Abstract
Proteins succumb to numerous post-translational modifications (PTMs). These relate to enzymatic or non-enzymatic reactions taking place in either the intracellular or extracellular compartment. While intracellular oxidative changes are mainly due to redox stress, extracellular PTMs may be induced in an inflammatory micro milieu that is rich in reactive species. The increasing recognition of oxidative modifications as a causing agent or side-effect of pathophysiological states and diseases puts oxidative PTMS (oxPTMs) into the spotlight of inflammation research. Pathological hyper-modification of proteins can lead to accumulation, aggregation, cell stress, altered antigenic peptides, and damage-associated molecular pattern (DAMP)-like recognition by host immunity. Such processes are linked to cardiovascular disease and autoinflammation. At the same time, a detailed understanding of the mechanisms governing inflammatory responses to oxPTMs may capitalize on new therapeutic routes for enhancing adaptive immune responses as needed, for instance, in oncology. We here summarize some of the latest developments of oxPTMs in disease diagnosis and therapy. Potential target proteins and upcoming technologies, such as gas plasmas, are outlined for future research that may aid in identifying the molecular basis of immunogenic vs. tolerogenic oxPTMs.
1. Introduction
Proteins are targets for immune cells, and modifications like aggregation are known to link to the pathogenesis of several diseases [1,2]. However, the cause of this protein aggregation is not always clear. Increasing evidence points to the relevance of post-translational modifications (PTMs) caused by not only enzymatic PTMs but also reactive oxygen and nitrogen species (here abbreviated as ROS, as RNS also include oxygen) [3,4]. Under physiological conditions, ROS are essential regulators for maintaining homeostasis and serve as signaling molecules [5]. Interestingly, cells of the innate immune system are major contributors to the inflammation and ROS production in such conditions [6,7]. Excessive inflammation leads to overshooting ROS production, which is often associated with autoimmunity and carcinogenesis [8,9]. The immune system is a double-edged sword in these two processes: In chronic inflammation and autoimmunity, cells of the adaptive immune system are major contributors to self-antigen-related tissue destruction and the production of autoantibodies targeted against the host [10,11]. Nonetheless, T cells directed against oncofetal or aberrant host proteins are a desired event in oncology and pertained in the clinic using checkpoint therapy to augment anti-tumor immunity [12,13]. Checkpoint therapy and other modulating immune effectors address such diseases, as they develop not only through the proteins and their PTMs but also because of the interaction between proteins and cells of the immune system.
Immune cells recognize structures on the cell surface to decide if a cell is healthy or not. The latter leads to immune cell activation and elimination of the infected, cancerogenic, or foreign cell. Not naturally occurring PTMs can mimic a non-self-structure and trigger an immune response, which can have advantages and disadvantages. On the one hand, this leads to an unwanted killing, as evident in neurodegenerative or autoimmune diseases. On the other hand, PTMs are promising as biomarkers for diagnosis and are increasingly recognized as novel agents for the treatment of cancer and allergies.
In this review, we describe various protein modifications that are linked to diseases: (I) the consequence of enzymatically driven changes, (II) the origin of non-enzymatic-induced PTMs and related disorders, (III) the immunogenicity of PTM-bearing epitopes, and (IV) the putative therapeutic use of PTMs. Finally, we outline novel tools for oxidative PTM research that not only rely on the generation of one kind of reactive species but a multitude of reactive species simultaneously, which aids in mimicking the inflammatory environment.
2. Overview of Enzymatic Post-Translational Modifications
Enzymatic PTMs, such as phosphorylation, acetylation, glycosylation, and ubiquitination, regulate protein function and interaction [14,15,16]. Enzymes (kinases, transferases) catalyze the covalent modification of an amino acid side chain and sometimes require co-substrates (e.g., ATP). While some enzymatic PTMs lead to activation or inhibition of intracellular proteins and are critical events in signal transduction [17,18,19,20,21], other enzymatic PTMs are essential for protein stability and regulation. For instance, the tumor suppressor p53 is stabilized by phosphorylation and regulated by an array of PTMs in more than 36 different amino acids [22,23,24]. Enzymatic PTMs transform the p53 protein folding, stability, and function. Although defective regulation leads to misfolding and malfunction of the protein, incorrect folding of p53 alone does not necessarily lead to cell cycle defects and tumor growth. Still, enzymatic PTMs are causative in the dysregulation of kinases, malfunction or misfolding of proteins, and aggregation of proteins that ultimately can cause diseases.
In Alzheimer’s disease (AD), aberrant enzymatic activity promotes hyperphosphorylation of the tubulin-associated unit (tau) protein leading to its oligomerization and aggregation [25]. However, aggregation of amyloid-β is a commonly assumed primary pathological process, as the amyloid hypothesis states that this aggregate promotes tau modification [26,27]. Amyloid-β aggregates enable redox-active metal ions to bind, which catalyzes the production of ROS [28]. The increased amount of intracellular ROS is then self-amplified via mitochondria. Hence, ROS and their subsequent non-enzymatic modifications on proteins causing, for instance, oxidation, chlorination, or nitration are intertwined with enzymatic PTMs processes.
3. Redox Stress and oxPTMs as the Cause of Different Diseases
Intracellular compartments, such as mitochondria, peroxisome, and endoplasmic reticulum, generate endogenous ROS. ROS can be classified in independently existing heavy reactive radicals and non-radical species. Radical species include superoxide (O2°−), oxygen radicals (O2°°), hydroxyl (OH°), alkoxy radical (RO°), peroxyl radical (ROO°), nitric oxide (NO°), and nitrogen dioxide (NO2°). Examples for non-radicals include hydrogen peroxide (H2O2), hypochlorous acid (HOCl), hypobromous acid (HOBr), ozone (O3), singlet oxygen (1O2), and peroxynitrite (ONOOH). ROS interact with intracellularly present proteins and induce modifications resulting in altered protein activity and function [29]. Although a balance of ROS level is sustained via enzymes and antioxidants, increased concentration of ROS is sometimes inevitable and leads to oxidative stress. Several diseases link to redox stress and ROS due to an unbalanced ROS homeostasis, such as diabetes, atherosclerosis, neurodegenerative diseases, macular degeneration, cancer, and others [30,31,32,33]. As a consequence, significant differences in enzyme activity lead to increased oxidative damage and lipid peroxidation [34,35,36,37]. Since many diseases can arise from redox stress, they show a specific fingerprint. Therefore, biomarkers of oxidative stress become increasingly relevant in the diagnosis of neurodegenerative diseases [38]. Oxidative stress, but also hypoxia, inflammation, and mutations in particular proteins, are just a few of the causes that lead to unfolded protein response (UPR). For example, tumor necrosis factor receptor-associated periodic syndrome is caused by UPR, and enhanced production of ROS in monocytes was determined in patients [39,40], suggesting a role for ROS-induced PTMs. For UPR, a somewhat more complex, but not negligible, correlation is shown between proteins and diseases [41,42,43,44]. UPR and redox stress are not only present in AD, atherosclerosis, diabetes, and inflammatory diseases, but also in asthma [45,46]. Evidence shows an increased amount of redox stress inhibitor hypoxia-inducible factor-1 during the development of allergic airway inflammation, and modulation of ROS improves the hyperresponsiveness suggesting ROS as a cause for asthma [47,48]. Since ROS can be scavenged and dampened by antioxidants, a number of therapies based on antioxidants have been postulated.
