The Link between Oral and Gut Microbiota in Inflammatory Bowel Disease and a Synopsis of Potential Salivary Biomarkers
Abstract
1. Introduction
2. Inflammatory Bowel Disease
3. Inflammatory Bowel Disease Pathogenesis
3.1. IBD Immune Dysregulation
3.2. IBD Genetic Predisposition
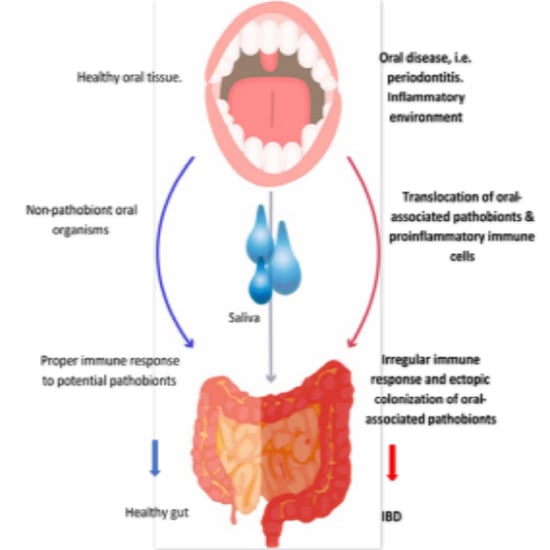
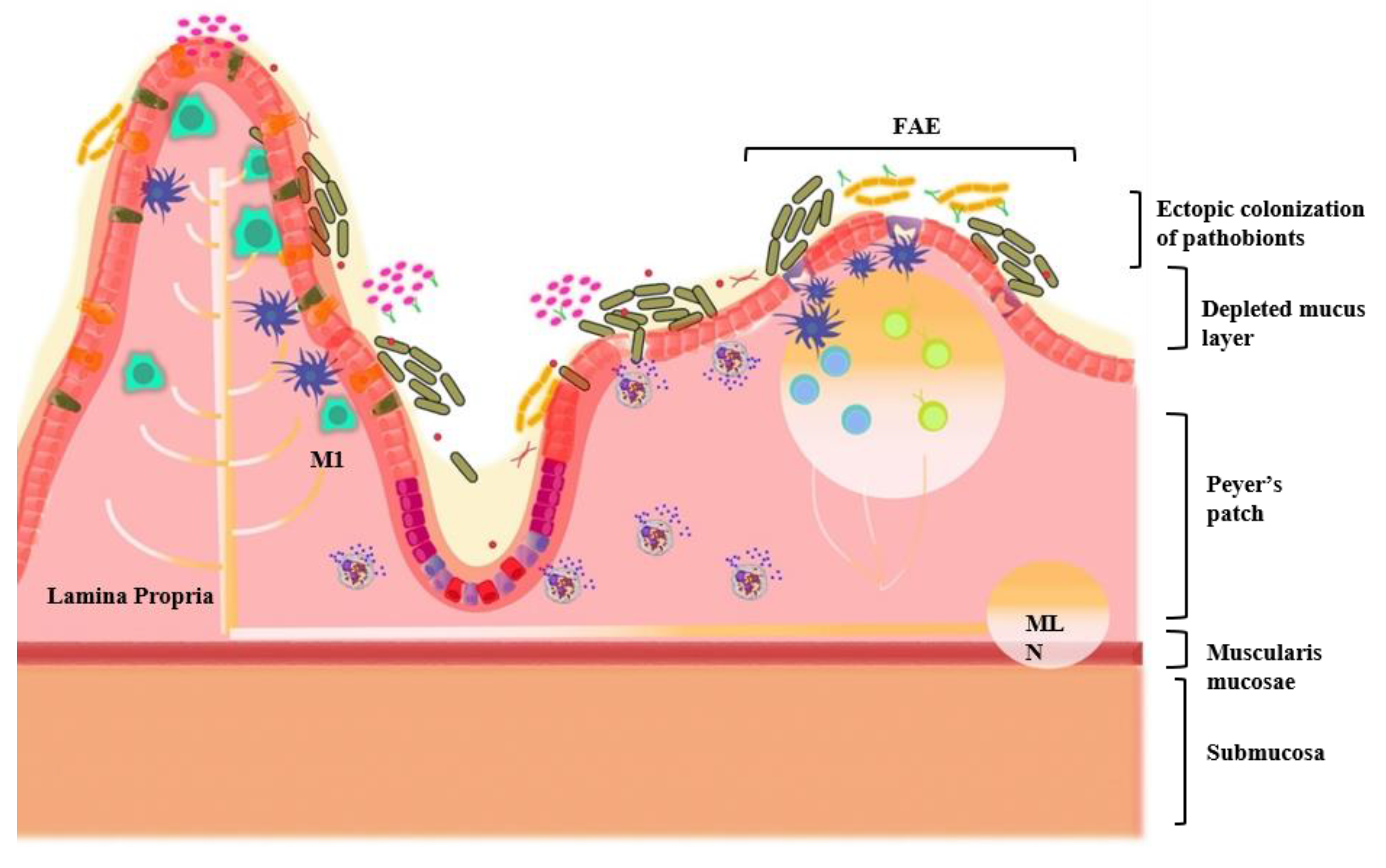
3.3. Oral–Gut Relationship in IBD Pathogenesis
4. The Gastrointestinal System
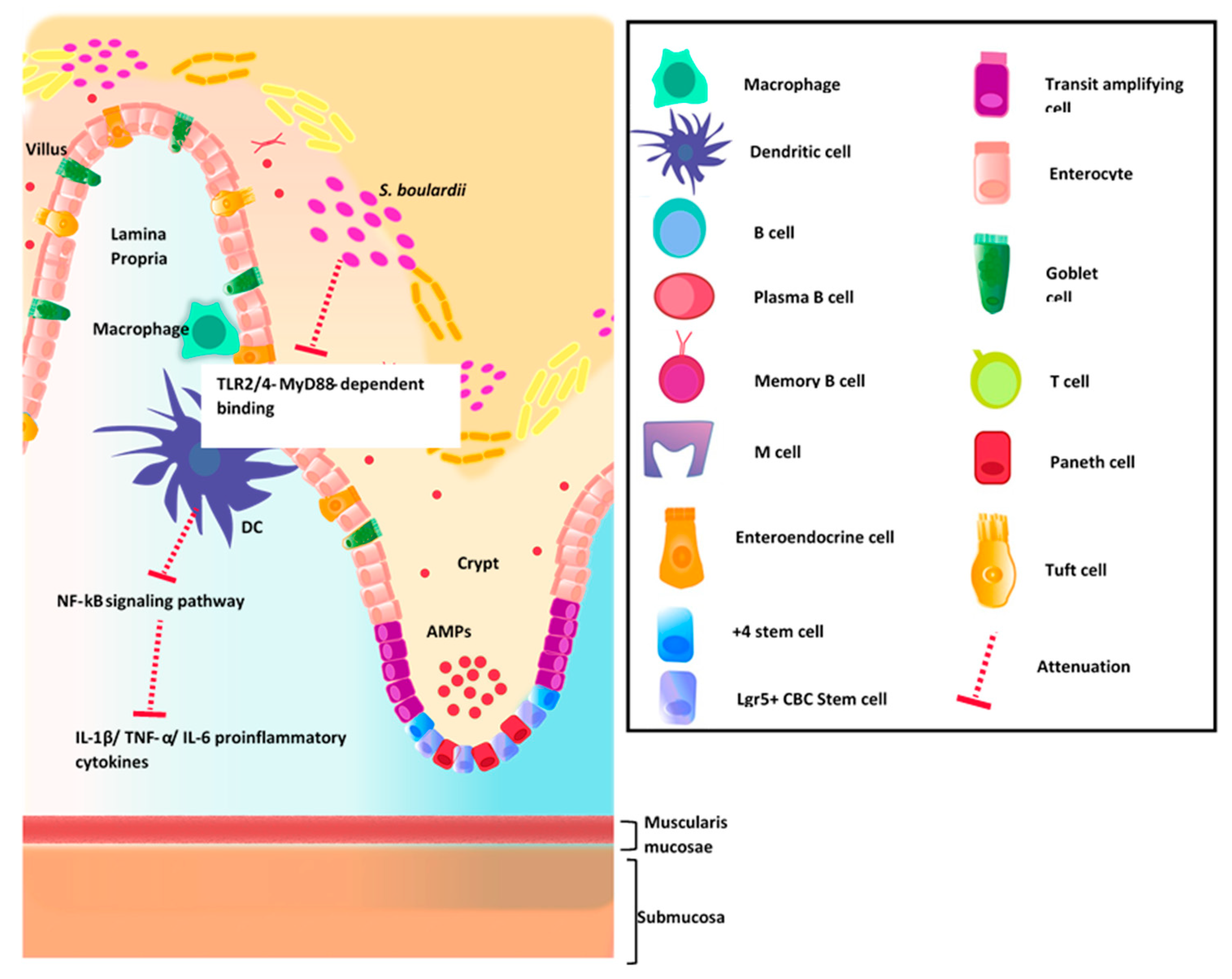
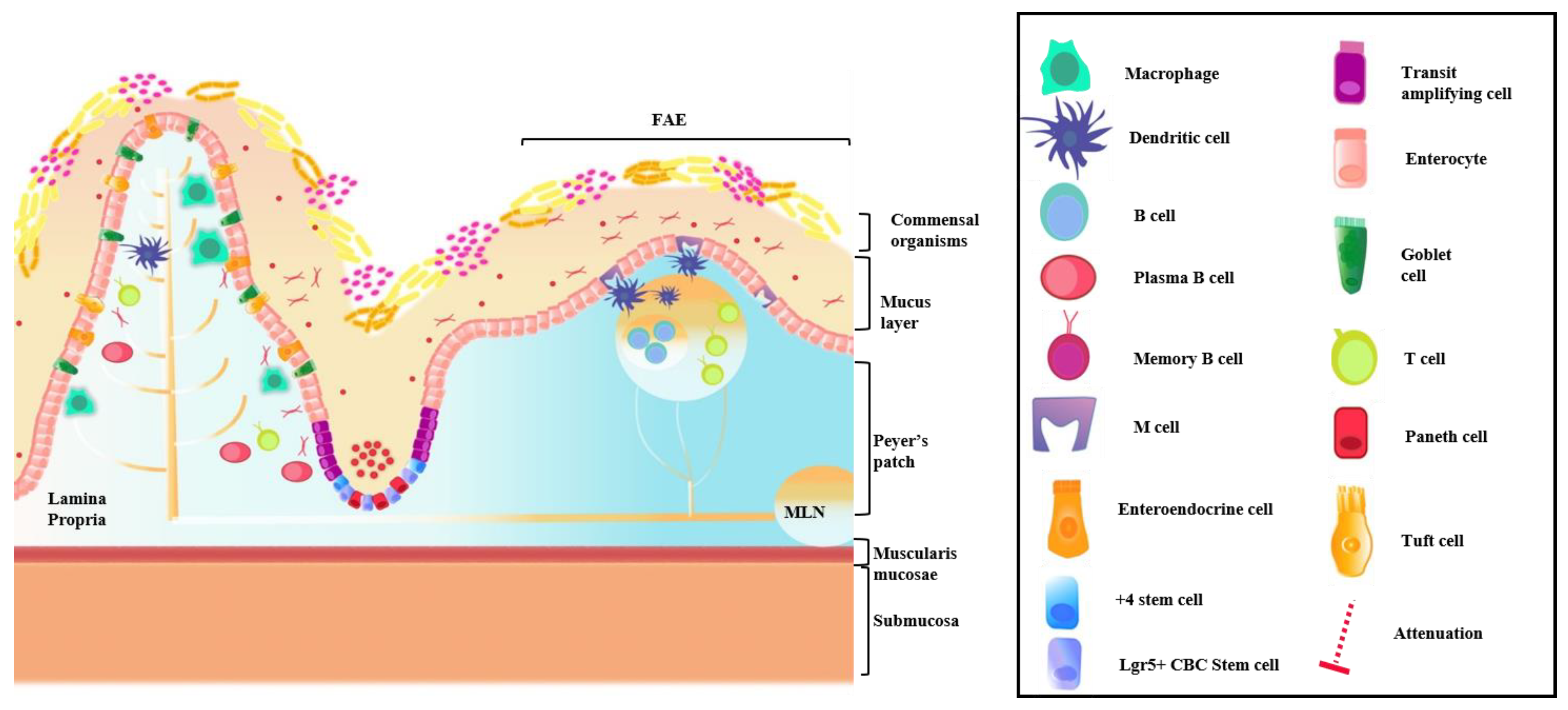
4.1. Intestinal Immune System
4.2. GI Host Defense
5. The Gut Microbiota in Health and IBD
Commensal Organisms and Epithelial Barrier Function
6. The Oral Microbiota in Health and IBD
7. The Oral Microbiota and Gut Microbiota Axis
7.1. Beneficial Gut Commensal Organisms
7.2. Beneficial Oral Commensal Organisms
8. Salivary Biomarkers for Monitoring Health Status in the Gut
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IBD | inflammatory bowel disease |
| UC | ulcerative colitis |
| CD | Crohn’s disease |
| IC | indeterminate colitis |
| GI | gastrointestinal |
| GWAS | genome-wide association study |
| NOD2 | nucleotide-binding oligomerization domain containing 2 |
| MDP | muramyl dipeptide |
| NF-kβ | nuclear factor-kappa beta |
| TLR | toll-like receptor |
| LPS | lipopolysaccharide |
| IL-17 | interleukin-17 |
| IL-23 | Interleukin 23 |
| IL-23R | IL-23 receptor |
| Th17 | T-helper 17 cells |
| FMT | fecal microbiota transplantation |
| FOS | fructo-oligosaccharides |
| R. bromii | Ruminococcus bromii |
| ROS | reactive oxygen species |
| S. Typhimurium | Salmonella enterica serotype Typhimurium |
| SHIME | simulator of the human intestinal microbial ecosystem |
| B. infantis | Bifidobacterium longum subspecies infantis |
| GCF | gingival crevicular fluid |
| TAP | Transit amplifying progenitor cells |
| ISCs | intestinal stem cells |
| AMPs | antimicrobial peptides |
| TGF-β1 | transforming growth factor beta-1 |
| Tregs | T-regulatory cells |
| SPF | specific pathogen free |
| pIgA | pentameric IgA |
| pIgR | polymeric immunoglobulin receptor |
| sIgA | secretory IgA |
| SCFAs | short-chain fatty acids |
| PRRs | Pattern recognition receptors |
| NODRs | NOD-like receptors |
| RLRs | RIG-I-like receptors |
| AhR | aryl hydrocarbon receptor |
| C. diff | Clostridium difficile |
| OTUs | operational taxonomic units |
| HOMD | human oral microbiome database |
| EPH | Ecological Plaque Hypothesis |
| KPH | Keystone-Pathogen Hypothesis |
| P. gingivalis | Porphyromonas gingivalis |
| AD | Alzheimer’s disease |
| HC | healthy controls |
| DSS | dextran sodium sulfate |
| LEfSe | linear discriminant analysis effect size |
| IFN-γ | interferon gamma |
| B6 | C57BL/6 |
| F. prausnitzii | Faecalibacterium prausnitzii |
| L. casei | Lactobacillus casei |
| L. rhamnosus | Lactobacillus rhamnosus |
| LcS | L. casei subspecies Shirota |
| CFUs | colony forming units |
| MUC2 | Mucin 2 |
| ZG16 | zymogen granule protein 16 |
| RELMβ | resistin-like molecule beta |
| ENS | enteric nervous system |
| IL-18 | interleukin-18 |
| ILFs | isolated lymphoid follicles |
| MLNs | mesenteric lymph nodes |
| TJs | tight junctions |
| GALT | gut-associated lymphoid tissue |
| 5FU | 5-fluorouracil |
| MAPK | mitogen-activated protein kinase |
| S. boulardii | Saccharomyces boulardii |
| FAE | follicle associated epithelium |
| IECs | intestinal epithelial cells |
| M | Microfold |
| GP2 | Glycoprotein 2 |
| FimH | fimbrin D-mannose specific adhesion |
| DCs | dendritic cells |
References
- Yu, Y.R.; Rodriguez, R.J. Clinical Presentation of Crohn’s, Ulcerative Colitis, and Indeterminate Colitis: Symptoms, Extraintestinal Manifestations, and Disease Phenotypes. Semin. Pediatric Surg. 2017, 26, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Guindi, M.; Riddell, R.H. Indeterminate colitis. J. Clin. Pathol. 2004, 57, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Fakhoury, M.; Negrulj, R.; Mooranian, A.; Al-Salami, H. Inflammatory bowel disease: Clinical aspects and treatments. J. Inflamm. Res. 2014, 7, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Eom, T.; Kim, Y.S.; Choi, C.H.; Sadowsky, M.J.; Unno, T. Current understanding of microbiota- and dietary-therapies for treating inflammatory bowel disease. J. Microbiol. 2018, 56, 189–198. [Google Scholar] [CrossRef]
