Abstract
Genetics research is increasingly focusing on mining fully sequenced genomes and their annotations to identify the causal genes associated with traits (phenotypes) of interest. However, a complex trait is typically associated with multiple quantitative trait loci (QTLs), each comprising many genes, that can positively or negatively affect the trait of interest. To help breeders in ranking candidate genes, we developed an analytical platform called pbg-ld that provides semantically integrated geno- and phenotypic data on Solanaceae species. This platform combines both unstructured data from scientific literature and structured data from publicly available biological databases using the Linked Data approach. In particular, QTLs were extracted from tables of full-text articles from the Europe PubMed Central (PMC) repository using QTLTableMiner++ (QTM), while the genomic annotations were obtained from the Sol Genomics Network (SGN), UniProt and Ensembl Plants databases. These datasets were transformed into Linked Data graphs, which include cross-references to many other relevant databases such as Gramene, Plant Reactome, InterPro and KEGG Orthology (KO). Users can query and analyze the integrated data through a web interface or programmatically via the SPARQL and RESTful services (APIs). We illustrate the usability of pbg-ld by querying genome annotations, by comparing genome graphs, and by two biological use cases in Jupyter Notebooks. In the first use case, we performed a comparative genomics study using pbg-ld to compare the difference in the genetic mechanism underlying tomato fruit shape and potato tuber shape. In the second use case, we developed a seamlessly integrated workflow that uses genomic data from pbg-ld knowledge graphs and prioritization pipelines to predict candidate genes within QTL regions for metabolic traits of tomato.
1. Introduction
The availability of annotated reference genome assemblies for several crop species including tomato [1], potato [2], brassica [3] and cucumber [4] has enabled plant breeders and researchers to elucidate a trait’s linkage to a genomic location(s). Mining genome annotations can help in identifying candidate genes that positively or negatively affect a trait of interest, which plant breeders aim to improve. However, genome annotations are commonly available across multiple databases and file formats (e.g., in the Generic Feature Format [GFF]), which hampers integrated data analyses.
Traditionally, plant breeders identified chromosomal regions using genetic markers that are statistically associated with traits of interest. These genomic regions are called quantitative trait loci (QTLs). A QTL region can easily contain thousands of genes including those that negatively influence the trait of interest [5]. Therefore, detecting the causative gene for breeding is of major importance. There are three major approaches to address the challenge of candidate gene prediction in crop species: (i) the analysis of gene expression data or co-expression networks [6], (ii) comparative genomics [7], and (iii) integrate information stored in scientific literature and in molecular biology databases such as the ELIXIR Core Data Resources [8] (including the European Nucleotide Archive (ENA) [9], Ensembl Plants [10] and UniProt [11]) and the Sol Genomics Network (SGN) [12]. To address the need for improved access to integrated plant data, we developed the Solanaceae Linked Data platform (pbg-ld) [13] that combines QTLs from scientific literature and genome annotations from public databases using the Linked Data approach [14]. Our approach is to create a semantic web of data rather than that of hypertext (HTML) documents using Uniform Resource Identifiers (URIs) and Resource Description Framework (RDF) [15]. A URI is an HTTP-based resource identifier assigned to an entity whereas RDF is a generic graph-based data model for describing entities and their relationships. In addition, publishing data according to FAIR Data Principles [15] further increases the degree of discoverability and (re-)usability of research data.
In plant sciences, several controlled vocabularies and ontologies have been developed to standardize domain-specific terms and/or represent the current knowledge of the domain in a machine-readable form. For example, the Solanaceae Phenotype Ontology (SPTO) [16], Crop Ontology [17], Plant Ontology (PO) [18], Phenotypic Quality Ontology (PATO) [19] and Trait Ontology (TO) [20] are used to identify plant-specific phenotypic information while Gene Ontology (GO) [21], Sequence Ontology (SO) [22] and Feature Annotation Location Description Ontology (FALDO) [23] are used to identify genotypic information. Similarly, the Chemical Entities of Biological Interest database/ontology (ChEBI) [24] is focused on small chemical compounds.
There are several plant-specific databases available that provide geno- and phenotypic data. For example, Ensembl Plants is a widely used integrated resource on plant genomes. Similarly, UniProt is a database of protein sequences, function annotations and proteomes of various species including plants. Both databases release their data in RDF-based format. The Arabidopsis Information Resource (TAIR) [25] is a resource to analyze and to compare molecular, biological, and genetical data of the model species Arabidopsis thaliana. Further, the Sol Genomics Network (SGN) [12] provides genomic, genetic and phenotypic information for members of the Solanaceae family. Plant Genome DataBase Japan (PGDBj) [26] is an integrated web resource for plant genome-related information from literature and public databases. However, the TAIR, SGN, and PGDBj do not distribute their data in a semantically interoperable (RDF) format. The Planteome [27] database provides gene annotations and phenotypes with the help of reference ontologies such as PO, TO, GO and ChEBI. Planteome is a user-friendly tool to query traits of interest, germplasm, and putative candidate genes. However, it lacks QTLs, genetic markers and links to publicly available databases such as Ensembl Plants.
We present the pbg-ld platform that provides semantically integrated geno- and phenotypic data on Solanaceae species such as the (wild) tomato and potato species. The resulting (linked) datasets are made available through a web interface or programmatic services (SPARQL and RESTful APIs). The use of these data access points is described in the results section. pbg-ld is an integrated plant resource that aids breeders in detecting candidate genes for complex traits using the knowledge available in the scientific literature and in public databases.
2. Data Generation and Ingestion Pipeline
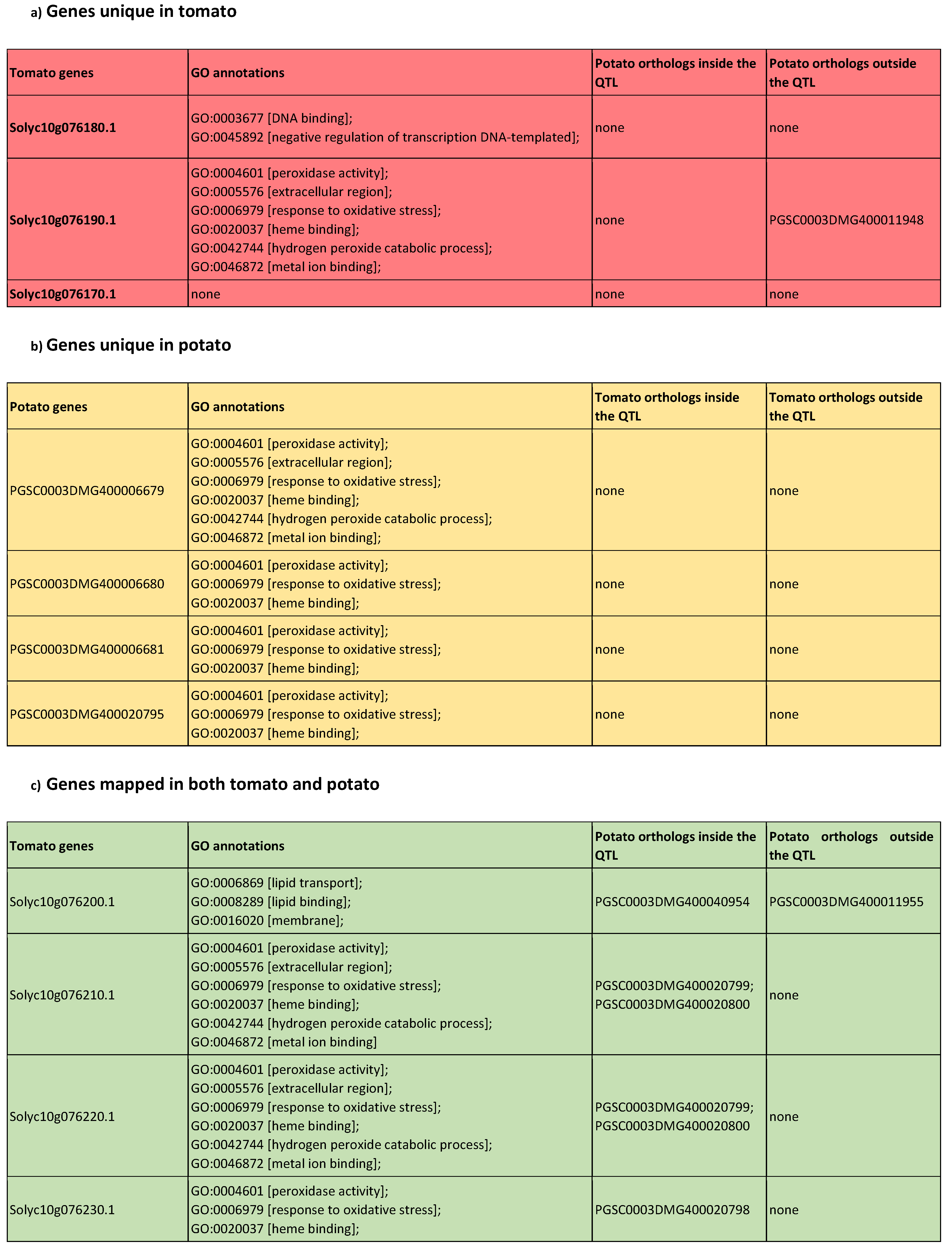
All data sets used in the creation of this linked data platform were initially classified as non-RDF based and RDF based data sources. The first step in our data ingestion pipeline was to convert non-RDF based data sources to RDF based data graphs. Later these RDF data graphs were integrated together and published as a linked data platform. Figure 1 illustrates the data generation and ingestion pipeline used by the pbg-ld platform. Geno- and phenotypic data from three Solanaceae species: (i) reference sequence tomato (S. lycopersicum), (ii) wild tomato (S. pennellii) and (iii) the reference sequence potato (S. tuberosum) is collected and integrated into this pipeline.
