Analysis of Random Dynamics of Cell Segmented by a Modified Active Contour Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Measurement of Particle/Cell Random Motion
2.3. Particle/Cell Tracking and Random Dynamics Analysis
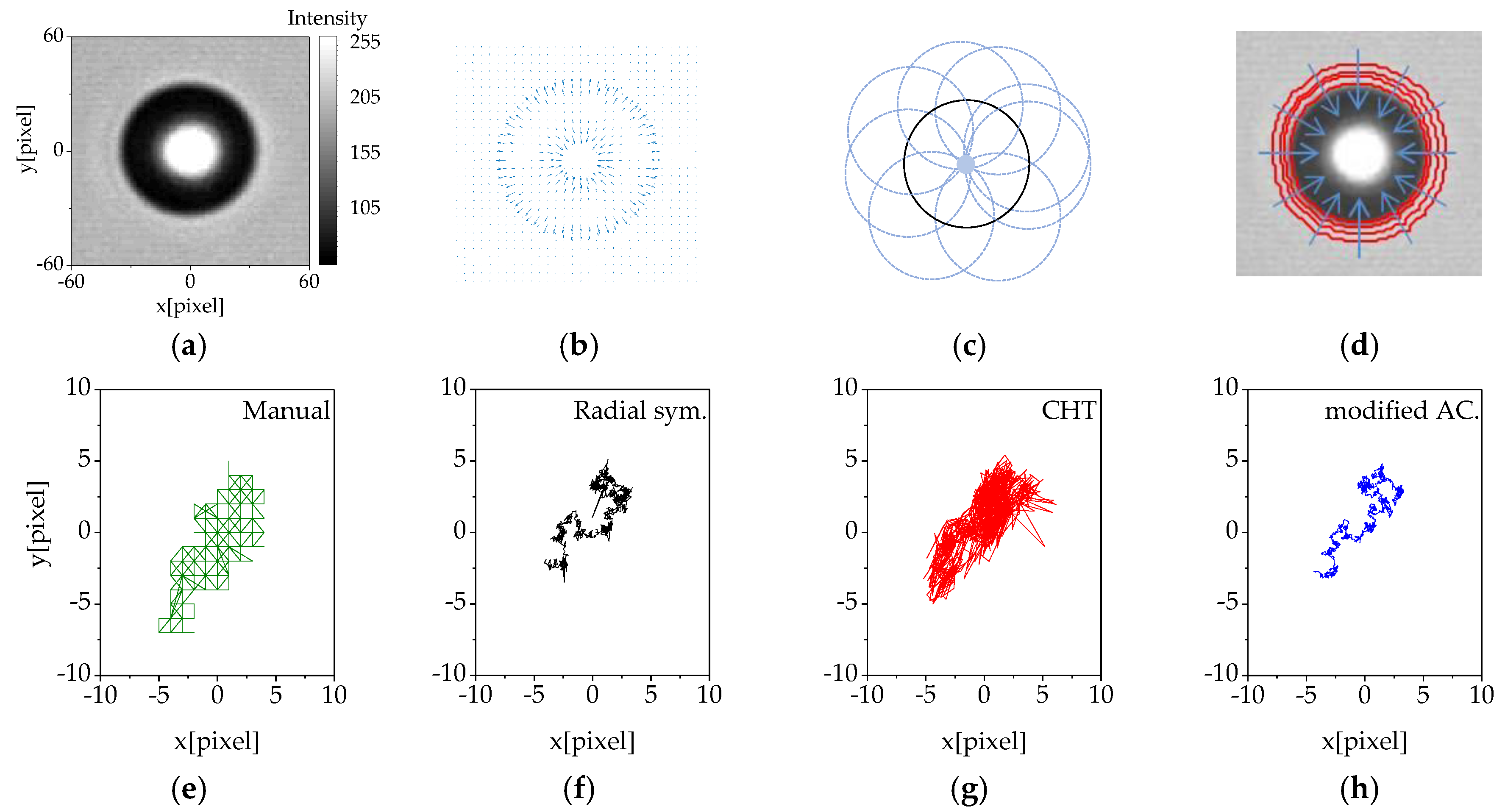
2.4. Modified Active Contour
3. Results & Discussion
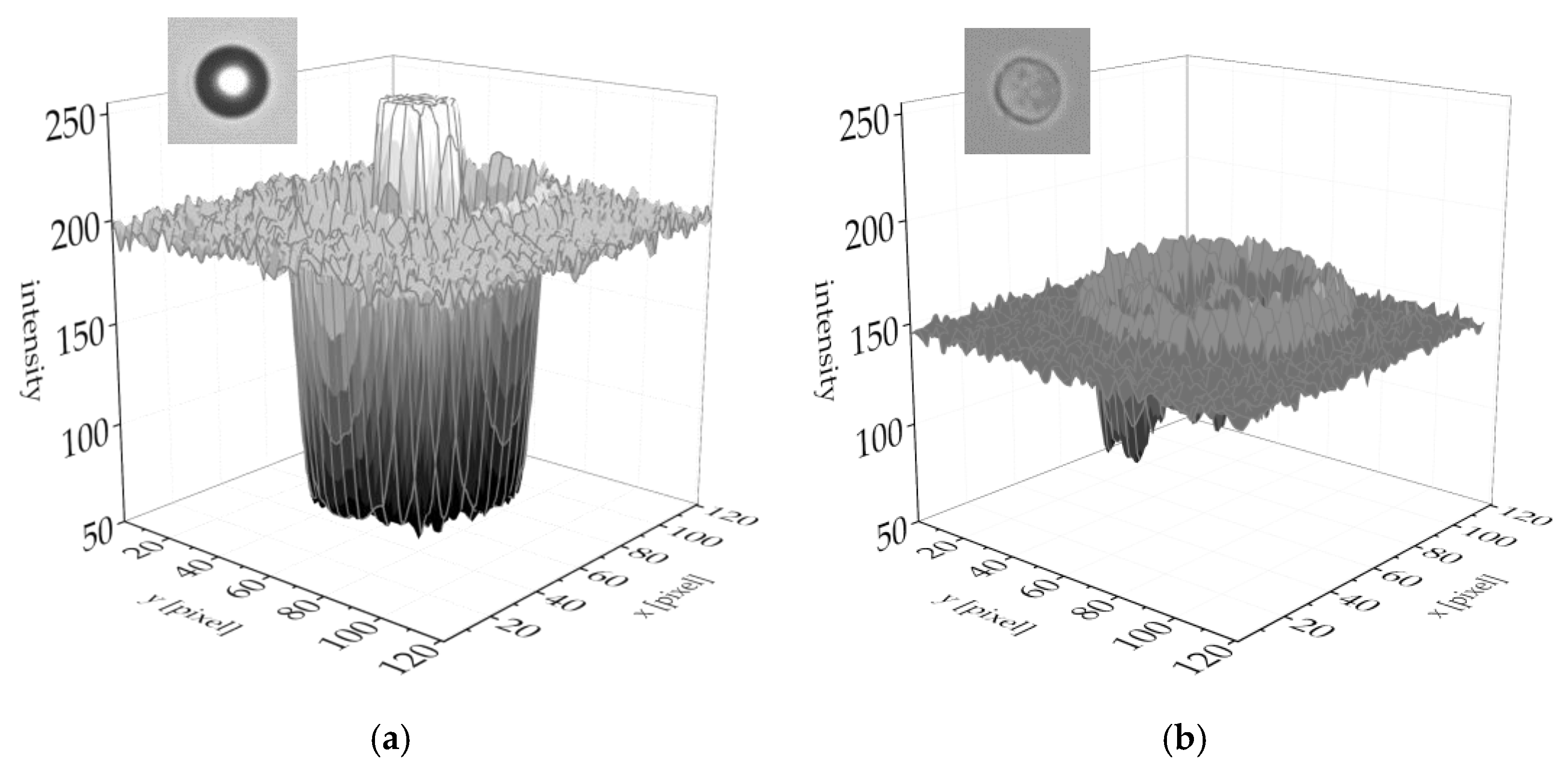
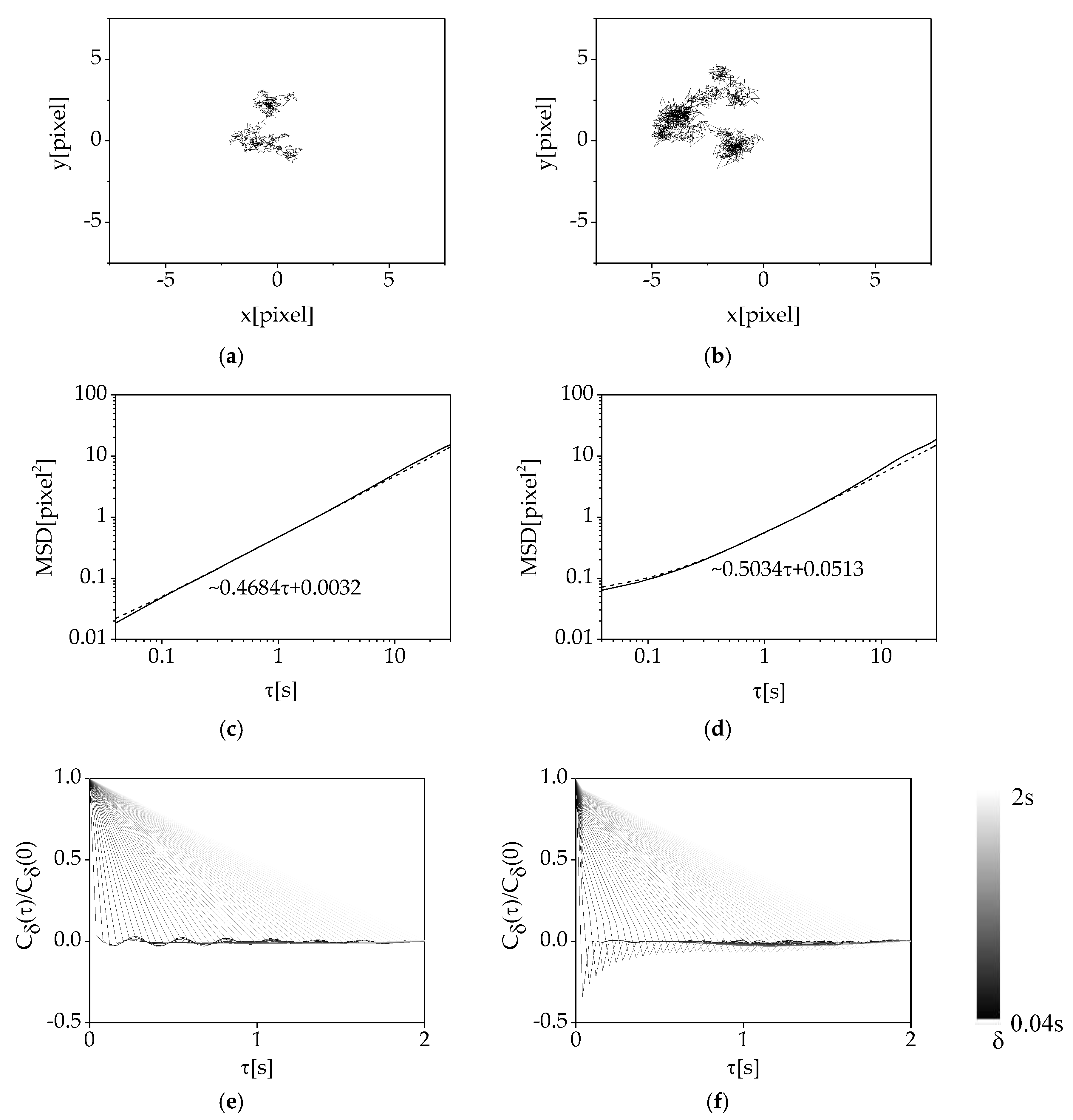
3.1. Difference of Trajectory between cPMS Particles and MCF-7 Cells
3.2. Comparison of Image Analysis Methods
3.3. Analysis of Cellular Random Motion by the Image Analysis Methods
3.4. Effects of Denoising
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gupton, S.L.; Anderson, K.L.; Kole, T.P.; Fischer, R.S.; Ponti, A.; Hitchcock-DeGregori, S.E.; Danuser, G.; Fowler, V.M.; Wirtz, D.; Hanein, D.; et al. Cell migration without a lamellipodium: Translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 2005, 168, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-H.; Giri, A.; Sun, S.X.; Wirtz, D. Three-dimensional cell migration does not follow a random walk. Proc. Natl. Acad. Sci. USA 2014, 111, 3949–3954. [Google Scholar] [CrossRef] [PubMed]
- Hyman, A.A.; White, J.G. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J. Cell Biol. 1987, 105, 2123–2135. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.I.; Bao, Z.; Boyle, T.J.; Waterston, R.H. The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat. Protoc. 2006, 1, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Koslover, E.F.; Chan, C.K.; Theriot, J.A. Cytoplasmic flow and mixing due to deformation of motile cells. Biophys. J. 2017, 113, 2077–2087. [Google Scholar] [CrossRef] [PubMed]
- Celebrano, M.; Kukura, P.; Renn, A.; Sandoghdar, V. Single-molecule imaging by optical absorption. Nat. Photonics 2011, 5, 95–98. [Google Scholar] [CrossRef]