- Cohen, L.J.; Cho, J.H.; Gevers, D.; Chu, H. Genetic Factors and the Intestinal Microbiome Guide Development of Microbe-Based Therapies for Inflammatory Bowel Diseases. Gastroenterology 2019, 156, 2174–2189. [Google Scholar] [CrossRef] [PubMed]
- Ellinghaus, D.; The International IBD Genetics Consortium (IIBDGC); Jostins, L.; Spain, S.L.; Cortes, A.; Bethune, J.; Han, B.; Park, Y.R.; Raychaudhuri, S.; Pouget, J.G.; et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 2016, 48, 510–518. [Google Scholar] [CrossRef]
- Annese, V. Genetics and epigenetics of IBD. Pharmacol. Res. 2020, 159, 104892. [Google Scholar] [CrossRef]
- Piovani, D.; Danese, S.; Peyrin-Biroulet, L.; Bonovas, S. Environmental, Nutritional, and Socioeconomic Determinants of IBD Incidence: A Global Ecological Study. J. Crohn’s Colitis 2020, 14, 323–331. [Google Scholar] [CrossRef]
- Sairenji, T.; Collins, K.L.; Evans, D.V. An Update on Inflammatory Bowel Disease. Prim Care Clin. Off. Pract. 2017, 44, 673–692. [Google Scholar] [CrossRef]
- Cury, D.; Oliveira, R.; Cury, M. Inflammatory bowel diseases: Time of diagnosis, environmental factors, clinical course, and management – a follow-up study in a private inflammatory bowel disease center (2003–2017). J. Inflamm. Res. 2019, 12, 127–135. [Google Scholar] [CrossRef]
- Cosnes, J.; Gower-Rousseau, C.; Seksik, P.; Cortot, A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011, 140, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2018, 390, 2769–2778. [Google Scholar] [CrossRef]
- Takahashi, L.A.R.; Cardial, D.T.; Argani, I.L.; Arnoni, L.R.R.; Cury, V.N.; Silva, L.F.A.C.C.; Júnior, A.O.; Borato, S.D.F. Human Development Index and Inflammatory Bowel Diseases. J. Adv. Med. Med Res. 2018, 26, 1–8. [Google Scholar] [CrossRef]
- Statovci, D.; Aguilera, M.; MacSharry, J.; Melgar, S. The Impact of Western Diet and Nutrients on the Microbiota and Immune Response at Mucosal Interfaces. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Rigottier-Gois, L. Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 2013, 7, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Pithadia, A.B.; Jain, S. Treatment of Inflammatory Bowel Disease (IBD). Pharmacol. Rep. 2011, 63, 294–295. [Google Scholar] [CrossRef]
- Celiberto, L.S.; Vallance, B.A.; Cavallini, D.C. Lactic Acid Bacteria: A Functional Approach; Taylor & Francis Group LLC: Boca Raton, FL, USA, 2020. [Google Scholar]
- Dang, X.; Xu, M.; Liu, D. Assessing the efficacy and safety of fecal microbiota transplantation and probiotic VSL#3 for active ulcerative colitis: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0228846. [Google Scholar]
- Imdad, A.; Nicholson, M.R.; Tanner-Smith, E.E.; Zackular, J.P.; Gómez-Duarte, O.G.; Beaulieu, D.B.; Acra, S. Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database Syst. Rev. 2018, 11, CD012774. [Google Scholar] [CrossRef]
- Bogach, J.; Pond, G.; Eskicioglu, C.; Seow, H. Age-Related Survival Differences in Patients With Inflammatory Bowel Disease-Associated Colorectal Cancer: A Population-Based Cohort Study. Inflamm. Bowel Dis. 2019, 25, 1957–1965. [Google Scholar] [CrossRef]
- Biancone, L.; Armuzzi, A.; Scribano, M.L.; Castiglione, F.; D’Incà, R.; Orlando, A.; Papi, C.; Daperno, M.; Vecchi, M.; Riegler, G.; et al. Cancer Risk in Inflammatory Bowel Disease: A 6-Year Prospective Multicenter Nested Case–Control IG-IBD Study. Inflamm. Bowel Dis. 2020, 26, 450–459. [Google Scholar] [CrossRef]
- Leone, V.; Chang, E.B.; Devkota, S. Diet, Microbes, and Host Genetics: The Perfect Storm in Inflammatory Bowel Diseases. J. Gastroenterol. 2013, 48, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X.; Luo, Z.; Ma, L.; Zhu, S.; Wang, Z.; Wen, J.; Cheng, S.; Gu, W.; Lian, Q.; et al. ECM1 is an essential factor for the determination of M1 macrophage polarization in IBD in response to LPS stimulation. Proc. Natl. Acad. Sci. USA 2020, 117, 3083–3092. [Google Scholar] [CrossRef] [PubMed]