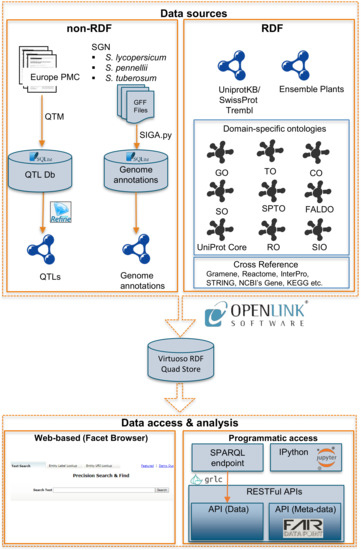
Figure 1.
Data generation and ingestion pipeline. All data originate from either non-RDF or RDF sources. Several tools are used to retrieve and transform non-RDF data into RDF graphs: QTLTableMiner++ (QTM) [28] is used to extract tomato and potato quantitative trait loci (QTLs) from Europe PubMed Central (PMC) articles; OpenRefine [29] is used to transform the QTLs into RDF according to the specified data model. Similarly, SIGA.py tool [30] converts the genome annotations, as provided by the Sol Genomics Network (SGN) in GFF files into RDF graphs with gene models and markers. In addition, the UniProt (proteomes) and Ensembl Plants (gene models) distribute their data in RDF format. All RDF graphs including domain-specific ontologies (in OWL) and database cross-references were stored and integrated with Virtuoso RDF Quad Store. The resulting linked datasets are made available for queries and analyses through data-access layer: (i) Linked data browser, (ii) SPARQL endpoint, (iii) grlc-based Web API [31] and (iv) FAIR Data Point (FDP) metadata service [32].
2.1. Data Sources
To facilitate the integration of geno- and pheno-typic data of Solanaceae species, we used data from (semi-)structured resources. Semi-structured data resources include scientific articles in XML file format obtained from Europe PMC [33] and the General Feature Format (GFF)-based text file [34]. Moreover, semi-structured data were classified as non-RDF data and were subsequently transformed into (structured) RDF data. On the contrary, structured data resources contained data in the form of RDF structure, for example, genome annotation in Ensemble Plants and Uniprot, as well as domain ontologies.
2.1.1. QTLs
QTL studies have widely been published in scientific articles, particularly in tables or supplementary materials. However, there is no established repository where experimental data on plant-specific QTL studies can be submitted. Therefore, QTL information was extracted from XML-based scientific literature and processed into RDF graphs [35] using the QTLTableMiner++ (QTM, v1.1.0) [28,36] and the OpenRefine software [29]. QTM extracted 237 QTLs from a total of 21 Solanaceae-specific full-text articles in the Europe PMC repository. 147 of these QTLs (i.e., 85 in tomato and 62 in potato) were associated with exact chromosomal locations based on peak/flanking markers while 108 of the QTLs (i.e., 77 in tomato and 31 in potato) were found to encompass one or more candidate genes.
2.1.2. SGN
SGN provides genome annotations in GFF files for Solanaceae species. The GFF files were transformed into RDF graphs [37] using the SIGA.py command-line tool (v0.5.1) [30] (Appendix A, Figure A1). The gene models and the genetic markers of (wild) tomato (S. lycopersicum and S. pennellii) and potato (S. tuberosum) were downloaded from the SGN’s FTP server (ftp://ftp.solgenomics.net/genomes/). For S. lycopersicum, the genome annotations comprising of gene models, SGN and SolCAP markers were taken from GFF files (ITAG 2.4, released on 23-02-2014) [38]. For S. pennelli, the genome annotations comprising of gene models (v2.0, released on 27-08-2014) and SGN markers (released on 10-08-2014) were taken as input [39]. Similarly, for S. tuberosum, the genome annotations of PGSC_DM (diploid/double monoploid, v4.03, released on 04-09-2013) were used [40].
2.1.3. Ensembl Plants and UniProt
Ensembl Plants is an genome-centeric integrated resource for plant sciences. Genome annotations of S. lycopersicum (release ITAG2.4 genome annotation based on SL2.50 genome assembly) [41] and S. tuberosum (release PGSC_DM 3.0 genome annotation based on SolTub3.0 genome assembly) were taken from the Ensembl Plants database (release 33) [42] in RDF format. The proteomes of S. lycopersicum [43] and S. tuberosum [44] were obtained from UniProt in the RDF/Turtle format.
2.2. Ontologies
pbg-ld makes use of the following domain-specific ontologies: Gene Ontology (GO) [45], Solanaceae Phenotype Ontology (SPTO) [16], Crop ontology (CO) [17], Sequence Ontology (SO) [46], Feature Annotation Location Description Ontology (FALDO) [47], Trait Ontology (TO) [20], UniProt Core [48], Semanticscience Integrated Ontology (SIO) [49], Relation Ontology (RO) [50], Plant Ontology (PO) [51], Phenotypic Quality Ontology (PATO) [52].
2.3. Linked Data Deployment
OpenLink’s Virtuoso Universal Server (version 7.20.3217, open-source edition) was used to store and to connect the data graphs in the RDF Quad Store. pbg-ld is made as a modular and re-deployable software with the help of Docker [53] and Ansible [54]. The pbg-ld platform including the associated RESTful web services, namely the grlc-based API for data and the FAIR Data Point API for metadata [55], can be deployed locally by the user.
2.4. Data Access & Analysis
pbg-ld provides access to the (meta)data through a web-based user interface (Virtuoso Faceted Browser) and programmatic interfaces such as SPARQL and RESTful APIs. Using the web-based user interface, a user can query the RDF triples in three different ways through (i) a free-text search box, (ii) an entity label search box or (iii) an entity URI search box. There is a SPARQL endpoint provided for a user to write and execute SPARQL queries on the RDF graphs available in the pbg-ld platform. Further, with the help of grlc tool [31], we published a customized RESTful API, built on the top of pbg-ld’s datasets to provide easy (programmatic) access based on the SPARQL endpoint. Data consumers who do not know the SPARQL query language can use this API to query the platform. This way grlc hides the complexities or intricacies of SPARQL. Appendix B, Table A1 provides a list of API endpoints available in pbg-ld. Lastly, the FAIR Data Point service is provided to expose machine-readable descriptions (metadata) about the pbg-ld datasets. To show a valuable use case of the pbg-ld platform, we have developed exemplary Jupyter (IPython) Notebooks [56,57].