In analogy to the phosphorylation system, oxidases and reductases regulate thiol groups on proteins for their activation or inactivation as signaling molecules, or transcription factors [5,49]. To control redox-signaling transmission, homeostasis of intracellular ROS levels requires enzymes and other antioxidant proteins [50,51,52]. The most important enzymes for ROS homeostasis are nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, xanthine oxidase, superoxide dismutase (SOD), nitric oxide synthase (NOS), NADPH oxidase (NOX), and myeloperoxidase (MPO) (Figure 1). An imbalance of the mentioned enzymes and their activity can lead to diseases.
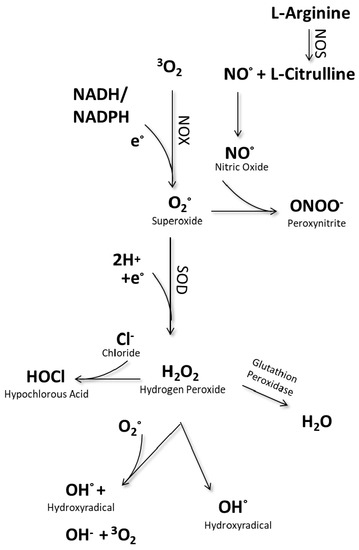
Figure 1.
Intracellular generation of reactive species.
For instance, SOD catalyzes the dismutation of radical superoxide anion (O2−) into hydrogen peroxide (H2O2) or ordinary molecular oxygen (O2). Dysregulation of SOD and total antioxidant capacity leads to oxidative stress and chronic hyperglycemia, as evident in diabetic disease [53,54,55]. Altered SOD levels, oxidative stress, and cytoplasmatic aggregates have also been shown in amyotrophic lateral sclerosis [56,57]. Another relevant protein, myeloperoxidase (MPO), catalyzes the oxidation of chloride ion and coverts thiocyanate to generate isocyanic acid. Isocyanic acid induces immunogenic PTMs as α-carbamylation and homocitrulline (ε-carbamyl-lysine) [58]. Carbamylated proteins accumulate in tissues and are mostly associated with aging [59,60], while citrullinated proteins are related to autoimmune diseases, such as rheumatoid arthritis (RA) (Table 1). The example of RA illustrates the complexity of protein modifications and their consequences: there are different oxPTMs, such as citrullination and glyoxidation, on both the proteins collagen and fibrinogen, and all can activate other immune cells. So far, a causative protein entity has not been identified, and no particular oxidative PTM has been found to induce RA disease. Additionally, RA is characterized by a heavy immuno-infiltrate that may contribute to disease pathogenesis. In RA, but also in other autoimmune disorders, oxidative PTMs play a critical role during development and progress [61]. Recent work increasingly focuses on the immunogenic properties of proteins and peptides that are not only present in the extracellular space as a consequence of tissue injury but also presented on the cell surface.

Table 1.
Protein post-translational modifications (PTMs) linked to the cause of disease.
4. Use of oxPTMs as Disease Biomarkers
Modified structures on the cell surface are characteristic footprints for some diseases and targets to trigger or block an immune response. Therefore, oxPTMs might be advantageous for disease diagnosis. For example, oxidation of low-density lipoprotein (LDL) has been proposed as a primary biomarker for diabetes [74] and cardiovascular diseases [75]. Furthermore, oxidation and chlorination of the extracellular matrix protein laminin were suggested as markers for cardiovascular disease and particularly atherosclerosis [73]. Just as LDL and laminin are anchored in the membrane and are accessible from outside, the major histocompatibility complex (MHC) is located on the cell surface to enable interaction with cells of the adaptive immune system. Each cell degrades intracellular proteins, loads the resulting fragments onto MHC, and presents it on the cell surface. Once a trafficking T cell recognizes the MHC structure, the receptor matches the MHC-bound protein fragment (epitope) as being of self- or non-self-origin. Epitopes of natural intracellular proteins are identified as “self”, and no immune response is triggered.
In contrast, PTMs on epitopes make them differ from their native forms and hence can generate foreign structures, allowing them to become a target of an immune response (Figure 2).
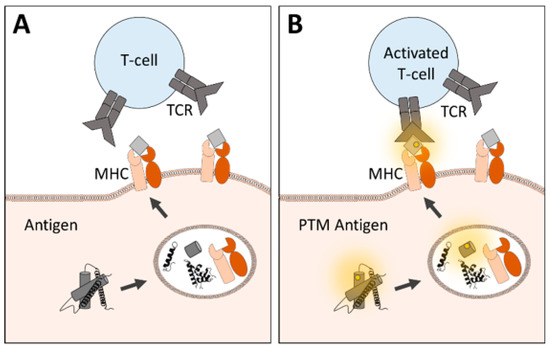
Figure 2.
Immunogenicity of native and modified peptides is presented via major histocompatibility complex (MHC). T cells express antigen-specific receptors (TCR) with a specific binding site for MHC-presented peptides. While (A) native structures may not induce T cell activation, (B) post-translational modified peptides enable an altered binding affinity leading to an immune response.
In some cases, such as cancers, PTMs on epitopes which differ from the current host proteins might even serve to amplify an immune response leading to the eradication of unwanted cells. However, in autoimmune diseases, immune cells falsely attack host structures because PTM epitopes are presented and can lead to a break of tolerance [76], as seen in diabetes type I [77]. Not only in diabetes but also in other autoimmune disorders, PTM epitopes are suspected. Since the direct detection of native and PTM epitopes is difficult due to the isolation and qualification, indirect evidence is usually provided. In RA, T cell activation is the consequence of PTMs, such as carbamylation, citrullination, deamidation, or acetylation [78,79,80,81]. Moreover, B cells release autoantibodies that are specific for PTMs, leading to the attack of host proteins and structures [64,82,83,84]. Targeting those antibodies with neutralizing citrullinated peptides became a pioneering therapy option [63,85], suggesting a role of oxPTMs as therapeutics (Table 2).

Table 2.
Using or targeting protein PTMs to control the disease.
