tracking with Deep Learning and the Viterbi Algorithm. In Proceedings of the 2018 International Conference on Manipulation, Automation and Robotics at Small Scales (MARSS), Nagoya, Japan, 4–8 July 2018; IEEE: Nagoya, Japan, 2018; pp. 1–7. [Google Scholar] [CrossRef]
- Narayanan, B.N.; Hardie, R. A Computationally Efficient U-Net Architecture for Lung Segmentation in Chest Radiographs (Preprint). In Proceedings of the 2019 National Aerospace and Electronics Conference (NAECON), Dayton, OH, USA, 15–19 July 2019. [Google Scholar]
- Liang, Y.; Tang, Z.; Yan, M.; Chen, J.; Xiang, Y. Comparison Detector: Convolutional Neural Networks for Cervical Cell Detection. arXiv 2019, arXiv:1810.05952. [Google Scholar]
- Al-Kofahi, Y.; Zaltsman, A.; Graves, R.; Marshall, W.; Rusu, M. A deep learning-based algorithm for 2-D cell segmentation in microscopy images. BMC Bioinform. 2018, 19, 365. [Google Scholar] [CrossRef] [PubMed]
- Nurzynska, K. Deep Learning as a Tool for Automatic Segmentation of Corneal Endothelium Images. Symmetry 2018, 10, 60. [Google Scholar] [CrossRef]
- Gupta, K.; Thapar, D.; Bhavsar, A.; Sao, A.K. Deep Metric Learning for Identification of Mitotic Patterns of HEp-2 Cell Images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 1 July 2019; pp. 1–7. [Google Scholar]
- Singh, S.; Janoos, F.; Pecot, T.; Caserta, E.; Leone, G.; Rittscher, J.; Machiraju, R. Identifying Nuclear Phenotypes Using Semi-supervised Metric Learning. In Proceedings of the Biennial International Conference on Information Processing in Medical Imaging, Kloster Irsee, Germany, 3–8 July 2011; pp. 398–410. [Google Scholar]
- Schroff, F.; Kalenichenko, D.; Philbin, J. FaceNet: A unified embedding for face recognition and clustering. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Chopra, S.; Hadsell, R.; LeCun, Y. Learning a similarity metric discriminatively, with application to face verification. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; pp. 1–8. [Google Scholar] [CrossRef]
- Dahia, G.; Segundo, M.P. Automatic Dataset Annotation to Learn CNN Pore Description for Fingerprint Recognition. arXiv 2018, arXiv:1809.10229. [Google Scholar]
- Redondo-Cabrera, C.; Lopez-Sastre, R. Unsupervised learning from videos using temporal coherency deep networks. Comput. Vis. Image Underst. 2019, 179, 79–89. [Google Scholar] [CrossRef]
- Zhang, S.; Gong, Y.; Wang, J. Deep Metric Learning with Improved Triplet Loss for Face Clustering in Videos. In Advances in Multimedia Information Processing; Springer: Cham, Switzerland, 2016; Volume 9916, pp. 497–508. [Google Scholar] [CrossRef]
- Ljosa, V.; Sokolnicki, K.L.; Carpenter, A.E. Annotated high-throughput microscopy image sets for validation. Nat. Methods 2012, 9, 637. [Google Scholar] [CrossRef]
- Paulauskaite-Taraseviciene, A.; Sutiene, K.; Valotka, J.; Raudonis, V.; Iesmantas, T. Deep learning-based detection of overlapping cells. In Machine Learning in Medical Imaging; Springer International Publishing: Cham, Switzerland, 2019; pp. 217–220. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Las Vegas, NV, USA, 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; IEEE: Honolulu, HI, USA, 2017; pp. 4700–4708. [Google Scholar] [CrossRef]