3. Results
3.1. Genome Annotations via the Faceted Browser
pbg-ld allows the user to access and analyze data with the help of the Faceted Browser. Figure 2 exemplifies a query for trait-gene associations using “fruit shape” as a search term. Here, this term (partially) matches several standardized trait names in the SPTO and TO ontologies (e.g., SP:0000038 and TO:0002628). By selecting either one, pbg-ld returns seven QTLs associated with the trait of interest (i.e., “fruit shape”). In Figure 2, one such a QTL is selected for further analysis, that is, QTL:4321030_4_14. QTM extracted this QTL from Table 4 of the Europe PMC article PMC4321030 [58]. This QTL is marked by flanking markers C2_At2g14260 and TG400 on chromosome 11, for which pbg-ld finds the list of all genes in this region. In Figure 2, one such gene (Solyc11g038340.1) in QTL:4321030_4_14 is selected. pbg-ld web interface contains direct links to allow the user to further browse the annotations, properties, and the sequence of this gene in the SGN, Ensembl Plants and UniProt databases. For example, Figure 2 shows the sequence of this selected gene in the SGN’s genome browser (JBrowser).
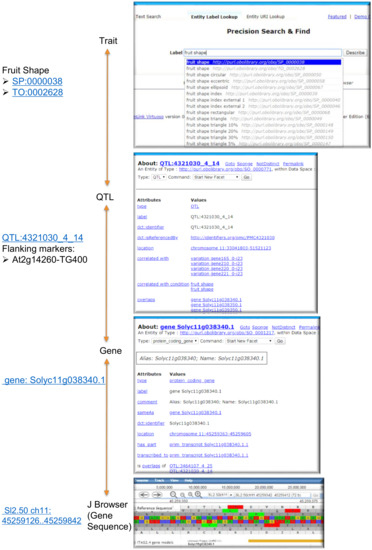
Figure 2.
Browsing trait-gene associations in the Faceted Browser for the example trait “fruit shape”.
3.2. Example Queries via SPARQL and RESTful API
- (I)
- SPARQL query to list QTLs, associated gene IDs and GO annotations related to an example trait “fruit shape” (SP:0000038).In addtion to the manual browsing via a web interface, Figure 3 exemplifies a programmatic way to query trait-gene associations. This query yields QTLs and candidate genes including GO terms (molecular function and biological process only) for the trait “fruit shape” (SP:0000038).
 Figure 3. Input and output of the SPARQL query to list QTLs, associated gene IDs, and Gene Ontology (GO) annotations for the trait “fruit shape” (SP:0000038).
Figure 3. Input and output of the SPARQL query to list QTLs, associated gene IDs, and Gene Ontology (GO) annotations for the trait “fruit shape” (SP:0000038). - (II)
- SPARQL query to list genes/proteins annotated with GO terms related to both “fruit” and “ripening”.SPARQL query in Figure 4 highlights a way to do textual search over the annotations of genes/proteins. With the bag-of-words expression, we query genes and proteins containing GO annotations with the words “fruit” and “ripening”. The resulting output is the list of genes/proteins involved in the biological process called “fruit ripening” (GO:0009835).
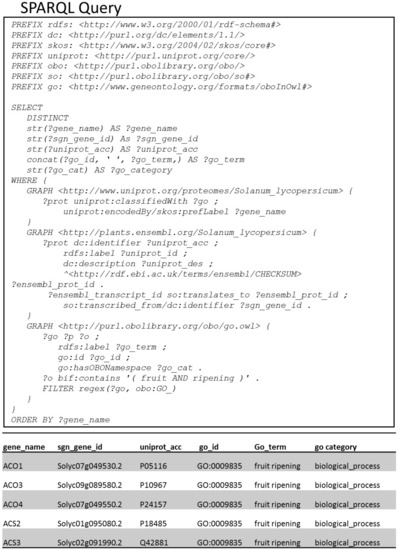 Figure 4. Input and output of the SPARQL query to list genes/proteins annotated with GO terms related to both “fruit” and “ripening”. Full-text search in the SPARQL query is done with the Virtuoso’s bif:contains predicate.
Figure 4. Input and output of the SPARQL query to list genes/proteins annotated with GO terms related to both “fruit” and “ripening”. Full-text search in the SPARQL query is done with the Virtuoso’s bif:contains predicate. - (III)
- Comparison of (wild) tomato genome graphs using the RESTful API.pbg-ld can be used to query genomic features across various biological databases. This can be done either by writing a SPARQL query against the endpoint or via the API call /countFeatures of pbg-ld, with the genomic graph as a parameter. For example, pbg-ld can count the genomic features annotated in the S. lycopersicum genome according to Ensembl Plants or SGN, and annotations in the S. pennelli genome according to SGN.We compared the differences between the genomic features of the tomato graphs in Figure 5. It is evident that there are a total of 33,785 protein coding genes in Ensemble Plants, whereas there are 34,725 protein coding genes in the SGN graph. There are 940 unique genes in the SGN database that are not mentioned in the Ensembl Plants database. Furthermore, the results also highlight that genetic markers are included in SGN but not in Ensembl Plants while the latter database contains RNAs. In addition, pbg-ld can also be used to compare genomic data of different species of the same family. S. pennellii is a wild tomato species that is relatively distant from the domesticated S. lycopersicum. Because of S. pennelli’s extreme stress tolerance, unusual morphology, and a genome sequence 119 Mb larger than S. lycopersicum, it is an important donor of germplasm for the cultivated tomato. While comparing the genomic features of S. lycopersicum vs. S. pennelli in SGN, the number of genomic features in S. pennellii is on average 1.5 times greater than that of S. lycopersicum (Figure 5).
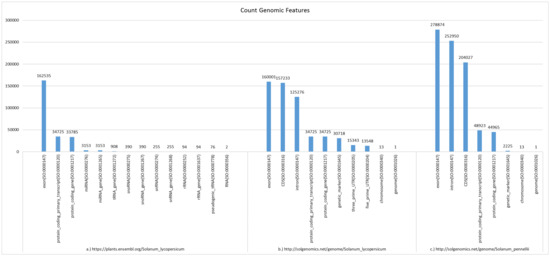 Figure 5. Bar charts of genomic feature counts for three Solanum genomes (graph IRIs): Ensembl: S. lycopersicum (http://plants.ensembl.org/Solanum_lycopersicum); SGN: S. lycopersicum (http://solgenomics.net/genome/Solanum_lycopersicum); SGN: S. pennellii (http://solgenomics.net/genome/Solanum_pennellii). The data were obtained through the pbg-ld Web API /countFeatures endpoint.
Figure 5. Bar charts of genomic feature counts for three Solanum genomes (graph IRIs): Ensembl: S. lycopersicum (http://plants.ensembl.org/Solanum_lycopersicum); SGN: S. lycopersicum (http://solgenomics.net/genome/Solanum_lycopersicum); SGN: S. pennellii (http://solgenomics.net/genome/Solanum_pennellii). The data were obtained through the pbg-ld Web API /countFeatures endpoint.