- Na, Y.R.; Stakenborg, M.; Seok, S.H.; Matteoli, G. Macrophages in intestinal inflammation and resolution: A potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.Y.; Wang, N.; Li, S.; Hong, M.; Wang, X.; Feng, Y. The Reactive Oxygen Species in Macrophage Polarization: Reflecting Its Dual Role in Progression and Treatment of Human Diseases. Oxidative Med. Cell. Longev. 2016, 2016, 2795090. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Eri, R.; Simms, L.A.; Florin, T.H.; Radford-Smith, G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm. Bowel Dis. 2009, 15, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Rogge, L. The IL-23/IL-17 pathway in human chronic inflammatory diseases—New insight from genetics and targeted therapies. Genes Immun. 2019, 20, 415–425. [Google Scholar] [CrossRef]
- Sandborn, W.J.; Ferrante, M.; Bhandari, B.R.; Berliba, E.; Feagan, B.G.; Hibi, T.; Tuttle, J.L.; Klekotka, P.; Friedrich, S.; Durante, M.; et al. Efficacy and Safety of Mirikizumab in a Randomized Phase 2 Study of Patients With Ulcerative Colitis. Gastroenterology 2020, 158, 537–549.e10. [Google Scholar] [CrossRef]
- Kitamoto, S.; Nagao-Kitamoto, H.; Jiao, Y.; Gillilland, M.G. The Intermucosal Connection between the Mouth and Gut in Commensal Pathobiont-Driven Colitis. Cell 2020, 182, 447–462.e14. [Google Scholar] [CrossRef]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A Frameshift Mutation in NOD2 Associated With Susceptibility to Crohn’s Disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef]
- Lauro, M.L.; D’Ambrosio, E.A.; Bahnson, B.J.; Grimes, C.L. The Molecular Recognition of Muramyl Dipeptide Occurs in the Leucine-rich Repeat Domain of Nod2. ACS Infect. Dis. 2017, 3, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef]
- Koch, B.E.V.; Yang, S.; Lamers, G.; Stougaard, J.; Spaink, H.P. Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88. Nature Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yeh, W.; Ohashi, P. LPS/TLR4 Signal Transduction Pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Nighot, M.; Al-Sadi, R.; Alhmoud, T.; Nighot, P.; Ma, T.Y. Lipopolysaccharide Regulation of Intestinal Tight Junction Permeability Is Mediated by TLR4 Signal Transduction Pathway Activation of FAK and MyD88. J. Immunol. 2015, 195, 4999–5010. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.Y.; Ko, H.J.; Vallance, B.A. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef]
- Granger, D.A.; Taylor, M.K. Salivary Bioscience—Foundations of Interdisciplinary Saliva Research and Applications; Granger, D.A., Ed.; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Umar, S. Intestinal Stem Cells. Curr. Gastroenterol. Rep. 2010, 12, 340–348. [Google Scholar] [CrossRef]
- Krasteva-Christ, G. Reference Module in Neuroscience and Biobehavioral Psychology. In Chapter: Extraoral Taste Receptors; Elsevier Inc.: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Troll, J.V.; Hamilton, M.K.; Abel, M.L.; Ganz, J.; Bates, J.M.; Stephens, W.Z.; Melancon, E.; Van Der Vaart, M.; Meijer, A.H.; Distel, M.; et al. Microbiota promote secretory cell determination in the intestinal epithelium by modulating host Notch signaling. Development 2018, 145, dev155317. [Google Scholar] [CrossRef]
- Banerjee, A.; McKinley, E.T.; von Moltke, J.; Coffey, R.J.; Lau, K.S. Interpreting heterogeneity in intestinal tuft cell structure and function. J. Clin. Investig. 2018, 128, 1711–1719. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.C.E.; Shanahan, M.T.; Singh, A.P.; Sethupathy, P. Gut Microbial Influences on the Mammalian Intestinal Stem Cell Niche. Stem Cells Int. 2017, 2017, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The Intestinal Crypt, a Prototype Stem Cell Compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Lueschow, S.R.; McElroy, S.J. The Paneth Cell: The Curator and Defender of the Immature Small Intestine. Front. Immunol. 2020, 11, 587. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Núñez, G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef]
- Kastl, A.J., Jr.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 33–45. [Google Scholar] [CrossRef]