- Xie, S.; Tu, Z. Holistically-Nested Edge Detection. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1395–1403. [Google Scholar] [CrossRef]
- Lin, T.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 42, 318–327. [Google Scholar] [CrossRef]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky Loss Function for Image Segmentation Using 3D Fully Convolutional Deep Networks. In Machine Learning in Medical Imaging; Wang, Q., Shi, Y., Suk, H.I., Suzuki, K., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 379–387. [Google Scholar]
- Suárez, J.L.; García, S.; Herrera, F. A Tutorial on Distance Metric Learning: Mathematical Foundations, Algorithms and Software. arXiv 2018, arXiv:1812.05944. [Google Scholar]
- Wang, J.; Zhou, F.; Wen, S.; Liu, X.; Lin, Y. Deep Metric Learning with Angular Loss. Comput. Vis. Pattern Recognit. 2017, arXiv:1708.01682. [Google Scholar]
- Kaya, M.; Bilge, H.S. Deep Metric Learning: A Survey. Symmetry 2019, 11, 66. [Google Scholar] [CrossRef]
- Hadsell, R.; Chopra, S.; LeCun, Y. Dimensionality Reduction by Learning an Invariant Mapping. In Proceedings of the 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, New York, NY, USA, 17–22 June 2006; IEEE Computer Society: Washington, DC, USA, 2006; pp. 1735–1742. [Google Scholar] [CrossRef]
- Wang, P.; Hu, X.; Li, Y.; Liu, Q.; Zhu, X. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process 2016, 122, 1–13. [Google Scholar] [CrossRef]
- Aswathy, M.A.; Jagannath, M. Detection of breast cancer on digital histopathology images: Present status and future possibilities. Inform. Med. Unlocked 2017, 8, 74–79. [Google Scholar] [CrossRef]
- Tosta, T.A.A.; Neves, L.A.; do Nascimento, M.Z. Segmentation methods of H&E-stained histological images of lymphoma: A review. Inform. Med. Unlocked 2017, 9, 35–43. [Google Scholar]
- Piórkowski, A.; Gertych, A. Color Normalization Approach to Adjust Nuclei Segmentation in Images of Hematoxylin and Eosin Stained Tissue. In Information Technology in Biomedicine; Pietka, E., Badura, P., Kawa, J., Wieclawek, W., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 393–406. [Google Scholar]
- Yi, F.; Huang, J.; Yang, L.; Xie, Y.; Xiao, G. Automatic extraction of cell nuclei from H&E-stained histopathological images. J. Med. Imaging 2017, 4, 027502. [Google Scholar] [CrossRef]
- Graham, S.; Vu, Q.D.; Raza, S.E.A.; Azam, A.; Tsang, Y.W.; Kwak, J.T.; Rajpoot, N. HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images. Med. Image Anal. 2019, 58, 101563. [Google Scholar] [CrossRef]
- Pan, X.; Li, L.; Yang, D.; He, Y.; Liu, Z.; Yang, H. An Accurate Nuclei Segmentation Algorithm in Pathological Image Based on Deep Semantic Network. IEEE Access 2019, 7, 110674–110686. [Google Scholar] [CrossRef]
- Khalili, E.; Hosseini, V.; Solhi, R.; Aminian, M. Rapid fluorometric quantification of bacterial cells using Redsafe nucleic acid stain. Iran. J. Microbiol. 2015, 7, 319–323. [Google Scholar]