3.3. Biological Use Case 1: Comparative Genomics to Study Tomato Fruit Shape and Potato Tuber Shape
For this use case, we used the pbg-ld endpoint (APIs) with a Jupyter Notebook [56] to study the difference in the genetic mechanism underlying fruit shape in tomatoes and tuber shape in potatoes. Both tomato fruits and potato tubers can have a genetically determined wide variation in their shape, for example elongated and round. The candidate gene Solyc10g076180 (SlOFP20 is a member of the OVATE Family Protein (OFP)) on chromosome 10 of the reference tomato genome (Heinz 1706) and is one of the determinants of round fruit shape. However, this gene does not have an ortholog in the reference potato genome (DM), which has very elongated tubers [59]. In this use case, first, we query and compare the QTL regions on chromosome 10 in tomato and in potato. This QTL regions are associated with round shape in tomato fruits and predominantly elongated shape in potatoes. We classify these genes in three categories (a) genes that are unique in tomato (b) genes that are unique in potato (c) genes that are common to both species (Table 1). Furthermore, we check the GO annotations as well as orthologs in all these genes. Three genes (Solyc10g076170.1, Solyc10g076190.1, Solyc10g076180.1) are present in class (a) which indicates that they are unique in tomato. Out of these genes, the Solyc10g076170.1 gene was removed from the latest UniProt release. Solyc10g076190.1 is a peroxidase gene (that is also common in class (b) and class (c)) in Table 1. Solyc10g076180.1 is the only unique member of the OVATE family. Genes in class (b) are all peroxidase genes whereas the class (c) contains several genes coding for peroxidases and one gene involved in lipid transport. According to our analysis the candidate gene Solyc10g076180.1 does not have a corresponding ortholog in the potato DM reference genome in the same QTL region on chromosome 10. Therefore, with our tool we explored this further, and retrieve a knowledge network based on homologs of the candidate gene. This network was retrieved with a nested query, in which we first located all paralogs of the Solyc10g076180.1 gene in the tomato genome and then we found orthologs of these genes in the potato genome (Figure 6). In this analysis, we found 10 OFP genes (green) in potato and 11 genes (red) in tomato. One less potato gene is due to the missing OFP20 gene in the studied QTL.

Table 1.
Table comparing genes in QTLs associated with (tomato) fruit shape and (potato) tuber shape. Three classes represent (a) genes unique in tomato; (b) genes unique in potato (c) genes mapped in both species. Each row contains a gene ID, GO annotations and orthologs inside/outside a QTL region. The query shows that only three genes are unique in tomato (i.e., Solyc10g076190.1, Solyc10g076170.1 and Solyc10g076180.1).
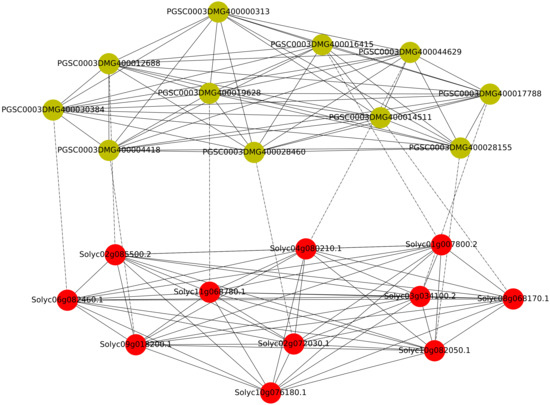
Figure 6.
A network of Solyc10g076180.1 homologs in tomato (red) and potato (green) including paralogous (solid) and orthologous relations (dotted). There is no ortholog of OFP20 gene in potato.
Out of these OFP genes in potato, PGSC0003DMG400028155 is located on chromosome 10 in the region 56030393-56031156. However, this region is 6.7Mb away from the studied QTL region, and thus seems unlikely to harbor the determinant candidate gene.
3.4. Biological Use Case 2: Prediction of Candidate Genes in Tomato QTL Regions with Functional Annotations and Evolutionary Analysis Using pbg-ld and QTLSearch
Predicting candidate genes for QTL regions is a key objective in plant genetics and breeding. However, a single QTL region can contain many genes. Mining candidate genes from such a QTL region could be done using existing knowledge of structural and functional gene annotations. In this use case, we aim to develop, illustrate, and analyze a seamlessly integrated workflow that uses linked genomic-data and prioritization pipelines to predict candidate genes within QTL regions for metabolic traits of tomato. Metabolic composition of a tomato is directly associated with its nutritional value, taste, aromas, and quality [60]. Metabolomic research studies in the past, have been able to predict functional characteristics of metabolites in the life-cycle of a plant. For example, volatiles are known to play an important role in the defense mechanism of plants against pathogens, where they serve as airborne signaling molecules to induce a defense response in other plant parts or neighboring plants [61]. Similarly, other metabolites like soluble solids (glucose, fructose, and sucrose) contribute to the sweetness of a fruit [62]. Lycopene is a carotenoid compound found in tomatoes which contributes to the nutritional value of a tomato and the red pigment in tomatoes responsible for fruit-color [63]. Similarly, terpenoids play a role in attracting pollinators [64].
Finding candidate genes within QTL regions for the trait of interest using computational approaches is a major challenge in plant bioinformatics. Several tools have been developed in the past that tried to prioritize candidate genes based on existing knowledge. QTLSearch is a software that searches for candidate causal genes in QTL studies by combining Gene Ontology annotations across many species and leveraging hierarchical orthologous groups [65]. QTG-Finder is a recently published tool that uses a machine learning model to prioritize candidate genes using function annotation, co-function network, and paralog copy number [66]. However, both tools have been developed in Arabidopsis thaliana and in rice (Oryza sativa), and are difficult to use and test in other species like tomato and potato. QTLSearch uses the HOGPROP algorithm that requires access to hierarchical orthologous groups available in OMA browser [67], to score candidate genes based on trait-related GO terms. As the OMA Browser data graph is cross-referenced in Uniprot, which is part of pbg-ld, the QTLSearch algorithm can be tested on tomato/potato data with the help of pbg-ld.
Figure 7 illustrates a candidate gene prediction workflow within a QTL region for the trait of interest, using function annotations and evolutionary genomics data. Input to this workflow is either a QTL region (containing physical location or a genetic location) or a trait of interest. If the input parameter is a trait of interest, the pbg-ld database retrieves all QTL locations for that trait in tomato. After receiving the QTL inputs, this workflow queries the set of all genes occurring within this QTL region. For every gene, this workflow retrieves a set of all GO terms as well as all orthologs and paralogs of these genes. These genome annotations are served as input to the QTLSearch pipeline, which uses the HOGPROP algorithm. This algorithm assigns scores to every gene within a QTL region. It uses GO terms which relate to the trait of interest and GO terms which relate to the genes within a QTL region to assess the distance of these functional annotations along gene phylogenies. This workflow has been developed in a Jupyter Notebook [56]. It is a modular framework in which data are fetched from multiple data sources and can also accommodate new analysis modules as they are being developed by our group or the scientific community.
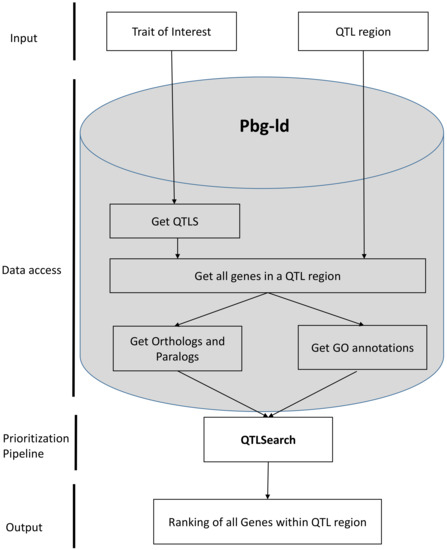
Figure 7.
A workflow to predict candidate genes in a QTL region for the traits of interest.
To test the usability of our workflow in predicting candidate genes for metabolic traits, we selected five QTLs for different metabolic traits, i.e, Brix/soluble solids, lycopene beta-cyclase activity and 2-phenylethanol/phenylacetaldehyde (Table 2). Out of the selected five QTLs for metabolic traits, three QTLs which relate to the following traits, soluble solids, lycopene beta-cyclase activity and phenolic compounds (2-phenylethanol and phenylacetaldehyde), have known candidates. Further, these candidate genes are already annotated with related GO terms in publicly available databases such as UniProt. While, for one of the QTL regions which relates to terpenoids, Terpene synthase is a known candidate gene, however, this synthase is not annotated with GO terms which show an association with the trait of interest (i.e., terpenoids). The two GO terms related to this gene are DNA binding and DNA methylation. Lastly, for the QTL region selected for volatile compounds (i.e., 3-methylbuthanal and 3-methylbuthanol) there are no well-known candidate genes in the QTLs that had experimentally proven significance.