- Azzouz, L.L.; Sharma, S. Physiology, Large Intestine; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Hiroto, K. Roles of the Gel-Forming MUC2 Mucin and Its O-glycosylation in the Protection Against Colitis and Colorectal Cancer. Biol. Pharm. Bull. 2012, 35, 1637–1641. [Google Scholar]
- Knoop, K.A.; Newberry, R.D. Goblet cells: Multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. 2018, 11, 1551–1557. [Google Scholar] [CrossRef]
- Birchenough, G.M.H.; Nyström, E.E.L.; Johansson, M.E.V.; Hansson, G.C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016, 352, 1535–1542. [Google Scholar] [CrossRef]
- Watanabe, K.; Itoh, K.; Park, S.-H.; Kaku, M.; Ishii, K.; Sasano, H.; Naitoh, T.; Unno, M.; Fukushima, K. Resistin-like Molecule Beta, a Colonic Epithelial Protein, Exhibits Antimicrobial Activity Against Staphylococcus Aureus Including Methicillin-Resistant Strains. Surg. Today 2020. [Google Scholar] [CrossRef]
- Jarret, A.; Jackson, R.; Duizer, C.; Healy, M.E.; Zhao, J.; Rone, J.M.; Bielecki, P.; Sefik, E.; Roulis, M.; Rice, T.; et al. Enteric Nervous System-Derived IL-18 Orchestrates Mucosal Barrier Immunity. Cell 2020, 180, 50–63.e12. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K. Unique Action of Interleukin-18 on T Cells and Other Immune Cells. Front. Immunol. 2018. [Google Scholar] [CrossRef]
- Strowig, T.; Henao-Mejia, J.; Elinav, E.; Flavell, R. Inflammasomes in health and disease. Nature 2012, 481, 278–286. [Google Scholar] [CrossRef]
- Pu, Z.; Che, Y.; Zhang, W.; Sun, H.; Meng, T.; Xie, H.; Cao, L.-J.; Hao, H. Dual roles of IL-18 in colitis through regulation of the function and quantity of goblet cells. Int. J. Mol. Med. 2019, 43, 2291–2302. [Google Scholar] [CrossRef]
- Neu, J. Gastroenterology and Nutrition: Neonatology Questions and Controversies; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Johansen, F.-E.; Yen, E.H.; Dickinson, B. Physiology of the Gastrointestinal Tract (Fourth Edition). In Chapter 43—Biology of Gut Immunoglobulins; Academic Press: Cambridge, MA, USA, 2006. [Google Scholar]
- Slifer, Z.M.; Blikslager, A.T. The Integral Role of Tight Junction Proteins in the Repair of Injured Intestinal Epithelium. Int. J. Mol. Sci. 2020, 21, 972. [Google Scholar] [CrossRef] [PubMed]
- Tulkens, J.; Vergauwen, G.; Van Deun, J.; Geeurickx, E.; Dhondt, B.; Lippens, L.; De Scheerder, M.-A.; Miinalainen, I.; Rappu, P.; De Geest, B.G.; et al. Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 2020, 69, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Justino, P.C.; Franco, A.X.; Pontier-Bres, R. Modulation of 5-fluorouracil Activation of toll-like/MyD88/NF-κB/MAPK Pathway by Saccharomyces Boulardii CNCM I-745 Probiotic. Cytokine 2020, 125, 154791. [Google Scholar] [CrossRef] [PubMed]
- Fagarasan, S.; Honjo, T. Intestinal IgA synthesis: Regulation of front-line body defences. Nat. Rev. Immunol. 2020, 3, 63–72. [Google Scholar] [CrossRef]
- Chang, J.E.; Buechler, M.B.; Gressier, E.; Turley, S.J.; Carroll, M.C. Mechanosensing by Peyer’s patch stroma regulates lymphocyte migration and mucosal antibody responses. Nat. Immunol. 2019, 20, 1506–1516. [Google Scholar] [CrossRef]
- McGhee, J.R.; Fujihashi, K. Inside the Mucosal Immune System. PLoS Biol. 2012, 10, e1001397. [Google Scholar] [CrossRef]
- Kanaya, T.; Williams, I.; Ohno, H. Intestinal M Cells: Tireless Samplers of Enteric Microbiota. Traffic (Copenhagen, Denmark) 2020, 21, 617. [Google Scholar] [CrossRef] [PubMed]
- Gebert, A.; Rothkotter, H.J.; Pabst, R. M cells in Peyer’s patches of the intestine. Int. Rev. Cytol. 1996, 167, 91–159. [Google Scholar]
- Beller, A.; Kruglov, A.; Durek, P.; von Goetze, V.; Werner, K.; Heinz, G.; et al. Specific Microbiota Enhances Intestinal IgA Levels by Inducing TGF-β in T Follicular Helper Cells of Peyer’s Patches in Mice. Eur. J. Immunol. 2020, 50, 783–794. [Google Scholar] [CrossRef]