Intracellular proteins with putative PTMs are degraded and loaded onto MHC, where they can serve as an antigenic or tolerogenic peptide. It would, therefore, come as no surprise if epitopes with PTMs were also found in Alzheimer’s patients since oxidatively modified proteins were identified in AD brains by proteomics analysis [95,96]. Since epitopes are structures that are recognized by immune cells, there are different therapeutic approaches to induce effective stimulation or inhibition of their recognition. Not only are endogenous proteins degraded to be presented as peptides via MHC, exogenous proteins are also taken up by antigen-presenting cells, and peptides derived from exogenous proteins also bind to MHC, allowing immune cells to become primed for specific antigens. Thus, vaccination and immunotherapy with oxPTM proteins might be an alternative to antibody therapy.
5. Therapeutic Use of oxPTMs
Although the cellular mechanism and immune cell interactions are partly incompletely understood, increased immunogenicity of oxPTMs has been shown already in model systems of inflammation [97], diabetes [67], and allergy [98]. In such a way, not only PTMs, but also protein stability, adjuvants, or other companion substances (e.g., carrier proteins, nanoparticles) can increase the success of immunomodulatory therapeutics. There is a correlation between protein stability and immunogenicity [99], and PTMs can alter protein structure [14,100]. Thus, PTMs can also act as an adjuvant to improve protective immune response, as was shown for therapeutic vaccination for cancer therapy. Oxidatively modified tumor lysate reduced tumor growth and was more effective than injection with native tumor lysate [93,101,102,103]. This ultimately led to an increase of the anti-tumor T cell receptor repertoire and more durable and clinically apparent anti-tumor effects [94] (Figure 3A). Although tumor lysates consist of a pool of proteins that may mask the immunogenicity of individual proteins, vaccination with tumor-associated proteins alone has so far only generated little success [104,105]. Instead, typical tumor vaccines consist of either peptides, DNA, or mRNA, or are viral/bacterial-based with different advantages and disadvantages [106,107,108,109,110]. Non-enzymatic, oxidative PTM proteins or peptides have not come into focus, despite their expected relevance, as observed in inflammatory disease. By contrast, therapeutic vaccination in AD patients with the modified tau peptide or tau peptide in combination with adjuvants has led to vaccine-induced antibodies potentially targeting tau proteins [89,111,112]. It appears feasible to use such an approach in oncology also.
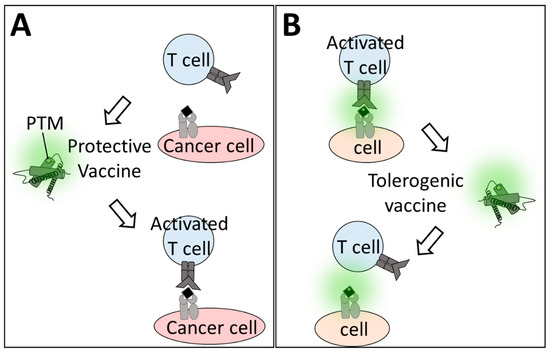
Figure 3.
Modified proteins as therapeutics. Vaccination with oxPTM proteins alters antigen-specific T cell activation. (A) Cancer cells evade immune response due to the expression of “self” antigens, and the oxPTM vaccine enlarges the T cell repertoire to enable T cell activation. (B) In oxPTM-induced diseases, vaccination with modified proteins can dampen by increasing the threshold that is needed for activation.
In allergy, the goal is not to amplify but to dampen antigen-specific immune responses. Antigen-specific immune cell tolerance is achieved via antigen-specific regulatory T cells (Tregs) [113,114] that, hence, are one target of tolerogenic vaccines for treating autoimmune diseases (e.g., diabetes type 1, multiple sclerosis) [91,115,116] (Figure 3B). Enzymatic or non-enzymatic PTMs may accelerate vaccination success since such vaccines might be able to address the T cells specific for PTM-carrying proteins that are not being targeted when using non-modified proteins or peptides. For example, immunotherapy with modified peanut extract reduced allergenicity in peanut-allergic mice [86,87]. Studies with modified allergens in patients are challenging and have not yet been completed [117,118]. Assuming that the modified-allergen vaccine reduces allergenicity, further research should be done to identify the mechanistic basis of immunogenic vs. tolerogenic non-enzymatic protein modifications.
6. Gas Plasma Technology as an Innovative Tool for oxPTM Research
Inflammation generates different types of ROS [119], such as nitric oxide, hypochlorous acid, nitrite, superoxide anion, hydrogen peroxide, peroxynitrite, and nitrite. Chronic inflammation leads to constant ROS production, which is associated with inflammatory disease and autoimmunity [61] as well as carcinogenesis [120]. As outlined above, there is ample evidence that oxPTMs are critical in these processes. Studying oxPTMs, however, is challenging because modeling the multi-ROS environment in inflammatory tissues is limited by chemical restrains. Gas plasma technology overcomes this hurdle by generating a multitude of ROS simultaneously. Using cysteine as a model molecule, we have provided evidence that the oxPTMs created with plasma treatment are the consequences of different reactive species. Moreover, the oxPTM pattern can be controlled by modulating the ROS-output of the plasma systems [121]. Hence, gas plasma technology is an ideal tool for studying the tolerogenic and immunogenic oxPTM patterns on target proteins.
7. Conclusions
Changes in the immunogenicity of proteins based on post-translational modifications can not only cause but also potentially control several diseases. Although PTMs are currently underrepresented in therapeutic schemes in inflammation, autoimmunity, and cancer, evidence points to their pivotal role and significance in disease modulation. In this regard, gas plasma technology might be a potent tool to generate oxPTM patterns on proteins, allowing studies to be done on their tolerogenic or immunogenic nature in many diseases in future research.
Author Contributions
Conceptualization, R.C. and S.B.; methodology, R.C.; writing—original draft preparation, R.C.; writing—review & editing, R.C., and S.B.; supervision, S.B.; funding acquisition, S.B. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge funding by the German Federal Ministry of Education and Research (BMBF), grant number 03Z22DN11.