Table 2.
A selected set of five QTLs for metabolic traits in tomatoes. These QTLs are used as test cases to analyse the prediction power of the underlying workflow.
The top three predicted candidate genes for each trait, with the help of our pipeline, are reported in Table 3.

Table 3.
Top three candidate genes found for the five selected metabolic traits (Brix/soluble solids, lycopene beta-cyclase activity and 2-phenylethanol/phenylacetaldehyde).
One of the most extensively studied metabolic traits in tomato is the total soluble solids content in fruits (i.e., TSS or Brix) [68]. We selected five GO terms related to these metabolites and Brix trait (see Table 3) and fed it to the QTLSearch pipeline in our workflow. Previously known studies have identified multiple QTLs that are associated with the Brix trait in tomatoes [58]. Out of the many known QTL locations, the most significant QTL is located on chromosome 9, containing Lin5 as the popularly known candidate gene for Brix trait [68]. Table 3 highlights the top three genes predicted from our workflow for the Brix QTL region from 3374710 to 3574710 on chromosome 9. Lin7 and Lin5 were the top predicted genes related to this trait. Both these genes are from a homologous family and are known to be associated with Brix. The scores of both genes are significantly higher than those of all other genes. We conclude that our pipeline performed well to predict the candidate genes for this QTL.
Carotenoid compounds are the primary determinants of tomato fruit color [71]. Carotenoids exert a broad range of functions which associate to photosynthesis, the formation of pigments, antioxidant activities, and being precursors to signaling molecules, including volatiles [72]. Lycopene is a major carotenoid in tomato [73]. Lycopene occurrence with other bioactive compounds, like vitamin C, vitamin E, other carotenoids (alpha-carotene, beta-carotene, gamma-carotene and lutein), and flavonoids is primarily associated with the color of a tomatoes. Lycopene beta-cyclase is a key enzyme occurring at the branch point of the carotenoid biosynthesis pathway and responsible for converting lycopene to beta-carotene. Lycopene beta-cyclase activity is also related to the total carotenoid content accumulated in the tomato fruit. The major QTL region which is related to Lycopene beta-cyclase activity is found to be located on chromosome 6 between the region 45280179–49150528 [74]. Here we analyzed the prediction of candidate genes for Lycopene beta-cyclase activity with the help of our developed workflow. Two GO terms that relate to lycopene beta-cyclase activity were selected for inclusion in our workflow. Previous study suggests that lycopene cyclase (LCY) is a known candidate gene related to this trait. In some databases, lycopene cyclase (LCY) is also annotated as neoxanthin synthase (NSY) as these are genes that are closely related carotenogenic enzymes belonging to the same family. Table 3 shows the results from the workflow, containing the top three genes predicted for this QTL region on chromosome 6. NSY was ranked at the top of the list and has a score significantly higher than all other genes. Here also, we can conclude that our workflow performed as expected.
Phenolic derivative compounds like 2-phenylethanol and phenylacetaldehyde have a great impact on the aroma of a tomato [70]. Several QTL locations related to phenolic compounds have been identified in the past out of which, a major QTL region on chromosome 8 mapped by the markers, TG330-CT77 and TG330-CT148 is associated with the accumulation of 2-phenylethanol and phenylacetaldehyde (having genomic coordinates 55068565-63267130) [75]. Additionally, two putative proteins, 2-phenylacetaldehyde reductases proteins (LePAR1 and LePAR2) are known candidates, which catalyze the conversion of 2-phenylacetaldehyde to 2-phenylethanol [76]. Both these proteins are members of a reductase/dehydrogenase family. Table 3 shows the top three genes predicted from our pipeline for these phenolic compounds on chromosome 8. Solyc08g068190.2 was the top predicted gene related to this trait. Although we are not sure if this gene is the same as LePAR1 and LePAR2, this gene belongs to the same aldehyde dehydrogenase family. Therefore, our workflow could detect the causal gene within this QTL region.
A major QTL related to Terpenoids has been mapped on chromoasome 1 with the genomic coordinates of 86142248–86467672 [77]. Proteins of the Terpene synthase (TPS) family and TPS gene are the expected candidate genes associated with Terpenoids. Five of the TPS-a subclade genes (TPS31, TPS32, TPS33, and TPS35) occur in close proximity within this QTL. Table 3 highlights the top tree genes predicted from our pipeline for this QTL. Our results suggest that the gene Solyc01g095030.2, which is a MYB transcription factor, is the causal gene for this QTL region. This is possibly the wrong prediction. The reason for our pipeline to give a wrong prediction here could be that there is no term present in the GO ontology that directly related to Terpenoids. Further, because of this missing GO annotation terms, Terpene synthase is not been well annotated with its function, which makes it difficult for our workflow to detect it as a high ranking gene for the Terpenoids trait.
Volatile compounds like 3-methylbuthanal and 3-methylbuthanol influence the flavor, sensory changes, and defense mechanism of tomato fruits [78]. A major QTL related to the volatile compounds (3-methylbuthanal, 3-methylbuthanol) has been mapped on chromosome 3 with the genomic coordinates of 69685329–71362039 [79]. However, it is not known which candidate gene in this QTL is responsible for changes in the concentration of these volatile compounds. Our results suggest that the lactate dehydrogenase (LDH) gene is possibly a candidate gene for this trait.
Out of the five QTLs, our workflow performed significantly well in detecting candidate genes for the QTLs of soluble solids, lycopene beta-cyclase activity, and phenolic compounds. Our workflow did not perform well in the detection of candidate genes within the QTL for terpenoids on chromosome 1. This is most probably due to the fact that this QTL region is not well annotated, and there are no GO terms related to Terpenoids. Lastly, our workflow predicts a candidate gene called LDH, for the previously unknown QTL region associated with volatile compounds.
QTLSearch, a prediction pipeline for candidate genes in QTL regions, is based on existing knowledge and evolutionary data (orthologs and paralogs). While the performance of QTLSearch is high with well-annotated data, it fails to perform well in detecting candidate genes for QTL regions where little is known. Hence, it is still very challenging to infer about candidate genes in a poorly annotated QTL region.
4. Discussion & Conclusions
The main objective of developing the pbg-ld platform was to improve the FAIRness of candidate gene identification in Solanaceae species by providing (semantically) integrated genomics and QTL datasets available in public resources (i.e., UniProt, Ensembl Plants, SGN and Europe PMC) from a central endpoint.
Genomic knowledge discovery is often confronted by the challenges of data integration from a multitude of independent databases and research articles. For discovering candidate genes with the help of large-scale data integration, there is a need to organize candidate data resources according to the FAIR data principles. The core development in this research is a Linked Data platform that semantically organizes and integrates genotypic and phenotypic data on Solanaceae species according to these principles. Hence, this progress in digital science helps genomic datasets to be more findable, accessible, interoperable and reusable.
After selecting various datasets and information relevant to candidate gene discovery, a critical step in the knowledge discovery process is the transformation of data into a suitable data infrastructure. Biological data are complex and highly connected, for example, there is ambiguity in the names of genes, proteins and transcripts, hence a semantic model with correct identifiers is required to differentiate them. pbg-ld addresses the challenges of providing a semantic layer over most used datasets for candidate gene discovery in tomato and potato. A critical step in our approach was the transformation of (semi-)structured or non-RDF data sources to inter-linked RDF graphs using existing and newly developed tools such as the QTM and SIGA.py. Further, FDP provides meta-data descriptions, which makes the user aware of the source graph(s) to perform queries and to interpret the results. Different data access points provide flexibility for users who wish to analyze and/or visualize data on this platform. Lastly, open accessibility of all the above mentioned tools used to generate and publish these data sets, offers the ability for the scientific community users to extend this tool for other crop species themselves.