- Ohno, H. Intestinal M cells. J. Biochem. 2016, 159, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Brandtzaeg, P. Secretory IgA: Designed for Anti-Microbial Defense. Front. Immunol. 2013, 4. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of Pathogens and Pathobionts by the Gut Microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Chow, J.; Tang, H.; Mazmanian, S.K. Pathobionts of the Gastrointestinal Microbiota and Inflammatory Disease. Curr. Opin. Immunol. 2011, 23, 473–480. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Rupnik, M.; Wilcox, M.H.; Gerding, D.N. Clostridium difficile infection: New developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 526–536. [Google Scholar] [CrossRef]
- van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; De Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef]
- Pascal, V.; Pozuelo, M.; Borruel, N.; Casellas, F.; Campos, D.; Santiago, A.; Martinez, X.; Varela, E.; Sarrabayrouse, G.; Machiels, K.; et al. A microbial signature for Crohn’s disease. Gut 2017, 66, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Silva, S.; Sabino, J.; Valles-Colomer, M.; Falony, G.; Kathagen, G.; Caenepeel, C.; Cleynen, I.; Van Der Merwe, S.; Vermeire, S.; Raes, J. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat. Microbiol. 2019, 4, 1826–1831. [Google Scholar] [CrossRef]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Avila, M.; Ojcius, D.M.; Yilmaz, O. The Oral Microbiota: Living with a Permanent Guest. DNA Cell Biol. 2009, 28, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.J.; Shah, N.; Valm, A.; Paster, B.; Dewhirst, F.; Inui, T.; Cisar, J.O. Interbacterial Adhesion Networks within Early Oral Biofilms of Single Human Hosts. Appl. Environ. Microbiol. 2017, 83, e00407-17. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.P.V.; Boches, S.K.; Cotton, S.L.; Goodson, J.M.; Kent, R.; Haffajee, A.D.; Socransky, S.S.; Hasturk, H.; Van Dyke, T.E.; Dewhirst, F.; et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 2009, 80, 1421–1432. [Google Scholar] [CrossRef]
- Zarco, M.F.; Vess, T.J.; Ginsburg, G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef] [PubMed]
- de Aquino, S.G.; Abdollahi-Roodsaz, S.; Koenders, M.I.; Van De Loo, F.A.J.; Pruijn, G.J.M.; Marijnissen, R.J.; Walgreen, B.; Helsen, M.M.; Bersselaar, L.A.V.D.; De Molon, R.S.; et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J. Immunol. 2014, 192, 4103–4111. [Google Scholar] [CrossRef] [PubMed]
- Arimatsu, K.; Yamada, H.; Miyazawa, H.; Minagawa, T.; Nakajima, M.; Ryder, M.I.; Gotoh, K.; Motooka, D.; Nakamura, S.; Iida, T.; et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci. Rep. 2014, 4, 4828. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef]
- Craig, S.; Blankenberg, D.; Parodi, A.C.L.; Paul, I.M.; Birch, L.L.; Savage, J.S.; Marini, M.E.; Stokes, J.L.; Nekrutenko, A.; Reimherr, M.L.; et al. Child Weight Gain Trajectories Linked To Oral Microbiota Composition. Sci. Rep. 2018, 8, 14030. [Google Scholar] [CrossRef]
- Rosier, B.T.; De Jager, M.; Zaura, E.; Krom, B.P. Historical and contemporary hypotheses on the development of oral diseases: Are we there yet? Front. Cell Infect. Microbiol. 2014, 4. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Waubant, E.; Chehoud, C.; Kuczynski, J.; DeSantis, T.Z.; Warrington, J.; Venkatesan, A.; Fraser, C.M.; Mowry, E.M. Gut microbiota in multiple sclerosis: Possible influence of immunomodulators. J. Investig. Med. 2015, 63, 729–734. [Google Scholar] [CrossRef]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef]
- Mikuls, T.R.; Payne, J.B.; Yu, F.; Thiele, G.M.; Reynolds, R.J.; Cannon, G.W.; Markt, J.; McGowan, D.; Kerr, G.S.; Redman, R.S.; et al. Periodontitis and Porphyromonas gingivalis in patients with rheumatoid arthritis. Arthritis Rheumatol. 2014, 66, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Oh, S.J.; Ahn, J.S.; Shin, Y.Y.; Yang, J.W.; Kim, H.S. Implication of Porphyromonas gingivalis in colitis and homeostasis of intestinal epithelium. Lab. Anim. Res. 2019, 35, 26. [Google Scholar] [CrossRef]
- Nakajima, M.; Arimatsu, K.; Kato, T.; Matsuda, Y.; Minagawa, T.; Takahashi, N.; Ohno, H.; Yamazaki, K. Oral Administration of P. gingivalis Induces Dysbiosis of Gut Microbiota and Impaired Barrier Function Leading to Dissemination of Enterobacteria to the Liver. PLoS ONE 2015, 10, e0134234. [Google Scholar] [CrossRef] [PubMed]