Acknowledgments
The ZIK plasmatis supported this work at Leibniz Insitute for Plasma Science and Technology Greifswald.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gao, Q. Oxidative Stress and Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 179–198. [Google Scholar] [CrossRef]
- Treweek, T.M.; Meehan, S.; Ecroyd, H.; Carver, J.A. Small heat-shock proteins: Important players in regulating cellular proteostasis. Cell. Mol. Life Sci. CMLS 2015, 72, 429–451. [Google Scholar] [CrossRef]
- Santos, A.L.; Lindner, A.B. Protein Posttranslational Modifications: Roles in Aging and Age-Related Disease. Oxidative Med. Cell. Longev. 2017, 2017, 5716409. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Yao, D.; Shi, Y.; Kabakoff, J.; Wu, W.; Reicher, J.; Ma, Y.; Moosmann, B.; Masliah, E.; Lipton, S.A.; et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol. Neurodegener. 2011, 6, 34. [Google Scholar] [CrossRef]
- Hanschmann, E.M.; Godoy, J.R.; Berndt, C.; Hudemann, C.; Lillig, C.H. Thioredoxins, glutaredoxins, and peroxiredoxins--molecular mechanisms and health significance: From cofactors to antioxidants to redox signaling. Antioxid. Redox Signal. 2013, 19, 1539–1605. [Google Scholar] [CrossRef] [PubMed]
- Obermayer, A.; Stoiber, W.; Grabcanovic-Musija, F.; Studnicka, M. Emerging Evidence Supports the Hypothesis that Neutrophil Extracellular Traps are a Major Factor in Genesis and Progression of Chronic Obstructive Pulmonary Disease. J. Immunol. Sci. 2018, 2, 31–37. [Google Scholar] [CrossRef][Green Version]
- Oishi, Y.; Manabe, I. Macrophages in age-related chronic inflammatory diseases. NPJ Aging Mech. Dis. 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Morgillo, F.; Dallio, M.; Della Corte, C.M.; Gravina, A.G.; Viscardi, G.; Loguercio, C.; Ciardiello, F.; Federico, A. Carcinogenesis as a Result of Multiple Inflammatory and Oxidative Hits: A Comprehensive Review from Tumor Microenvironment to Gut Microbiota. Neoplasia 2018, 20, 721–733. [Google Scholar] [CrossRef]
- Liu, Y.; Meyer, C.; Xu, C.; Weng, H.; Hellerbrand, C.; ten Dijke, P.; Dooley, S. Animal models of chronic liver diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G449–G468. [Google Scholar] [CrossRef] [PubMed]
- Querol, L.; Devaux, J.; Rojas-Garcia, R.; Illa, I. Autoantibodies in chronic inflammatory neuropathies: Diagnostic and therapeutic implications. Nat. Rev. Neurol. 2017, 13, 533–547. [Google Scholar] [CrossRef]
- Pugliese, A. Autoreactive T cells in type 1 diabetes. J. Clin. Investig. 2017, 127, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Kuen, D.S.; Chung, Y. Future prospects of immune checkpoint blockade in cancer: From response prediction to overcoming resistance. Exp. Mol. Med. 2018, 50, 109. [Google Scholar] [CrossRef]
- Torphy, R.J.; Schulick, R.D.; Zhu, Y. Newly Emerging Immune Checkpoints: Promises for Future Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 2642. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.M.; Hogg, P.J. Post-translational control of protein function by disulfide bond cleavage. Antioxid. Redox Signal. 2013, 18, 1987–2015. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Walther, D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 2015, 11, e1004049. [Google Scholar] [CrossRef]
- Khoury, G.A.; Baliban, R.C.; Floudas, C.A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011, 1. [Google Scholar] [CrossRef]
- Deribe, Y.L.; Pawson, T.; Dikic, I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010, 17, 666–672. [Google Scholar] [CrossRef]
- Nguyen, L.K.; Kolch, W.; Kholodenko, B.N. When ubiquitination meets phosphorylation: A systems biology perspective of EGFR/MAPK signalling. Cell Commun. Signal. 2013, 11, 52. [Google Scholar] [CrossRef]
- Biggar, K.K.; Li, S.S. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015, 16, 5–17. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Guan, K.L. Mechanisms of Hippo pathway regulation. Genes. Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef]
- Fang, D.; Hawke, D.; Zheng, Y.; Xia, Y.; Meisenhelder, J.; Nika, H.; Mills, G.B.; Kobayashi, R.; Hunter, T.; Lu, Z. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J. Biol. Chem. 2007, 282, 11221–11229. [Google Scholar] [CrossRef] [PubMed]
- Loughery, J.; Meek, D. Switching on p53: An essential role for protein phosphorylation? Biodiscovery 2013. [Google Scholar] [CrossRef]
- Ashcroft, M.; Kubbutat, M.H.; Vousden, K.H. Regulation of p53 function and stability by phosphorylation. Mol. Cell Biol. 1999, 19, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Kruse, J.P.; Gu, W. Modes of p53 regulation. Cell 2009, 137, 609–622. [Google Scholar] [CrossRef]
- Simic, G.; Babic Leko, M.; Wray, S.; Harrington, C.; Delalle, I.; Jovanov-Milosevic, N.; Bazadona, D.; Buee, L.; de Silva, R.; Di Giovanni, G.; et al. Tau Protein Hyperphosphorylation and Aggregation in Alzheimer’s Disease and Other Tauopathies, and Possible Neuroprotective Strategies. Biomolecules 2016, 6, 6. [Google Scholar] [CrossRef]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef]
- Cheignon, C.; Tomas, M.; Bonnefont-Rousselot, D.; Faller, P.; Hureau, C.; Collin, F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox. Biol. 2018, 14, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Protein oxidation and peroxidation. Biochem. J. 2016, 473, 805–825. [Google Scholar] [CrossRef]
- Forstermann, U.; Xia, N.; Li, H. Roles of Vascular Oxidative Stress and Nitric Oxide in the Pathogenesis of Atherosclerosis. Circ. Res. 2017, 120, 713–735. [Google Scholar] [CrossRef]