- Lukashevich, M.; Starovoitov, V. Cell Nuclei Counting and Segmentation for Histological Image Analysis. In Pattern Recognition and Information Processing; Ablameyko, S.V., Krasnoproshin, V.V., Lukashevich, M.M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 86–97. [Google Scholar]
| Resnet-104 | ResNet-68 | DenseNet-71 | ||||
|---|---|---|---|---|---|---|
| Input layer: 256×256×1 grayscale image | ||||||
| L1 | Conv1 | Conv1 | Conv1 | |||
| L2 | Conv2: 3*2=6 layers | Conv2: 3*1=3 layers | Conv2: 2*2=4 layers | |||
| Conv2+1 layer. Max pool | ||||||
| L3 | Conv3: 3*2=6 layers | Conv3: 3*1=3 layers | Conv3: 2*2=4 layers | |||
| Conv3+1 layer. Max pool | ||||||
| L4 | Conv4: 3*4=12 layers | Conv4: 3*2=6 layers | Conv4: 2*4=8 layers | |||
| Conv4+1 layer. Max pool | ||||||
| L5 | Conv5: 3*4=12 layers | Conv5: 3*2=6 layers | Conv5: 2*4=8 layers | |||
| Conv5+1 layer. Max pool | ||||||
| L6 | Conv6: 3*6=18 layers | Conv6: 3*3=9 layers | Conv6: 2*6=12 layers | |||
| Conv6+1 layer. Max pool | ||||||
| L7 | Conv7: 3*6=18 layers | Conv7: 3*3=9 layers | Conv7: 3 layers | |||
| Upsampling layers | ||||||
| L8 | Deconvolution . Add L6 output before max pooling . 1+4 = 5 conv layers | |||||
| L9 | Deconvolution . Add L5 output before max pooling . 1+4 = 5 conv layers. | |||||
| L10 | Deconvolution . Add L4 output before max pooling . 1+4 = 5 conv layers | |||||
| L11 | Deconvolution . Add L3 output before max pooling . 1+4 = 5 conv layers | |||||
| L12 | Deconvolution . Add L2 output before max pooling . 1+4 = 5 conv layers. Metrics are included. | |||||
| L13 | Softmax. 1 convolutional layer | |||||
| ∑ | 104 | 68 | 71 | |||
| DenseNet-71 | ResNet-68 | ResNet-104 | |
|---|---|---|---|
| No metric learning | 0.97952 ± 0.00198 | 0.97176 ± 0.00394 | 0.98312 ± 0.00272 |
| Contrastive loss | 0.98703 ± 0.00458 | 0.98678 ± 0.00351 | 0.98873 ± 0.00199 |
| Triplet Loss | 0.98817 ± 0.00225 | 0.98902 ± 0.00229 | 0.99089 ± 0.00150 |
| DenseNet-71 | ResNet-68 | ResNet-104 | |
|---|---|---|---|
| No metric learning | 0.8118 ± 0.0043 | 0.8109 ± 0.0057 | 0.8175 ± 0.0098 |
| Contrastive loss | 0.8233 ± 0.0083 | 0.8327 ± 0.0048 | 0.8401 ± 0.0027 |
| Triplet Loss | 0.8308 ± 0.0065 | 0.8357 ± 0.0029 | 0.8461 ± 0.0034 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iesmantas, T.; Paulauskaite-Taraseviciene, A.; Sutiene, K. Enhancing Multi-tissue and Multi-scale Cell Nuclei Segmentation with Deep Metric Learning. Appl. Sci. 2020, 10, 615. https://doi.org/10.3390/app10020615
Iesmantas T, Paulauskaite-Taraseviciene A, Sutiene K. Enhancing Multi-tissue and Multi-scale Cell Nuclei Segmentation with Deep Metric Learning. Applied Sciences. 2020; 10(2):615. https://doi.org/10.3390/app10020615
Chicago/Turabian StyleIesmantas, Tomas, Agne Paulauskaite-Taraseviciene, and Kristina Sutiene. 2020. "Enhancing Multi-tissue and Multi-scale Cell Nuclei Segmentation with Deep Metric Learning" Applied Sciences 10, no. 2: 615. https://doi.org/10.3390/app10020615
APA StyleIesmantas, T., Paulauskaite-Taraseviciene, A., & Sutiene, K. (2020). Enhancing Multi-tissue and Multi-scale Cell Nuclei Segmentation with Deep Metric Learning. Applied Sciences, 10(2), 615. https://doi.org/10.3390/app10020615







