Classification of Dermoscopy Skin Lesion Color-Images Using Fractal-Deep Learning Features
Abstract
Featured Application
Abstract
1. Introduction
2. Related Works
3. Background
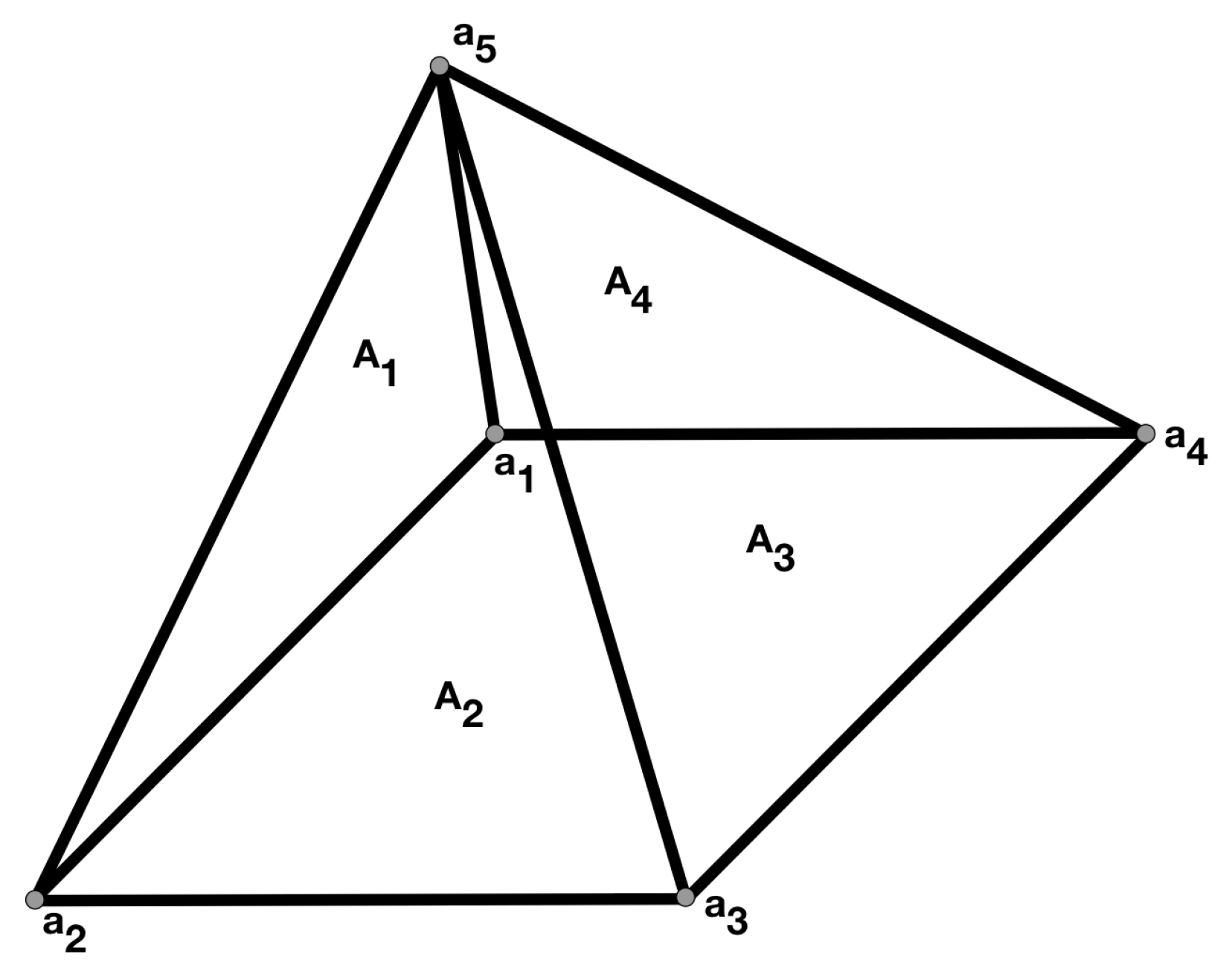
The Fractal Signature of Texture Images
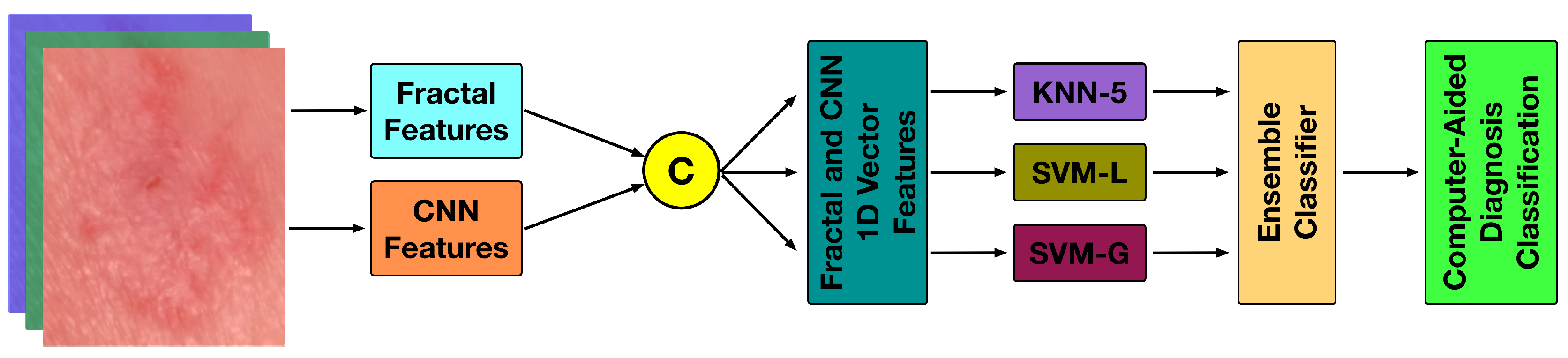
4. The Proposed Methodology