- Witherick, J.; Wilkins, A.; Scolding, N.; Kemp, K. Mechanisms of oxidative damage in multiple sclerosis and a cell therapy approach to treatment. Autoimmune Dis. 2010, 2011, 164608. [Google Scholar] [CrossRef] [PubMed]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging. 2018, 13, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, F.; Pupo, G.; Tramutola, A.; Giorgi, A.; Schinina, M.E.; Coccia, R.; Head, E.; Butterfield, D.A.; Perluigi, M. Redox proteomics analysis of HNE-modified proteins in Down syndrome brain: Clues for understanding the development of Alzheimer disease. Free Radic. Biol. Med. 2014, 71, 270–280. [Google Scholar] [CrossRef]
- Barone, E.; Head, E.; Butterfield, D.A.; Perluigi, M. HNE-modified proteins in Down syndrome: Involvement in development of Alzheimer disease neuropathology. Free Radic. Biol. Med. 2017, 111, 262–269. [Google Scholar] [CrossRef]
- McGrath, L.T.; McGleenon, B.M.; Brennan, S.; McColl, D.; Mc, I.S.; Passmore, A.P. Increased oxidative stress in Alzheimer’s disease as assessed with 4-hydroxynonenal but not malondialdehyde. QJM 2001, 94, 485–490. [Google Scholar] [CrossRef]
- Staron, A.; Makosa, G.; Koter-Michalak, M. Oxidative stress in erythrocytes from patients with rheumatoid arthritis. Rheumatol. Int. 2012, 32, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Maciejczyk, M.; Zalewska, A.; Gerreth, A.K. Salivary Redox Biomarkers in Selected Neurodegenerative Diseases. J. Clin. Med. 2020, 9, 497. [Google Scholar] [CrossRef]
- Dickie, L.J.; Aziz, A.M.; Savic, S.; Lucherini, O.M.; Cantarini, L.; Geiler, J.; Wong, C.H.; Coughlan, R.; Lane, T.; Lachmann, H.J.; et al. Involvement of X-box binding protein 1 and reactive oxygen species pathways in the pathogenesis of tumour necrosis factor receptor-associated periodic syndrome. Ann. Rheum. Dis. 2012, 71, 2035–2043. [Google Scholar] [CrossRef]
- Bachetti, T.; Ceccherini, I. Tumor necrosis factor receptor-associated periodic syndrome as a model linking autophagy and inflammation in protein aggregation diseases. J. Mol. Med. 2014, 92, 583–594. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, R.J. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 2002, 110, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Kim, D.I.; Kang, M.R.; Lee, K.S.; Park, S.Y.; Jeong, J.S.; Lee, Y.C. Endoplasmic reticulum stress influences bronchial asthma pathogenesis by modulating nuclear factor kappaB activation. J. Allergy Clin. Immunol. 2013, 132, 1397–1408. [Google Scholar] [CrossRef]
- Pathinayake, P.S.; Hsu, A.C.; Waters, D.W.; Hansbro, P.M.; Wood, L.G.; Wark, P.A.B. Understanding the Unfolded Protein Response in the Pathogenesis of Asthma. Front Immunol. 2018, 9, 175. [Google Scholar] [CrossRef]
- Huerta-Yepez, S.; Baay-Guzman, G.J.; Bebenek, I.G.; Hernandez-Pando, R.; Vega, M.I.; Chi, L.; Riedl, M.; Diaz-Sanchez, D.; Kleerup, E.; Tashkin, D.P.; et al. Hypoxia inducible factor promotes murine allergic airway inflammation and is increased in asthma and rhinitis. Allergy 2011, 66, 909–918. [Google Scholar] [CrossRef]
- Nesi, R.T.; Barroso, M.V.; Souza Muniz, V.; de Arantes, A.C.; Martins, M.A.; Brito Gitirana, L.; Neves, J.S.; Benjamim, C.F.; Lanzetti, M.; Valenca, S.S. Pharmacological modulation of reactive oxygen species (ROS) improves the airway hyperresponsiveness by shifting the Th1 response in allergic inflammation induced by ovalbumin. Free Radic. Res. 2017, 51, 708–722. [Google Scholar] [CrossRef]
- Vara, D.; Watt, J.M.; Fortunato, T.M.; Mellor, H.; Burgess, M.; Wicks, K.; Mace, K.; Reeksting, S.; Lubben, A.; Wheeler-Jones, C.P.D.; et al. Direct Activation of NADPH Oxidase 2 by 2-Deoxyribose-1-Phosphate Triggers Nuclear Factor Kappa B-Dependent Angiogenesis. Antioxid. Redox Signal. 2018, 28, 110–130. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, X.; Cueto, R.; Effi, C.; Zhang, Y.; Tan, H.; Qin, X.; Ji, Y.; Yang, X.; Wang, H. Biochemical basis and metabolic interplay of redox regulation. Redox Biol. 2019, 26, 101284. [Google Scholar] [CrossRef]
- Panieri, E.; Santoro, M.M. ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 2016, 7, e2253. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.; Noonepalle, S.; Lu, Q.; Srivastava, A.; Zemskov, E.; Black, S.M. ROS Signaling in the Pathogenesis of Acute Lung Injury (ALI) and Acute Respiratory Distress Syndrome (ARDS). Adv. Exp. Med. Biol. 2017, 967, 105–137. [Google Scholar] [CrossRef]
- Ganjifrockwala, F.A.; Joseph, J.T.; George, G. Serum total superoxide dismutase enzyme activity in type 2 diabetic patients with retinopathy in Mthatha region of the Eastern Cape Province of South Africa. Biomed. Res-India 2017, 28, 532–538. [Google Scholar]
- Zhao, J.S.; Jin, H.X.; Gao, J.L.; Pu, C.; Zhang, P.; Huang, J.J.; Cheng, L.; Feng, G. Serum Extracellular Superoxide Dismutase Is Associated with Diabetic Retinopathy Stage in Chinese Patients with Type 2 Diabetes Mellitus. Dis. Markers 2018, 2018, 8721379. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.; Rao, A.; Prasad, B.R.; Kumari, S. Serum levels of antioxidants and superoxide dismutase in periodontitis patients with diabetes type 2. J. Indian Soc. Periodontol. 2014, 18, 451–455. [Google Scholar] [CrossRef]
- Cereda, C.; Leoni, E.; Milani, P.; Pansarasa, O.; Mazzini, G.; Guareschi, S.; Alvisi, E.; Ghiroldi, A.; Diamanti, L.; Bernuzzi, S.; et al. Altered intracellular localization of SOD1 in leukocytes from patients with sporadic amyotrophic lateral sclerosis. PLoS ONE 2013, 8, e75916. [Google Scholar] [CrossRef][Green Version]
- Ayers, J.I.; Diamond, J.; Sari, A.; Fromholt, S.; Galaleldeen, A.; Ostrow, L.W.; Glass, J.D.; Hart, P.J.; Borchelt, D.R. Distinct conformers of transmissible misfolded SOD1 distinguish human SOD1-FALS from other forms of familial and sporadic ALS. Acta Neuropathol. 2016, 132, 827–840. [Google Scholar] [CrossRef]