- Rautava, J.; Pinnell, L.J.; Vong, L.; Akseer, N.; Assa, A.; Sherman, P.M. Oral microbiome composition changes in mouse models of colitis. J. Gastroenterol. Hepatol. 2015, 30, 521–527. [Google Scholar] [CrossRef]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Santos, L. Intestinal microbiota regulates tryptophan metabolism following oral infection with Toxoplasma gondii—Santos—Parasite Immunology—Wiley Online Library. Parasite Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Denson, L.; Vlamakis, H.; Franzosa, E.; Thomas, S.; Gotman, N.M.; Rufo, P.; Baker, S.S.; Sauer, C.; Markowitz, J.; et al. Compositional and Temporal Changes in the Gut Microbiome of Pediatric Ulcerative Colitis Patients Are Linked to Disease Course. Cell Host Microbe 2018, 24, 600–610.e4. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Freire, M.O.; Moustafa, A.; Harkins, D.M.; Torralba, M.G.; Zhang, Y.; Leong, P.; Saffery, R.; Bockmann, M.; Kuelbs, C.; Hughes, T.; et al. Longitudinal Study of Oral Microbiome Variation in Twins. Sci. Rep. 2020, 10, 7954. [Google Scholar] [CrossRef]
- Bailey, M.T.; Dowd, S.E.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef]
- Liu, R.T. The microbiome as a novel paradigm in studying stress and mental health. Am. Psychol. 2017, 72, 655–667. [Google Scholar] [CrossRef]
- Docktor, M.J.; Paster, B.J.; Abramowicz, S.; Ingram, J.; Wang, Y.E.; Correll, M.; Jiang, H.; Cotton, S.L.; Kokaras, A.S.; Bousvaros, A. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 935–942. [Google Scholar] [CrossRef]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 18, 13780–13785. [Google Scholar] [CrossRef]
- Lucas Lopez, R.; Grande Burgos, M.J.; Galvez, A.; Perez Pulido, R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: A state of the science review. Apmis 2017, 125, 3–10. [Google Scholar] [CrossRef]
- Salim, S.Y.; Soderholm, J.D. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflamm. Bowel Dis. 2011, 17, 362–381. [Google Scholar] [CrossRef]
- Hasslöf, P.; Stecksén-Blicks, C. Chapter 10, Probiotic Bacteria and Dental Caries. Fluoride Oral Environ. 2020, 28, 99–107. [Google Scholar]
- Derrien, M.; van Passel, M.W.; van de Bovenkamp, J.H.; Schipper, R.G.; de Vos, W.M.; Dekker, J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 2010, 1, 254–268. [Google Scholar] [CrossRef]
- Fourie, N.H.; Wang, D.; Abey, S.K.; Sherwin, L.B.; Joseph, P.V.; Rahim-Williams, B.; Ferguson, E.G.; Henderson, W.A. The microbiome of the oral mucosa in irritable bowel syndrome. Gut Microbes 2016, 7, 286–301. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, N.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Said, H.; Suda, W.; Nakagome, S.; Chinen, H.; Oshima, K.; Kim, S.; Kimura, R.; Iraha, A.; Ishida, H.; Fujita, J.; et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014, 21, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Yost, S.; Duran-Pinedo, A.E.; Krishnan, K.; Frias-Lopez, J. Potassium is a key signal in host-microbiome dysbiosis in periodontitis. PLoS Pathog. 2017, 13, e1006457. [Google Scholar] [CrossRef] [PubMed]
- Salonen, A.; de Vos, W.M. Impact of Diet on Human Intestinal Microbiota and Health. Annu. Rev. 2014, 5, 239–262. [Google Scholar] [CrossRef]
- Derwa, Y.; Gracie, D.J.; Hamlin, P.J.; Ford, A.C. Systematic review with meta-analysis: The efficacy of probiotics in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2017, 46, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Dickson, I. Gut microbiota: Oral bacteria: A cause of IBD? Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 4–5. [Google Scholar] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; Preter, V.D.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Borchers, A.T.; Selmi, C.; Meyers, F.J.; Keen, C.L.; Gershwin, M.E. Probiotics and immunity. J. Gastroenterol. 2009, 44, 26–46. [Google Scholar] [CrossRef]
- Hill, D.; Sugrue, I.; Tobin, C.; Hill, C.; Stanton, C.; Ross, R.P. The Lactobacillus casei Group: History and Health Related Applications. Front. Microbiol. 2018, 9, 2107. [Google Scholar] [CrossRef]