- Kukura, P.; Celebrano, M.; Renn, A.; Sandoghdar, V. Single-molecule sensitivity in optical absorption at room temperature. J. Phys. Chem. Lett. 2010, 1, 3323–3327. [Google Scholar] [CrossRef]
- Wieser, S.; Schütz, G.J. Tracking single molecules in the live cell plasma membrane—Do’s and Don’t’s. Methods 2008, 46, 131–140. [Google Scholar] [CrossRef]
- Cheezum, M.K.; Walker, W.F.; Guilford, W.H. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys. J. 2001, 81, 2378–2388. [Google Scholar] [CrossRef]
- Rogers, S.S.; Waigh, T.A.; Zhao, X.; Lu, J.R. Precise particle tracking against a complicated background: Polynomial fitting with Gaussian weight. Phys. Biol. 2007, 4, 220–227. [Google Scholar] [CrossRef]
- Thompson, R.E.; Larson, D.R.; Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002, 82, 2775–2783. [Google Scholar] [CrossRef]
- Manzo, C.; Garcia-Parajo, M.F. A review of progress in single particle tracking: From methods to biophysical insights. Rep. Prog. Phys. 2015, 78, 124601. [Google Scholar] [CrossRef] [PubMed]
- Crocker, J.C.; Grier, D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996, 179, 298–310. [Google Scholar] [CrossRef]
- Liu, Z.; Lavis, L.D.; Betzig, E. Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell 2015, 58, 644–659. [Google Scholar] [CrossRef] [PubMed]
- Press, W.H.; Flannery, B.P.; Teukolsky, S.A.; Vetterling, W.T. Numerical Recipes; Cambridge University Press: Cambridge, UK, 1989. [Google Scholar]
- Shen, H.; Tauzin, L.J.; Baiyasi, R.; Wang, W.; Moringo, N.; Shuang, B.; Landes, C.F. Single particle tracking: From theory to biophysical applications. Chem. Rev. 2017, 117, 7331–7376. [Google Scholar] [CrossRef]
- Berglund, A.J.; McMahon, M.D.; McClelland, J.J.; Liddle, J.A. Fast, bias-free algorithm for tracking single particles with variable size and shape. Opt. Express 2008, 16, 14064–14075. [Google Scholar] [CrossRef]
- Parthasarathy, R. Rapid, accurate particle tracking by calculation of radial symmetry centers. Nat. Methods 2012, 9, 724–726. [Google Scholar] [CrossRef]