- Jaisson, S.; Pietrement, C.; Gillery, P. Protein Carbamylation: Chemistry, Pathophysiological Involvement, and Biomarkers. Adv. Clin. Chem. 2018, 84, 1–38. [Google Scholar] [CrossRef]
- Gorisse, L.; Pietrement, C.; Vuiblet, V.; Schmelzer, C.E.; Kohler, M.; Duca, L.; Debelle, L.; Fornes, P.; Jaisson, S.; Gillery, P. Protein carbamylation is a hallmark of aging. Proc. Natl. Acad. Sci. USA 2016, 113, 1191–1196. [Google Scholar] [CrossRef]
- Desmons, A.; Okwieka, A.; Doue, M.; Gorisse, L.; Vuiblet, V.; Pietrement, C.; Gillery, P.; Jaisson, S. Proteasome-dependent degradation of intracellular carbamylated proteins. Aging (Albany NY) 2019, 11, 3624–3638. [Google Scholar] [CrossRef]
- Di Dalmazi, G.; Hirshberg, J.; Lyle, D.; Freij, J.B.; Caturegli, P. Reactive oxygen species in organ-specific autoimmunity. Auto Immun. Highlights 2016, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Grimsrud, P.A.; Picklo, M.J., Sr.; Griffin, T.J.; Bernlohr, D.A. Carbonylation of adipose proteins in obesity and insulin resistance: Identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol. Cell Proteom. 2007, 6, 624–637. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Cerqueira, C.; Ossipova, E.; Gunasekera, S.; Hansson, M.; Mathsson, L.; Catrina, A.I.; Sommarin, Y.; Klareskog, L.; Lundberg, K.; Ronnelid, J.; et al. Targeting of anti-citrullinated protein/peptide antibodies in rheumatoid arthritis using peptides mimicking endogenously citrullinated fibrinogen antigens. Arthritis Res. Ther. 2015, 17, 155. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Ge, C.; Lonnblom, E.; Lin, X.; Feng, H.; Xiao, L.; Bai, J.; Ayoglu, B.; Nilsson, P.; Nandakumar, K.S.; et al. The autoantibody response to cyclic citrullinated collagen type II peptides in rheumatoid arthritis. Rheumatology 2019, 58, 1623–1633. [Google Scholar] [CrossRef]
- Pruijn, G.J. Citrullination and carbamylation in the pathophysiology of rheumatoid arthritis. Front Immunol. 2015, 6, 192. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, K.; Barker, K.; Tang, Y.; Kahn, P.; Wiktor, P.; Brunner, A.; Knabben, V.; Takulapalli, B.; Buckner, J.; Nepom, G.; et al. A Contra Capture Protein Array Platform for Studying Post-translationally Modified (PTM) Auto-antigenomes. Mol. Cell Proteom. 2016, 15, 2324–2337. [Google Scholar] [CrossRef]
- Delong, T.; Baker, R.L.; He, J.; Barbour, G.; Bradley, B.; Haskins, K. Diabetogenic T-cell clones recognize an altered peptide of chromogranin A. Diabetes 2012, 61, 3239–3246. [Google Scholar] [CrossRef]
- Rodriguez-Rodriguez, P.; Sandebring-Matton, A.; Merino-Serrais, P.; Parrado-Fernandez, C.; Rabano, A.; Winblad, B.; Avila, J.; Ferrer, I.; Cedazo-Minguez, A. Tau hyperphosphorylation induces oligomeric insulin accumulation and insulin resistance in neurons. Brain 2017, 140, 3269–3285. [Google Scholar] [CrossRef]
- Gratuze, M.; Julien, J.; Petry, F.R.; Morin, F.; Planel, E. Insulin deprivation induces PP2A inhibition and tau hyperphosphorylation in hTau mice, a model of Alzheimer’s disease-like tau pathology. Sci. Rep. 2017, 7, 46359. [Google Scholar] [CrossRef]
- Boullier, A.; Bird, D.A.; Chang, M.K.; Dennis, E.A.; Friedman, P.; Gillotre-Taylor, K.; Horkko, S.; Palinski, W.; Quehenberger, O.; Shaw, P.; et al. Scavenger receptors, oxidized LDL, and atherosclerosis. Ann. N. Y. Acad. Sci. 2001, 947, 214–222; discussion 222–213. [Google Scholar] [CrossRef]
- Kita, T.; Kume, N.; Minami, M.; Hayashida, K.; Murayama, T.; Sano, H.; Moriwaki, H.; Kataoka, H.; Nishi, E.; Horiuchi, H.; et al. Role of oxidized LDL in atherosclerosis. Ann. N. Y. Acad. Sci. 2001, 947, 199–205; discussion 205–196. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Goyal, T.; Mehta, J.L. Oxidized LDL, LOX-1 and atherosclerosis. Cardiovasc. Drugs Ther. 2011, 25, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Nybo, T.; Dieterich, S.; Gamon, L.F.; Chuang, C.Y.; Hammer, A.; Hoefler, G.; Malle, E.; Rogowska-Wrzesinska, A.; Davies, M.J. Chlorination and oxidation of the extracellular matrix protein laminin and basement membrane extracts by hypochlorous acid and myeloperoxidase. Redox. Biol. 2019, 20, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Urbina, A.; Benitez, S.; Perez, A.; Sanchez-Quesada, J.L. Modified low-density lipoproteins as biomarkers in diabetes and metabolic syndrome. Front Biosci. 2018, 23, 1220–1240. [Google Scholar] [CrossRef]
- Trpkovic, A.; Resanovic, I.; Stanimirovic, J.; Radak, D.; Mousa, S.A.; Cenic-Milosevic, D.; Jevremovic, D.; Isenovic, E.R. Oxidized low-density lipoprotein as a biomarker of cardiovascular diseases. Crit. Rev. Clin. Lab. Sci. 2015, 52, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Raposo, B.; Merky, P.; Lundqvist, C.; Yamada, H.; Urbonaviciute, V.; Niaudet, C.; Viljanen, J.; Kihlberg, J.; Kyewski, B.; Ekwall, O.; et al. T cells specific for post-translational modifications escape intrathymic tolerance induction. Nat. Commun. 2018, 9, 353. [Google Scholar] [CrossRef]
- Sidney, J.; Vela, J.L.; Friedrich, D.; Kolla, R.; von Herrath, M.; Wesley, J.D.; Sette, A. Low HLA binding of diabetes-associated CD8+ T-cell epitopes is increased by post translational modifications. BMC Immunol. 2018, 19, 12. [Google Scholar] [CrossRef]
- Marre, M.L.; McGinty, J.W.; Chow, I.T.; DeNicola, M.E.; Beck, N.W.; Kent, S.C.; Powers, A.C.; Bottino, R.; Harlan, D.M.; Greenbaum, C.J.; et al. Modifying Enzymes Are Elicited by ER Stress, Generating Epitopes That Are Selectively Recognized by CD4(+) T Cells in Patients With Type 1 Diabetes. Diabetes 2018, 67, 1356–1368. [Google Scholar] [CrossRef]