Data sets are not static and constantly emerging over time. pbg-ld combines open data from different third party resources such as Europe PMC, SGN, UniProt and Ensemble Plants. As these data sets are not static, a significant improvement in pbg-ld could be to automate the process of regular updates of the data sets. For example, QTL information in pbg-ld is extracted from tables of scientific articles in Europe PMC using the QTM tool. However, QTL data graphs are static and require manual updates. Ideally, the QTL data graph should be updated automatically, whenever a new QTL study is published. A researcher is interested in retrieving the most newly published scientific articles in the domain of his/her research interests. Another improvement to the pbg-ld platform could be to enable (interactive) visualization of the knowledge graphs.
Complementary to our developments, some similar plant-specific software and databases provide genotypic and phenotypic data sets in a semantically integrated way. For example, KNETMiner is an open source software that integrates plant-specific biological data sets into a knowledge graph [80]. These biological data sets contain information related to genes, biological pathways, phenotypes and publications for many important crop species like wheat, rice, sorgum, potato, tomato, and so forth. Additionally, KNETMiner has an evidence-based gene ranking algorithm that ranks and visualizes this integrated data based on gene annotations. However, the current version of KNETMiner [81] provides free-access to integrated data sets only for some important crops like wheat (Triticum aestivum), rice (Oryza sativa), Arabidopsis thaliana. The integrated data set for other crop species like, tomato (S. lycopersicum), potato (S. tuberosum), and sorgum (Sorgum bicolor) is are not openly accessible. Similarly, Planteome database [27] provides gene annotations and phenotypes with the help of reference ontologies such as PO, TO, GO and ChEBI. Planteome is a user-friendly tool to query traits of interest, germplasm, and putative candidate genes. However, it lacks QTLs, genetic markers and links to publicly available databases such as Ensembl Plants. Therefore, our Linked Data platform is a unique resource for Solanaceae species that provides access to available knowledge about genome annotations in public databases and scientific literature.
pbg-ld currently contains the gene models and the genetic markers based GFF files of the reference tomato genome (S. lycopersicum), wild tomato (S pennellii), and the reference potato genome (S. tuberosum). However, other than these genomes, the SGN database, for instance also includes GFF files from other Solanaceae and closely related genomes species, such as reference genome of pepper (Capsicum annuum) [82], eggplant (Solanum melongena L.) [83] and so forth. Converting these GFF files to RDF graphs and adding them to the pbg-ld would improve analyses based on comparative genomics. Nevertheless, a big challenge in doing this type of analysis based on knowledge graphs is to have the mappings of biological entities (genes, proteins, etc.) across multiple species. There is a chance of having different identifiers in two related species, for example, the genomes of the reference sequence potato S. tuberosum and wild type species M6 have both been sequenced, however, both use different sets of identifiers and the mapping of genes between these genotypes is not available. However, having the data about cross references in these genes for both the genomes can give us better insights into underpinning certain gene functions with comparative studies.
To conclude, pbg-ld is an integrated resource for Solanaceae species that provides access to available knowledge about genome annotations in public databases and scientific literature. This resource aids in the identification of candidate genes for complex traits using available knowledge in the databases and literature.
Author Contributions
Conceptualization: G.S., R.F., A.K., R.G.F.V.; methodology: G.S., A.K., R.F.; software: A.K., M.B., G.S., C.M.-O.; validation: G.S., C.W.B.B., Y.M.T., A.G.B.; formal analysis: G.S., A.K.; investigation: G.S., A.K., C.W.B.B.; resources: A.K., M.B., G.S.; data curation: A.K., G.S., M.B.; writing–original draft preparation: G.S., A.K.; writing–review and editing: G.S., A.K., R.G.F.V., C.W.B.B., R.F.; visualization: G.S., A.K., M.B.; supervision: R.G.F.V., C.W.B.B., A.K., R.F.; project administration: R.G.F.V., A.K., G.S., R.F.; funding acquisition: R.G.F.V., R.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Netherlands Organisation for Scientific Research (NWO) via the Netherlands eScience Center (grant number: 27014204).
Acknowledgments
We thank Lars Ridder for stimulating discussions and guidance. This article is based on the results of one of the chapters of the Ph.D thesis of the first author Gurnoor Singh entitled “Genomics data integration for knowledge discovery using genome annotations from molecular databases and scientific literature” delivered at Wageningen University [84]. This work was carried out on the Dutch national e-infrastructure with the support of SURF Foundation.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Figure A1.
SIGA.py software architecture.
Appendix B

Table A1.
List of RESTful API endpoints of pbg-ld generated by the grlc service (deployed on http://localhost:8088).
Table A1.
List of RESTful API endpoints of pbg-ld generated by the grlc service (deployed on http://localhost:8088).
| Number | Endpoint (Path) | Description | Input Parameters | Response Fields |
|---|---|---|---|---|
| 1. | /countFeatures | Count genomic features. | graph - genome graph IRI, endpoint - SPARQL endpoint | feature_id, feature_name, n |
| 2. | /getFeatureLocation | Get the genomic location of a feature. | featureid - feature ID, endpoint | feature_id, feature_name, chrom, begin_pos, end_pos, taxon_id |
| 3. | /getFeaturesInInterval | Get genomic features given an interval. | graph, chrom, begin, end, feature - feature type, endpoint | feature_id, feature_name, chrom, begin_pos, end_pos |
| 4. | /getGeneAnnotations | Get annotations from SGN, Ensembl and UniProt given a gene ID. | geneid - gene ID, endpoint | gene_id, gene_name, transcript_id, sgn_des, uniprot_acc, uniprot_reviewed, uniprot_existence, uniprot_des, uniprot_goa |
| 5. | /getGenesInQTL | Get genes that overlap with a QTL identified by ID. | qtlid - QTL ID (see QTM), endpoint | gene_id |
| 6. | /getOrthologs | Get orthologs of a gene identified by ID. | geneid, endpoint | gene_id, ortholog_id |
| 7. | /getParalogs | Get paralogs of a gene identified by ID.. | geneid, endpoint | gene_id, paralog_id |
| 8. | /getQTLs | Get QTLs associated with a trait identified by ID. | traitid - trait ID (e.g. using SPTO or TO), endpoint | qtl_id |
| 9. | /getQTLsPerArticle | Get QTLs described in an article identified by ID. | pmcid - Pubmed Central ID, endpoint | qtl_id |
| 10. | /getTraitIds | Get term ID(s) for a trait defined in several trait ontologies. | trait - trait term(s) (incl. boolean operators), endpoint | trait_id, trait_term |
| 11. | /sumQTLs | Summarize QTLs extracted from articles. | endpoint | taxon_id, n_articles, n_qtls, n_qtls_with_loc, n_qtls_with_genes |
References
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef]
- Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275. [Google Scholar] [CrossRef]
- Chibon, P.Y.; Schoof, H.; Visser, R.G.; Finkers, R. Marker2sequence, mine your QTL regions for candidate genes. Bioinformatics 2012, 28, 1921–1922. [Google Scholar] [CrossRef]
- Astola, L.; Stigter, H.; van Dijk, A.D.; van Daelen, R.; Molenaar, J. Inferring the gene network underlying the branching of tomato inflorescence. PLoS ONE 2014, 9, e89689. [Google Scholar] [CrossRef] [PubMed]
- Shinozuka, H.; Cogan, N.O.; Spangenberg, G.C.; Forster, J.W. Quantitative Trait Locus (QTL) meta-analysis and comparative genomics for candidate gene prediction in perennial ryegrass (Lolium perenne L.). BMC Genet. 2012, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- Durinx, C.; McEntyre, J.; Appel, R.; Apweiler, R.; Barlow, M.; Blomberg, N.; Cook, C.; Gasteiger, E.; Kim, J.H.; Lopez, R.; et al. Identifying ELIXIR Core Data Resources. F1000Research 2016, 5, 1–16. [Google Scholar] [CrossRef]
- Harrison, P.W.; Alako, B.; Amid, C.; Cerdeño-Tárraga, A.; Cleland, I.; Holt, S.; Hussein, A.; Jayathilaka, S.; Kay, S.; Keane, T.; et al. The European Nucleotide Archive in 2018. Nucleic Acids Res. 2018, 47, D84–D88. [Google Scholar] [CrossRef] [PubMed]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating tools for visualizing, mining, and analyzing plant genomic data. In Plant Genomics Databases; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–31. [Google Scholar]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar]
- Mueller, L.A.; Solow, T.H.; Taylor, N.; Skwarecki, B.; Buels, R.; Binns, J.; Lin, C.; Wright, M.H.; Ahrens, R.; Wang, Y.; et al. The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005, 138, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Kuzniar, A. pbg-ld. zenodo 2019. [Google Scholar] [CrossRef]
- Berners-Lee, T. Linked Data. 2006. Available online: https://www.w3.org/DesignIssues/LinkedData.html (accessed on 1 July 2020).