- Veiga, P.; Gallini, C.A.; Beal, C.; Michaud, M.; Delaney, M.L.; Dubois, A.; Khlebnikov, A.; Vlieg, J.E.V.H.; Punit, S.; Glickman, J.N.; et al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. USA 2010, 107, 18132–18137. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F., 3rd; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015, 29, 1395–1403. [Google Scholar] [CrossRef]
- Ou, Y.; Chen, S.; Ren, F.; Zhang, M.; Ge, S.; Guo, H.; Zhang, H.; Zhao, L. Lactobacillus casei Strain Shirota Alleviates Constipation in Adults by Increasing the Pipecolinic Acid Level in the Gut. Front. Microbiol. 2019, 10, 324. [Google Scholar] [CrossRef]
- Liu, M.-L.; Ding, J.; Zhang, H.; Shen, J.; Hao, Y.; Zhang, X.; Qi, W.; Luo, X.; Zhang, T.; Wang, N. Lactobacillus casei LH23 modulates the immune response and ameliorates DSS-induced colitis via suppressing JNK/p-38 signal pathways and enhancing histone H3K9 acetylation. Food Funct. 2020, 11, 5473–5485. [Google Scholar] [CrossRef] [PubMed]
- Hulston, C.J.; Churnside, A.A.; Venables, M.C. Probiotic supplementation prevents high-fat, overfeeding-induced insulin resistance in human subjects. Br. J. Nutr. 2015, 113, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Haukioja, A. Probiotics and Oral Health. Eur. J. Dent. 2010, 4, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Moman, R.; O’Neill, C.A.; Ledder, R. Mitigation of the Toxic Effects of Periodontal Pathogens by Candidate Probiotics in Oral Keratinocytes, and in an Invertebrate Model. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Shimauchi, H.; Mayanagi, G.; Nakaya, S.; Minamibuchi, M.; Ito, Y.; Yamaki, K.; Hirata, H. Improvement of Periodontal Condition by Probiotics With Lactobacillus Salivarius WB21, A Randomized, Double-Blind, Placebo-Controlled Study. J. Clin. Periodontol. 2008, 35, 897–905. [Google Scholar] [CrossRef]
- Edgar, W. Saliva: Its secretion, composition and functions. Br. Dent. J. 1992, 172, 305–312. [Google Scholar] [CrossRef]
- Ahmadi-Motamayel, F.; Falsafi, P.; Goodarzi, M.; Poorolaja, L. Evaluation of Salivary Catalase, Vitamin C, and Alpha-Amylase in Smokers and Non-Smokers: A Retrospective Cohort Study. J. Oral Pathol. Med. 2017, 46, 377–380. [Google Scholar] [CrossRef]
- Van Nieuw Amerongen, A.; Bolscher, J.; Veerman, E. Salivary Proteins: Protective and Diagnostic Value in Cariology? Caries Res. 2004, 38, 247–253. [Google Scholar] [CrossRef]
- Humphrey, S.; Williamson, R. A Review of Saliva: Normal Composition, Flow, and Function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef]
- Zalewska, A.; Zwierz, K.; Zółkowski, K.; Gindzieński, A. Structure and Biosynthesis of Human Salivary Mucins. Acta Biochim. Pol. 2000, 47, 1067–1079. [Google Scholar] [CrossRef]
- Holsinger, C.F.; Bui, D.T. Anatomy, Function, and Evaluation of the Salivary Glands. In Salivary Gland Disorders; SpringerLink: Berlin/Heidelberg, Germany, 2007; pp. 1–16. [Google Scholar]
- Kaczor-Urbanowicz, K.; Martin Carreras-Presas, C.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D. Saliva Diagnostics—Current Views and Directions. Exp. Biol. Med. (Maywood, NJ) 2017, 242, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.W.; Omaye, S.T. Use of Saliva Biomarkers to Monitor Efficacy of Vitamin C in Exercise-Induced Oxidative Stress. Antioxidants (Basel) 2017, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Gisbert, J.P. Helicobacter Pylori-Associated Diseases. Gastroenterol. Hepatol. 2015, 38 (Suppl. 1), 15–26. [Google Scholar]
- Gholami Parizad, E.; Khosravi, A.; Amraei, M.; Valizadeh, A.; Davoudian, A. Comparing HBV Viral Load in Serum, Cerumen, and Saliva and Correlation With HBeAg Serum Status in Patients With Chronic Hepatitis B Infection. Zahedan J. Res. Med. Sci. 2016, 16. [Google Scholar] [CrossRef]
- Khadse, S.V.; Bajaj, G.; Vibhakar, P.; Nainani, P.; Ahuja, R.; Deep, G. Evaluation of Specificity and Sensitivity of Oral Fluid for Diagnosis of Hepatitis, B.J. Clin. Diagn. Res. 2016, 10, BC12–BC14. [Google Scholar]
- Leon, L.A.A.; De Almeida, A.J.; De Paula, V.S.; Tourinho, R.S.; Villela, D.A.M.; Gaspar, A.M.C.; Lewis-Ximenez, L.L.; Pinto, M.A. Longitudinal Study of Hepatitis A Infection by Saliva Sampling: The Kinetics of HAV Markers in Saliva Revealed the Application of Saliva Tests for Hepatitis A Study. PLoS ONE 2015, 10, e0145454. [Google Scholar]