- Nguyen, H.; James, E.A. Immune recognition of citrullinated epitopes. Immunology 2016, 149, 131–138. [Google Scholar] [CrossRef]
- Ospelt, C.; Bang, H.; Feist, E.; Camici, G.; Keller, S.; Detert, J.; Kramer, A.; Gay, S.; Ghannam, K.; Burmester, G.R. Carbamylation of vimentin is inducible by smoking and represents an independent autoantigen in rheumatoid arthritis. Ann. Rheum. Dis. 2017, 76, 1176–1183. [Google Scholar] [CrossRef]
- McLaughlin, R.J.; de Haan, A.; Zaldumbide, A.; de Koning, E.J.; de Ru, A.H.; van Veelen, P.A.; van Lummel, M.; Roep, B.O. Human islets and dendritic cells generate post-translationally modified islet autoantigens. Clin. Exp. Immunol. 2016, 185, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, H.; Sehnert, B.; Bockermann, R.; Engstrom, A.; Kalden, J.R.; Holmdahl, R. Humoral immune response to citrullinated collagen type II determinants in early rheumatoid arthritis. Eur. J. Immunol. 2005, 35, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Masson-Bessiere, C.; Sebbag, M.; Girbal-Neuhauser, E.; Nogueira, L.; Vincent, C.; Senshu, T.; Serre, G. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the alpha- and beta-chains of fibrin. J. Immunol. 2001, 166, 4177–4184. [Google Scholar] [CrossRef]
- Vossenaar, E.R.; Despres, N.; Lapointe, E.; van der Heijden, A.; Lora, M.; Senshu, T.; van Venrooij, W.J.; Menard, H.A. Rheumatoid arthritis specific anti-Sa antibodies target citrullinated vimentin. Arthritis Res. Ther. 2004, 6, R142–R150. [Google Scholar] [CrossRef]
- Gunasekera, S.; Fernandes-Cerqueira, C.; Wennmalm, S.; Wahamaa, H.; Sommarin, Y.; Catrina, A.I.; Jakobsson, P.J.; Goransson, U. Stabilized Cyclic Peptides as Scavengers of Autoantibodies: Neutralization of Anticitrullinated Protein/Peptide Antibodies in Rheumatoid Arthritis. ACS Chem. Biol. 2018, 13, 1525–1535. [Google Scholar] [CrossRef]
- van der Kleij, H.P.M.; Warmenhoven, H.J.M.; van Ree, R.; Versteeg, S.A.; Pieters, R.H.H.; Dreskin, S.C.; Knulst, A.C.; van Hoffen, E.; Opstelten, D.J.E.; Koppelman, S.J.; et al. Chemically modified peanut extract shows increased safety while maintaining immunogenicity. Allergy 2019, 74, 986–995. [Google Scholar] [CrossRef]
- Bannon, G.A.; Cockrell, G.; Connaughton, C.; West, C.M.; Helm, R.; Stanley, J.S.; King, N.; Rabjohn, P.; Sampson, H.A.; Burks, A.W. Engineering, characterization and in vitro efficacy of the major peanut allergens for use in immunotherapy. Int. Arch. Allergy Immunol. 2001, 124, 70–72. [Google Scholar] [CrossRef]
- Novak, P.; Schmidt, R.; Kontsekova, E.; Zilka, N.; Kovacech, B.; Skrabana, R.; Vince-Kazmerova, Z.; Katina, S.; Fialova, L.; Prcina, M.; et al. Safety and immunogenicity of the tau vaccine AADvac1 in patients with Alzheimer’s disease: A randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Neurol. 2017, 16, 123–134. [Google Scholar] [CrossRef]
- Kontsekova, E.; Zilka, N.; Kovacech, B.; Novak, P.; Novak, M. First-in-man tau vaccine targeting structural determinants essential for pathological tau-tau interaction reduces tau oligomerisation and neurofibrillary degeneration in an Alzheimer’s disease model. Alzheimers Res. Ther. 2014, 6, 44. [Google Scholar] [CrossRef]
- Orban, T.; Farkas, K.; Jalahej, H.; Kis, J.; Treszl, A.; Falk, B.; Reijonen, H.; Wolfsdorf, J.; Ricker, A.; Matthews, J.B.; et al. Autoantigen-specific regulatory T cells induced in patients with type 1 diabetes mellitus by insulin B-chain immunotherapy. J. Autoimmun. 2010, 34, 408–415. [Google Scholar] [CrossRef]
- Serr, I.; Furst, R.W.; Achenbach, P.; Scherm, M.G.; Gokmen, F.; Haupt, F.; Sedlmeier, E.M.; Knopff, A.; Shultz, L.; Willis, R.A.; et al. Type 1 diabetes vaccine candidates promote human Foxp3(+)Treg induction in humanized mice. Nat. Commun. 2016, 7, 10991. [Google Scholar] [CrossRef] [PubMed]
- Strollo, R.; Ponchel, F.; Malmstrom, V.; Rizzo, P.; Bombardieri, M.; Wenham, C.Y.; Landy, R.; Perret, D.; Watt, F.; Corrigall, V.M.; et al. Autoantibodies to posttranslationally modified type II collagen as potential biomarkers for rheumatoid arthritis. Arthritis Rheum. 2013, 65, 1702–1712. [Google Scholar] [CrossRef]
- Chiang, C.L.; Kandalaft, L.E.; Tanyi, J.; Hagemann, A.R.; Motz, G.T.; Svoronos, N.; Montone, K.; Mantia-Smaldone, G.M.; Smith, L.; Nisenbaum, H.L.; et al. A dendritic cell vaccine pulsed with autologous hypochlorous acid-oxidized ovarian cancer lysate primes effective broad antitumor immunity: From bench to bedside. Clin. Cancer Res. 2013, 19, 4801–4815. [Google Scholar] [CrossRef] [PubMed]
- Tanyi, J.L.; Bobisse, S.; Ophir, E.; Tuyaerts, S.; Roberti, A.; Genolet, R.; Baumgartner, P.; Stevenson, B.J.; Iseli, C.; Dangaj, D.; et al. Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Perluigi, M.; Sultana, R. Oxidative stress in Alzheimer’s disease brain: New insights from redox proteomics. Eur. J. Pharmacol. 2006, 545, 39–50. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Sultana, R.; Poon, H.F. Redox proteomics: A new approach to investigate oxidative stress in Alzheimer’s disease. Neurodegener. Disord. Aging Antioxid. N. Y. Marcel Dekker Inc. 2006. [Google Scholar] [CrossRef]
- Biedron, R.; Konopinski, M.K.; Marcinkiewicz, J.; Jozefowski, S. Oxidation by neutrophils-derived HOCl increases immunogenicity of proteins by converting them into ligands of several endocytic receptors involved in antigen uptake by dendritic cells and macrophages. PLoS ONE 2015, 10, e0123293. [Google Scholar] [CrossRef]
- Marcinkiewicz, J.; Chain, B.M.; Olszowska, E.; Olszowski, S.; Zgliczynski, J.M. Enhancement of Immunogenic Properties of Ovalbumin as a Result of Its Chlorination. Int. J. Biochem. 1991, 23, 1393–1395. [Google Scholar] [CrossRef]