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Solanaceae Phenotype Ontology (SPTO). Available online: http://bioportal.bioontology.org/ontologies/SPTO (accessed on 2 September 2019).
- Shrestha, R.; Matteis, L.; Skofic, M.; Portugal, A.; McLaren, G.; Hyman, G.; Arnaud, E. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Physiol. 2012, 3, 326. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L.; Walls, R.L.; Elser, J.; Gandolfo, M.A.; Stevenson, D.W.; Smith, B.; Preece, J.; Athreya, B.; Mungall, C.J.; Rensing, S.; et al. The Plant Ontology as a Tool for Comparative Plant Anatomy and Genomic Analyses. Plant Cell Physiol. 2013, 54, e1. [Google Scholar] [CrossRef]
- Walls, R.L.; Athreya, B.; Cooper, L.; Elser, J.; Gandolfo, M.A.; Jaiswal, P.; Mungall, C.J.; Preece, J.; Rensing, S.; Smith, B.; et al. Ontologies as integrative tools for plant science. Am. J. Bot. 2012, 99, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Trait Ontology (TO). Available online: http://purl.obolibrary.org/obo/to.owl (accessed on 2 September 2019).
- The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef]
- Eilbeck, K.; Lewis, S.E.; Mungall, C.J.; Yandell, M.; Stein, L.; Durbin, R.; Ashburner, M. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol. 2005, 6, R44. [Google Scholar] [CrossRef]
- Bolleman, J.T.; Mungall, C.J.; Strozzi, F.; Baran, J.; Dumontier, M.; Bonnal, R.J.P.; Buels, R.; Hoehndorf, R.; Fujisawa, T.; Katayama, T.; et al. FALDO: A semantic standard for describing the location of nucleotide and protein feature annotation. J. Biomed. Semantics 2016, 7, 39. [Google Scholar] [CrossRef]
- Hastings, J.; de Matos, P.; Dekker, A.; Ennis, M.; Harsha, B.; Kale, N.; Muthukrishnan, V.; Owen, G.; Turner, S.; Williams, M.; et al. The ChEBI reference database and ontology for biologically relevant chemistry: Enhancements for 2013. Nucleic Acids Res. 2013, 41, D456–D463. [Google Scholar] [CrossRef]
- Garcia-Hernandez, M.; Berardini, T.; Chen, G.; Crist, D.; Doyle, A.; Huala, E.; Knee, E.; Lambrecht, M.; Miller, N.; Mueller, L.A.; et al. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genom. 2002, 2, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, A.; Ichihara, H.; Asamizu, E.; Shirasawa, S.; Nakamura, Y.; Tabata, S.; Hirakawa, H. Plant genome database Japan (PGDBj). In Plant Genomics Databases; Springer: Berlin/Heidelberg, Germany, 2017; pp. 45–77. [Google Scholar]
- Cooper, L.; Meier, A.; Laporte, M.A.; Elser, J.L.; Mungall, C.; Sinn, B.T.; Cavaliere, D.; Carbon, S.; Dunn, N.A.; Smith, B.; et al. The Planteome database: An integrated resource for reference ontologies, plant genomics and phenomics. Nucleic Acids Res. 2018, 46, D1168–D1180. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kuzniar, A.; van Mulligen, E.M.; Gavai, A.; Bachem, C.W.; Visser, R.G.F.; Finkers, R. QTLTableMiner++: Semantic mining of QTL tables in scientific articles. BMC Bioinform. 2018, 19, 183. [Google Scholar] [CrossRef] [PubMed]
- OpenRefine. Available online: https://openrefine.org/ (accessed on 17 September 2020).
- Kuzniar, A. SIGA.py. zenodo 2019. [Google Scholar] [CrossRef]
- Meroño-Peñuela, A.; Hoekstra, R. grlc Makes GitHub Taste Like Linked Data APIs; The Semantic Web, Sack, H., Rizzo, G., Steinmetz, N., Mladenić, D., Auer, S., Lange, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 342–353. [Google Scholar]
- Da Silva Santos, L.B.; Wilkinson, M.D.; Kuzniar, A.; Kaliyaperumal, R.; Thompson, M.; Dumontier, M.; Burger, K. FAIR Data Points supporting big data interoperability. In Enterprise Interoperability in the Digitized and Networked Factory of the Future; ISTE: Guimaraes, Portugal, 2016; pp. 270–279. [Google Scholar]
- Europe PMC Consortium. Europe PMC: A full-text literature database for the life sciences and platform for innovation. Nucleic Acids Res. 2015, 43, D1042–D1048. [Google Scholar] [CrossRef] [PubMed]
- Generic Feature Format version 3 (GFF3). Available online: https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md (accessed on 17 September 2020).
- Kuzniar, A.; Singh, G. Quantitative Trait Loci of Solanaceae species. zenodo 2019. [Google Scholar] [CrossRef]
- Singh, G.; Kuzniar, A. QTLTableMiner++. zenodo 2019. [Google Scholar] [CrossRef]
- Kuzniar, A. Genome annotations of Solanaceae species. zenodo 2019. [Google Scholar] [CrossRef]
- SGN: Solanum lycopersicum (ITAG2.4). Available online: ftp://ftp.solgenomics.net/genomes/Solanumlycopersicum/annotation/ITAG2.4release/ (accessed on 2 September 2019).
- SGN: Solanum pennellii (v2). Available online: ftp://ftp.solgenomics.net/genomes/Solanumpennellii. (accessed on 2 September 2019).
- SGN: Solanum tuberosum (PGSC v4.03). Available online: ftp://ftp.solgenomics.net/genomes/Solanumtuberosum/annotation/PGSC4.03/ (accessed on 2 September 2019).
- Ensembl Plants: Solanum lycopersicum. Available online: http://plants.ensembl.org/Solanum_lycopersicum (accessed on 2 September 2019).
- Ensembl Plants: Solanum tuberosum. Available online: http://plants.ensembl.org/Solanum_tuberosum (accessed on 2 September 2019).
- UniProt: Solanum lycopersicum. Available online: https://www.uniprot.org/proteomes/UP000004994 (accessed on 2 September 2019).
- UniProt: Solanum tuberosum. Available online: https://www.uniprot.org/proteomes/UP000011115 (accessed on 2 September 2019).
- Gene Ontology (GO). Available online: http://purl.obolibrary.org/obo/go.owl (accessed on 2 September 2019).
- Sequence Ontology (SO). Available online: http://purl.obolibrary.org/obo/so.owl (accessed on 2 September 2019).
- Feature Annotation Location Description Ontology (FALDO). Available online: http://biohackathon.org/resource/faldo.rdf (accessed on 2 September 2019).
- UniProt RDF Schema Ontology (UniProt Core). Available online: https://www.uniprot.org/core/ (accessed on 2 September 2019).