- Smith, C.S.; Joseph, N.; Rieger, B.; Lidke, K.A. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods 2010, 7, 373–375. [Google Scholar] [CrossRef]
- Mashanov, G.; Molloy, J. Automatic detection of single fluorophores in live cells. Biophys. J. 2007, 92, 2199–2211. [Google Scholar] [CrossRef]
- Lee, S.U.; Chung, S.Y.; Park, R.H. A comparative performance study of several global thresholding techniques for segmentation. Comput. Vis. Graph. Image Process. 1990, 52, 171–190. [Google Scholar] [CrossRef]
- Sezgin, M.; Sankur, B. Survey over image thresholding techniques and quantitative performance evaluation. J. Electron. Imaging 2004, 13, 146–166. [Google Scholar]
- Demou, Z.N.; McIntire, L.V. Fully automated three-dimensional tracking of cancer cells in collagen gels: Determination of motility phenotypes at the cellular level. Cancer Res. 2002, 62, 5301–5307. [Google Scholar] [PubMed]
- Thurston, G.; Jaggi, B.; Palcic, B. Measurement of cell motility and morphology with an automated microscope system. Cytometry 1988, 9, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Kachouie, N.N.; Fieguth, P.; Ramunas, J.; Jervis, E. Probabilistic model-based cell tracking. Int. J. Biomed. Imaging 2006, 2006, 012186. [Google Scholar] [CrossRef]
- Young, D.; Glasbey, C.; Gray, A.; Martin, N. Towards automatic cell identification in DIC microscopy. J. Microsc. 1998, 192, 186–193. [Google Scholar] [CrossRef]
- Nandy, K.; Gudla, P.R.; Amundsen, R.; Meaburn, K.J.; Misteli, T.; Lockett, S.J. Automatic segmentation and supervised learning-based selection of nuclei in cancer tissue images. Cytometry Part A 2012, 81, 743–754. [Google Scholar] [CrossRef]
- Wählby, C.; Sintorn, I.M.; Erlandsson, F.; Borgefors, G.; Bengtsson, E. Combining intensity, edge and shape information for 2D and 3D segmentation of cell nuclei in tissue sections. J. Microsc. 2004, 215, 67–76. [Google Scholar] [CrossRef]