- Scheiblhofer, S.; Laimer, J.; Machado, Y.; Weiss, R.; Thalhamer, J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Exp. Rev. Vaccines 2017, 16, 479–489. [Google Scholar] [CrossRef]
- Xin, F.; Radivojac, P. Post-translational modifications induce significant yet not extreme changes to protein structure. Bioinformatics 2012, 28, 2905–2913. [Google Scholar] [CrossRef]
- Vandenberk, L.; Garg, A.D.; Verschuere, T.; Koks, C.; Belmans, J.; Beullens, M.; Agostinis, P.; De Vleeschouwer, S.; Van Gool, S.W. Irradiation of necrotic cancer cells, employed for pulsing dendritic cells (DCs), potentiates DC vaccine-induced antitumor immunity against high-grade glioma. Oncoimmunology 2016, 5, e1083669. [Google Scholar] [CrossRef]
- Lin, A.; Gorbanev, Y.; De Backer, J.; Van Loenhout, J.; Van Boxem, W.; Lemiere, F.; Cos, P.; Dewilde, S.; Smits, E.; Bogaerts, A. Non-Thermal Plasma as a Unique Delivery System of Short-Lived Reactive Oxygen and Nitrogen Species for Immunogenic Cell Death in Melanoma Cells. Adv. Sci. 2019, 6, 1802062. [Google Scholar] [CrossRef]
- Bekeschus, S.; Clemen, R.; Niessner, F.; Sagwal, S.K.; Freund, E.; Schmidt, A. Medical Gas Plasma Jet Technology Targets Murine Melanoma in an Immunogenic Fashion. Adv. Sci. 2020, 7, 1903438. [Google Scholar] [CrossRef] [PubMed]
- Bowen, W.S.; Svrivastava, A.K.; Batra, L.; Barsoumian, H.; Shirwan, H. Current challenges for cancer vaccine adjuvant development. Exp. Rev. Vaccines 2018, 17, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Finn, O.J. The dawn of vaccines for cancer prevention. Nat. Rev. Immunol. 2018, 18, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Vandermeulen, G.; Preat, V. Cancer DNA vaccines: Current preclinical and clinical developments and future perspectives. J. Exp. Clin. Cancer Res. 2019, 38, 146. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.W.; Tsao, Y.P.; Kung, J.T.; Ding, Y.A.; Sytwu, H.K.; Xiao, X.; Chen, S.L. Recombinant adeno-associated virus expressing human papillomavirus type 16 E7 peptide DNA fused with heat shock protein DNA as a potential vaccine for cervical cancer. J. Virol. 2000, 74, 2888–2894. [Google Scholar] [CrossRef]
- Miao, L.; Li, L.; Huang, Y.; Delcassian, D.; Chahal, J.; Han, J.; Shi, Y.; Sadtler, K.; Gao, W.; Lin, J.; et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019, 37, 1174–1185. [Google Scholar] [CrossRef]
- Hutchins, L.F.; Makhoul, I.; Emanuel, P.D.; Pennisi, A.; Siegel, E.R.; Jousheghany, F.; Guo, X.; Pashov, A.D.; Monzavi-Karbassi, B.; Kieber-Emmons, T. Targeting tumor-associated carbohydrate antigens: A phase I study of a carbohydrate mimetic-peptide vaccine in stage IV breast cancer subjects. Oncotarget 2017, 8, 99161–99178. [Google Scholar] [CrossRef]
- Londhe, V.Y.; Date, V. Personalized neoantigen vaccines: A glimmer of hope for glioblastoma. Exp. Rev. Vaccines 2020, 19, 407–417. [Google Scholar] [CrossRef]
- Novak, P.; Schmidt, R.; Kontsekova, E.; Kovacech, B.; Smolek, T.; Katina, S.; Fialova, L.; Prcina, M.; Parrak, V.; Dal-Bianco, P.; et al. FUNDAMANT: An interventional 72-week phase 1 follow-up study of AADvac1, an active immunotherapy against tau protein pathology in Alzheimer’s disease. Alzheimers Res. Ther. 2018, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Maphis, N.M.; Peabody, J.; Crossey, E.; Jiang, S.; Jamaleddin Ahmad, F.A.; Alvarez, M.; Mansoor, S.K.; Yaney, A.; Yang, Y.; Sillerud, L.O.; et al. Qss Virus-like particle-based vaccine induces robust immunity and protects against tauopathy. NPJ. Vaccines 2019, 4, 26. [Google Scholar] [CrossRef]
- Domogalla, M.P.; Rostan, P.V.; Raker, V.K.; Steinbrink, K. Tolerance through Education: How Tolerogenic Dendritic Cells Shape Immunity. Front Immunol. 2017, 8, 1764. [Google Scholar] [CrossRef] [PubMed]
- Vigario, F.L.; Kuiper, J.; Slutter, B. Tolerogenic vaccines for the treatment of cardiovascular diseases. EBioMedicine 2020, 57, 102827. [Google Scholar] [CrossRef] [PubMed]
- Roep, B.O.; Wheeler, D.C.S.; Peakman, M. Antigen-based immune modulation therapy for type 1 diabetes: The era of precision medicine. Lancet Diabetes Endocrinol. 2019, 7, 65–74. [Google Scholar] [CrossRef]
- Zubizarreta, I.; Florez-Grau, G.; Vila, G.; Cabezon, R.; Espana, C.; Andorra, M.; Saiz, A.; Llufriu, S.; Sepulveda, M.; Sola-Valls, N.; et al. Immune tolerance in multiple sclerosis and neuromyelitis optica with peptide-loaded tolerogenic dendritic cells in a phase 1b trial. Proc. Natl. Acad. Sci. USA 2019, 116, 8463–8470. [Google Scholar] [CrossRef] [PubMed]
- Jongejan, L.; van Ree, R.; Poulsen, L.K. Hypoallergenic molecules for subcutaneous immunotherapy. Exp. Rev. Clin. Immunol. 2016, 12, 5–7. [Google Scholar] [CrossRef]
- Wood, R.A.; Sicherer, S.H.; Burks, A.W.; Grishin, A.; Henning, A.K.; Lindblad, R.; Stablein, D.; Sampson, H.A. A phase 1 study of heat/phenol-killed, E. coli-encapsulated, recombinant modified peanut proteins Ara h 1, Ara h 2, and Ara h 3 (EMP-123) for the treatment of peanut allergy. Allergy 2013, 68, 803–808. [Google Scholar] [CrossRef]
- Blaser, H.; Dostert, C.; Mak, T.W.; Brenner, D. TNF and ROS Crosstalk in Inflammation. Trends Cell Biol. 2016, 26, 249–261. [Google Scholar] [CrossRef]
- El-Kenawi, A.; Ruffell, B. Inflammation, ROS, and Mutagenesis. Cancer Cell 2017, 32, 727–729. [Google Scholar] [CrossRef]
- Lackmann, J.W.; Wende, K.; Verlackt, C.; Golda, J.; Volzke, J.; Kogelheide, F.; Held, J.; Bekeschus, S.; Bogaerts, A.; Schulz-von der Gathen, V.; et al. Chemical fingerprints of cold physical plasmas-an experimental and computational study using cysteine as tracer compound. Sci. Rep. 2018, 8, 7736. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).