- Semanticscience Integrated Ontology (SIO). Available online: http://semanticscience.org/ontology/sio.owl (accessed on 2 September 2019).
- Relation Ontology (RO). Available online: http://purl.obolibrary.org/obo/ro.owl (accessed on 2 September 2019).
- Plant Ontology (PO). Available online: http://purl.obolibrary.org/obo/po.owl (accessed on 2 September 2019).
- Phenotype Quality Ontology (PATO). Available online: http://purl.obolibrary.org/obo/pato.owl (accessed on 2 September 2019).
- Boettiger, C. An introduction to Docker for reproducible research. ACM SIGOPS Oper. Syst. Rev. 2015, 49, 71–79. [Google Scholar] [CrossRef]
- Ansible. Available online: https://www.ansible.com/ (accessed on 1 July 2020).
- Kuzniar, A.; Kaliyaperumal, R. FAIR Data Point. zenodo 2017. [Google Scholar] [CrossRef]
- Jupyter Notebooks for the biological use cases. Available online: https://github.com/candYgene/notebooks/ (accessed on 17 September 2020).
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.E.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.B.; Grout, J.; Corlay, S.; et al. Jupyter Notebooks-a publishing format for reproducible computational workflows. In Proceedings of the ELPUB, Göttingen, Germany, 7–9 June 2016; pp. 87–90. [Google Scholar]
- Haggard, J.E.; Johnson, E.B.; Clair, D.A.S. Multiple QTL for horticultural traits and quantitative resistance to Phytophthora infestans linked on Solanum habrochaites chromosome 11. G3 (Bethesda) 2015, 5, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.; Díaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.; Tikunov, Y.; Molthoff, J.; Grandillo, S.; Viquez-Zamora, M.; de Vos, R.; de Maagd, R.; van Heusden, S.; Bovy, A. Identification of Loci Affecting Accumulation of Secondary Metabolites in Tomato Fruit of a Solanum lycopersicum X Solanum chmielewskii Introgression Line Population. Front. Plant Sci. 2016, 7, 1428. [Google Scholar] [CrossRef] [PubMed]
- Shulaev, V.; Silverman, P.; Raskin, I. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 1997, 385, 718. [Google Scholar] [CrossRef]
- Luengwilai, K.; Fiehn, O.E.; Beckles, D.M. Comparison of leaf and fruit metabolism in two tomato (Solanum lycopersicum L.) genotypes varying in total soluble solids. J. Agric. Food Chem. 2010, 58, 11790–11800. [Google Scholar] [CrossRef]
- Di Mascio, P.; Kaiser, S.; Sies, H. Lycopene as the most efficient biological carotenoid singlet oxygen quencher. Arch. Biochem. Biophys. 1989, 274, 532–538. [Google Scholar] [CrossRef]
- Falara, V.; Akhtar, T.A.; Nguyen, T.T.; Spyropoulou, E.A.; Bleeker, P.M.; Schauvinhold, I.; Matsuba, Y.; Bonini, M.E.; Schilmiller, A.L.; Last, R.L.; et al. The tomato terpene synthase gene family. Plant Physiol. 2011, 157, 770–789. [Google Scholar] [CrossRef]
- Warwick Vesztrocy, A.; Dessimoz, C.; Redestig, H. Prioritising candidate genes causing QTL using hierarchical orthologous groups. Bioinformatics 2018, 34, i612–i619. [Google Scholar] [CrossRef]
- Lin, F.; Fan, J.; Rhee, S.Y. QTG-Finder: A machine-learning based algorithm to prioritize causal genes of quantitative trait loci in Arabidopsis and rice. bioRxiv 2019, 484204. [Google Scholar] [CrossRef]
- Schneider, A.; Dessimoz, C.; Gonnet, G.H. OMA Browser—exploring orthologous relations across 352 complete genomes. Bioinformatics 2007, 23, 2180–2182. [Google Scholar] [CrossRef]
- Fridman, E.; Liu, Y.; Carmel-Goren, L.; Gur, A.; Shoresh, M.; Pleban, T.; Eshed, Y.; Zamir, D. Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol. Genet. Genomics 2002, 266, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; D’Harlingue, A.; Backhaus, R.A.; Kumagai, M.H.; Camara, B. Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur. J. Biochem. 2000, 267, 6346–6352. [Google Scholar] [CrossRef] [PubMed]
- Tadmor, Y.; Fridman, E.; Gur, A.; Larkov, O.; Lastochkin, E.; Ravid, U.; Zamir, D.; Lewinsohn, E. Identification of malodorous, a wild species allele affecting tomato aroma that was selected against during domestication. J. Agric. Food Chem. 2002, 50, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Marti, R.; Rosello, S.; Cebolla-Cornejo, J. Tomato as a source of carotenoids and polyphenols targeted to cancer prevention. Cancers 2016, 8, 58. [Google Scholar] [CrossRef]
- Giuliano, G. Plant carotenoids: Genomics meets multi-gene engineering. Curr. Opin. Plant Biol. 2014, 19, 111–117. [Google Scholar] [CrossRef]
- Shi, J.; Kakuda, Y.; Yeung, D. Antioxidative properties of lycopene and other carotenoids from tomatoes: Synergistic effects. Biofactors 2004, 21, 203–210. [Google Scholar] [CrossRef]
- Cunningham, F.X.; Pogson, B.; Sun, Z.; McDonald, K.A.; DellaPenna, D.; Gantt, E. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 1996, 8, 1613–1626. [Google Scholar] [CrossRef]
- Rousseaux, M.C.; Jones, C.M.; Adams, D.; Chetelat, R.; Bennett, A.; Powell, A. QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor. Appl. Genet. 2005, 111, 1396–1408. [Google Scholar] [CrossRef]
- Tieman, D.M.; Loucas, H.M.; Kim, J.Y.; Clark, D.G.; Klee, H.J. Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry 2007, 68, 2660–2669. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Xu, Y.; Liang, J.; Chang, P.; Yan, F.; Li, M.; Liang, Y.; Zou, Z. Genome-wide association mapping for tomato volatiles positively contributing to tomato flavor. Front. Plant Sci. 2015, 6, 1042. [Google Scholar] [CrossRef]
- Socaci, S.A.; Socaciu, C.; Mureşan, C.; Fărcaş, A.; Tofană, M.; Vicaş, S.; Pintea, A. Chemometric discrimination of different tomato cultivars based on their volatile fingerprint in relation to lycopene and total phenolics content. Phytochem. Anal. 2014, 25, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Rambla, J.L.; Medina, A.; Fernandez-del Carmen, A.; Barrantes, W.; Grandillo, S.; Cammareri, M.; Lopez-Casado, G.; Rodrigo, G.; Alonso, A.; Garcia-Martinez, S.; et al. Identification, introgression, and validation of fruit volatile QTLs from a red-fruited wild tomato species. J. Exp. Bot. 2016, 68, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Hassani-Pak, K.; Rawlings, C. Knowledge discovery in biological databases for revealing candidate genes linked to complex phenotypes. J. Integr. Bioinform. 2017, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- KNETMiner. Available online: https://knetminer.com/resources (accessed on 31 August 2020).
- Hulse-Kemp, A.M.; Maheshwari, S.; Stoffel, K.; Hill, T.A.; Jaffe, D.; Williams, S.R.; Weisenfeld, N.; Ramakrishnan, S.; Kumar, V.; Shah, P.; et al. Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 2018, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Shirasawa, K.; Miyatake, K.; Nunome, T.; Negoro, S.; Ohyama, A.; Yamaguchi, H.; Sato, S.; Isobe, S.; Tabata, S.; et al. Draft genome sequence of eggplant (Solanum melongena L.): The representative solanum species indigenous to the old world. DNA Res. 2014, 21, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Singh, G. Genomics Data Integration for Knowledge Discovery Using Genome Annotations from Molecular Databases and Scientific Literature. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlads, 2019. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).