- Delgado-Gonzalo, R.; Uhlmann, V.; Schmitter, D.; Unser, M. Snakes on a Plane: A perfect snap for bioimage analysis. IEEE Signal Process. Mag. 2015, 32, 41–48. [Google Scholar] [CrossRef]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Dzyubachyk, O.; Van Cappellen, W.A.; Essers, J.; Niessen, W.J.; Meijering, E. Advanced level-set-based cell tracking in time-lapse fluorescence microscopy. IEEE Trans. Med. Imaging 2010, 29, 852–867. [Google Scholar] [CrossRef]
- Sethian, J.A. Level Set Methods and Fast Marching Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science; Cambridge University Press: Cambridge, UK, 1999; Volume 3. [Google Scholar]
- Yang, F.; Mackey, M.A.; Ianzini, F.; Gallardo, G.; Sonka, M. Cell segmentation, tracking, and mitosis detection using temporal context. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Palm Springs, CA, USA, 26–29 October 2005; pp. 302–309. [Google Scholar]
- Reif, F. Fundamentals of Statistical and Thermal Physics; Waveland Press: Long Grove, IL, USA, 2009. [Google Scholar]
- Saxton, M.J.; Jacobson, K. Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 373–399. [Google Scholar] [CrossRef]
- Hyun, J.; Kim, S.; Kim, D.; Choi, S.; Key, J.; Kim, Y.; Lee, S.; Lee, S. Comparison of the abnormal diffusion characteristics of tumor cells. Microfluid. Nanofluid. 2019, 23, 119. [Google Scholar] [CrossRef]
- Yuen, H.; Princen, J.; Illingworth, J.; Kittler, J. Comparative study of Hough transform methods for circle finding. Image Vis. Comput. 1990, 8, 71–77. [Google Scholar] [CrossRef]
- Xu, J.; Chutatape, O.; Chew, P. Automated optic disk boundary detection by modified active contour model. IEEE Trans. Biomed. Eng. 2007, 54, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Kanopoulos, N.; Vasanthavada, N.; Baker, R.L. Design of an image edge detection filter using the Sobel operator. IEEE J. Solid State Circuits 1988, 23, 358–367. [Google Scholar] [CrossRef]