- Ghosh, M.; Nandi, S.; Dutta, S.; Saha, M. Detection of Hepatitis B Virus Infection: A Systematic Review. World J. Hepatol. 2015, 7, 2482–2491. [Google Scholar] [CrossRef]
- Bonaldo, M.C.; Ribeiro, I.P.; Lima, N.S.; Dos Santos, A.A.C.; Menezes, L.S.R.; Da Cruz, S.O.D.; Da Mello, I.S.; Furtado, N.D.; De Moura, E.E.; Damasceno, L.; et al. Isolation of Infective Zika Virus From Urine and Saliva of Patients in Brazil. PLoS Negl. Trop. Dis. 2016, 10, 045443. [Google Scholar] [CrossRef]
- Weerasekera, M.; Sissons, C.; Wong, L.; Anderson, S.; Holmes, A.; Cannon, R. Use of Denaturing Gradient Gel Electrophoresis for the Identification of Mixed Oral Yeasts in Human Saliva. J. Med. Microbiol. 2013, 62, 319–330. [Google Scholar] [CrossRef]
- Ozbay, Y.; Aydin, S.; Dagli, A.F.; Akbulut, M.; Dagli, N.; Kilic, N.; Rahman, A.; Sahin, I.; Polat, V.; Ozercan, H.I.; et al. Obestatin is present in saliva: Alterations in obestatin and ghrelin levels of saliva and serum in ischemic heart disease. BMB Rep. 2008, 41, 55–61. [Google Scholar] [CrossRef]
- Chatterton, R.; Vogelsong, K.; Lu, Y.; Ellman, A.; Hudgens, G. Salivary Alpha-Amylase as a Measure of Endogenous Adrenergic Activity. Clin. Physiol. (Oxf., Engl.) 1996, 16, 433–448. [Google Scholar] [CrossRef]
- Indira, M.; Chandrashekar, P.; Kattappagari, K.; Chandra, L.; Chitturi, R.; Bv, R. Evaluation of Salivary Glucose, Amylase, and Total Protein in Type 2 Diabetes Mellitus Patients. Indian J. Dent. Res. 2015, 26, 271. [Google Scholar] [PubMed]
- Evans, L.W.; Zhang, F.; Omaye, S.T. Vitamin C Supplementation Reduces Exercise-Induced Oxidative Stress and Increases Peak Muscular Force. Food Nutr. Sci. 2017, 8, 812. [Google Scholar] [CrossRef]
- Owen, R.; Griffiths, J.L.; Ruth, E.; Coxon, K.M.; Philip James, R.A. Chapter Five—Inflammatory Bowel Disease and Targeted Oral Anti-TNFα Therapy: Advances in Protein Chemistry and Structural Biology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, F.; Zhang, Q.; Liu, Y.; You, P.; Sun, S.; Lin, J.; Chen, N. Salivary exosomal PSMA7, A promising biomarker of inflammatory bowel disease. Protein Cell 2017, 8, 686–695. [Google Scholar] [CrossRef]
| Phylum | Genus | Species | Morphology | Reference |
|---|---|---|---|---|
| Proteobacteria | Klebsiella | Klebsiella pneumoniae | Nonmotile, encapsulated, rod-shaped bacilli, facultative anaerobic | [111] |
| Proteobacteria | Enterobacter | Enterobacter spp. | Rod-shaped bacilli, non-spore-forming, motile, facultative anaerobic | [49] |
| Proteobacteria | Neisseria | Neisseria spp. | Lipooligosaccharide (LOS), strict aerobe | [114] |
| Bacteroidetes | Prevotella | Prevotella nigrescens | Nonmotile, rod-shaped bacilli, anaerobic | [115] |
| Fusobacteria | Fusobacterium | Fusobacterium spp. | Non-spore forming, rod-shaped bacilli, potent LPS, anaerobic | [115] |
| Bacteroidetes | Porphyromonas | Porphyromonas gingivalis | Nonmotile, Gram-negative, anaerobic, rod-shaped | [108] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartlett, A.; Gullickson, R.G.; Singh, R.; Ro, S.; Omaye, S.T. The Link between Oral and Gut Microbiota in Inflammatory Bowel Disease and a Synopsis of Potential Salivary Biomarkers. Appl. Sci. 2020, 10, 6421. https://doi.org/10.3390/app10186421
Bartlett A, Gullickson RG, Singh R, Ro S, Omaye ST. The Link between Oral and Gut Microbiota in Inflammatory Bowel Disease and a Synopsis of Potential Salivary Biomarkers. Applied Sciences. 2020; 10(18):6421. https://doi.org/10.3390/app10186421
Chicago/Turabian StyleBartlett, Allison, Robert G. Gullickson, Rajan Singh, Seungil Ro, and Stanley T. Omaye. 2020. "The Link between Oral and Gut Microbiota in Inflammatory Bowel Disease and a Synopsis of Potential Salivary Biomarkers" Applied Sciences 10, no. 18: 6421. https://doi.org/10.3390/app10186421
APA StyleBartlett, A., Gullickson, R. G., Singh, R., Ro, S., & Omaye, S. T. (2020). The Link between Oral and Gut Microbiota in Inflammatory Bowel Disease and a Synopsis of Potential Salivary Biomarkers. Applied Sciences, 10(18), 6421. https://doi.org/10.3390/app10186421