- Weber, S.C.; Thompson, M.A.; Moerner, W.; Spakowitz, A.J.; Theriot, J.A. Analytical tools to distinguish the effects of localization error, confinement, and medium elasticity on the velocity autocorrelation function. Biophys. J. 2012, 102, 2443–2450. [Google Scholar] [CrossRef]
- Savin, T.; Doyle, P.S. Static and dynamic errors in particle tracking microrheology. Biophys. J. 2005, 88, 623–638. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]



| Method | Radial sym. | CHT | Modified AC | |||
|---|---|---|---|---|---|---|
| Raw | Denoised | Raw | Denoised | Raw | Denoised | |
| 4D | 0.503 | 0.555 | 0.554 | 0.568 | 0.545 | 0.550 |
| σ2 | 0.051 | 0.017 | 0.558 | 0.507 | 0.013 | 0.011 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hyun, J.Y.; Ha, S.; Baek, J.; Han, J.; An, H.; Woo, S.-H.; Kim, Y.S.; Lee, S.W.; Yang, S.; Lee, S.Y. Analysis of Random Dynamics of Cell Segmented by a Modified Active Contour Method. Appl. Sci. 2020, 10, 6806. https://doi.org/10.3390/app10196806
Hyun JY, Ha S, Baek J, Han J, An H, Woo S-H, Kim YS, Lee SW, Yang S, Lee SY. Analysis of Random Dynamics of Cell Segmented by a Modified Active Contour Method. Applied Sciences. 2020; 10(19):6806. https://doi.org/10.3390/app10196806
Chicago/Turabian StyleHyun, Ji Yeon, Seungeon Ha, Jongmin Baek, Junghun Han, Honggi An, Sung-Hun Woo, Yoon Suk Kim, Sang Woo Lee, Sejung Yang, and Sei Young Lee. 2020. "Analysis of Random Dynamics of Cell Segmented by a Modified Active Contour Method" Applied Sciences 10, no. 19: 6806. https://doi.org/10.3390/app10196806
APA StyleHyun, J. Y., Ha, S., Baek, J., Han, J., An, H., Woo, S.-H., Kim, Y. S., Lee, S. W., Yang, S., & Lee, S. Y. (2020). Analysis of Random Dynamics of Cell Segmented by a Modified Active Contour Method. Applied Sciences, 10(19), 6806. https://doi.org/10.3390/app10196806