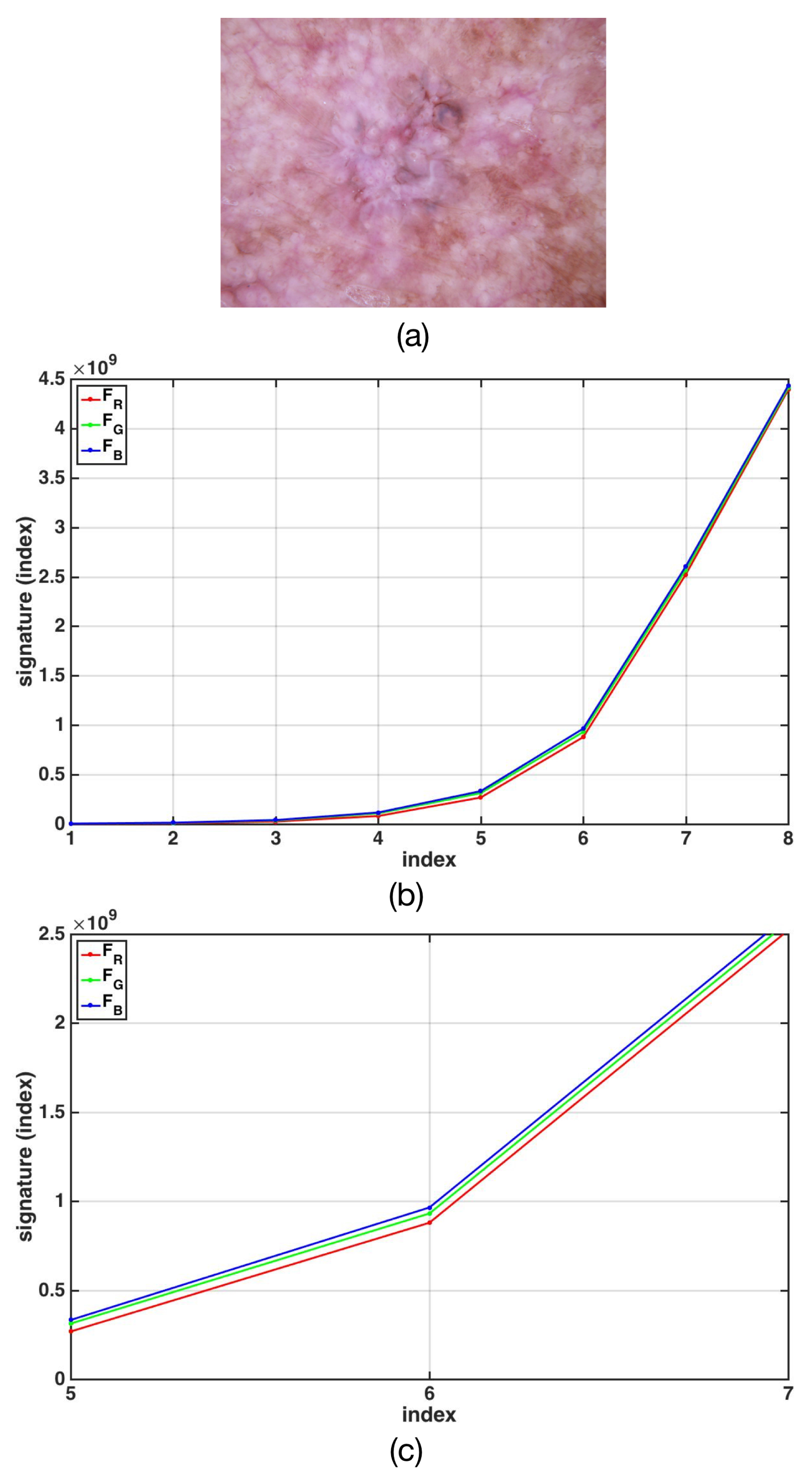
4.1. The Fractal Signature Features
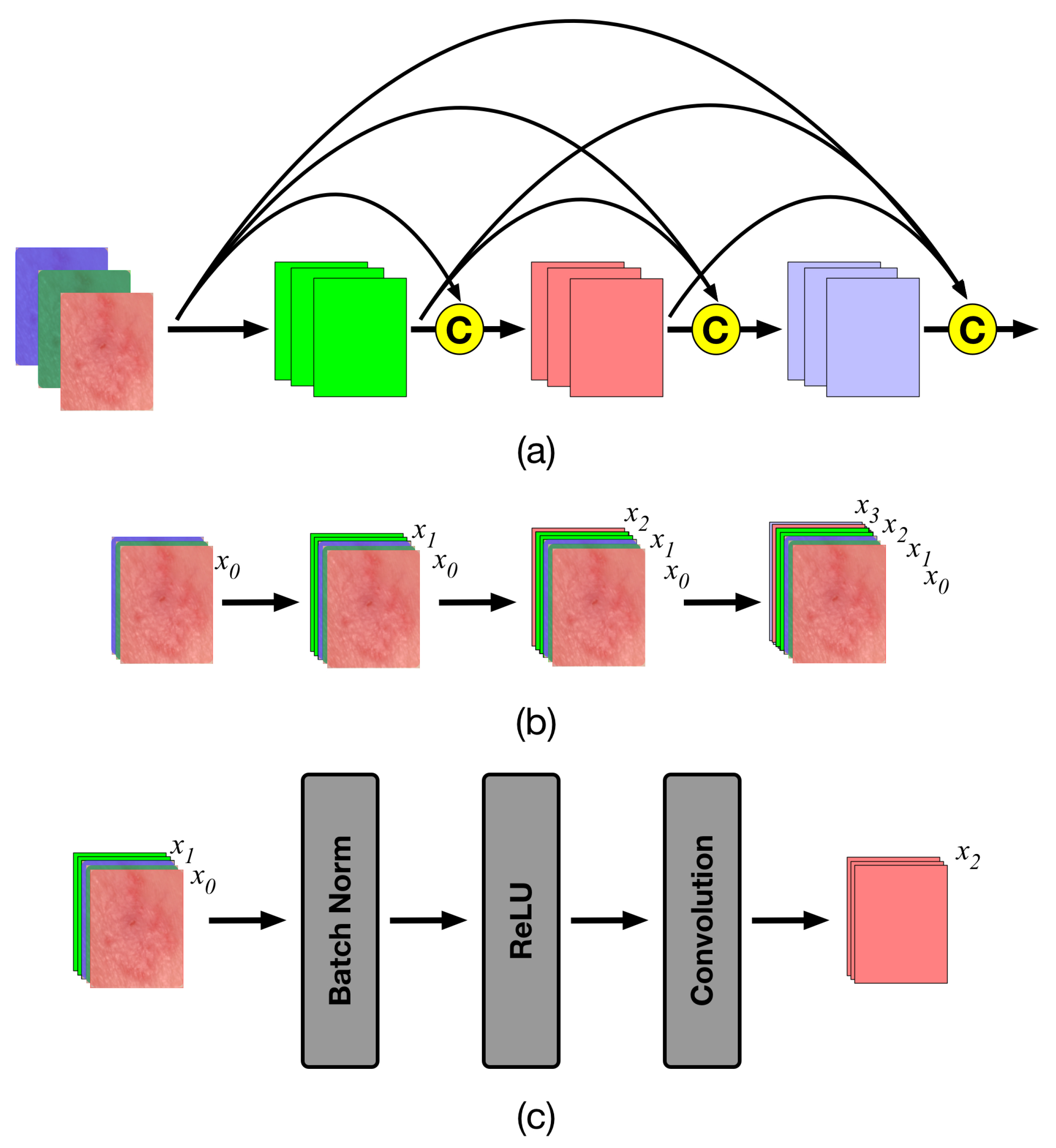
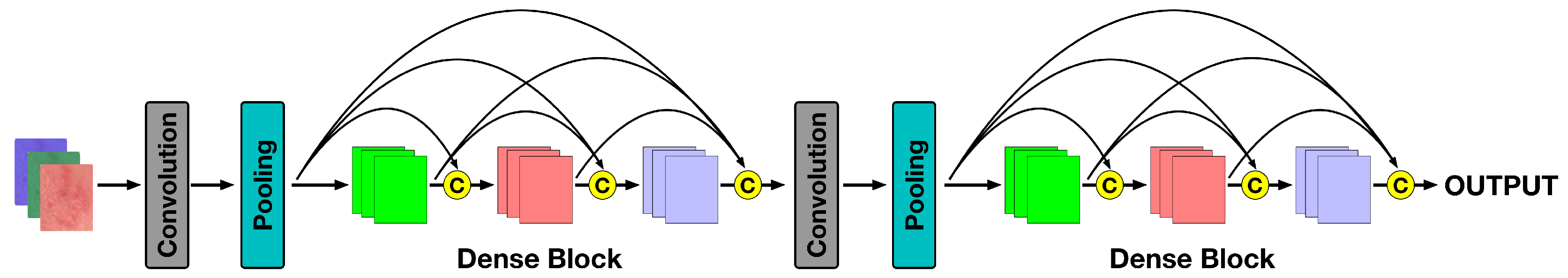
4.2. The Densenet Features
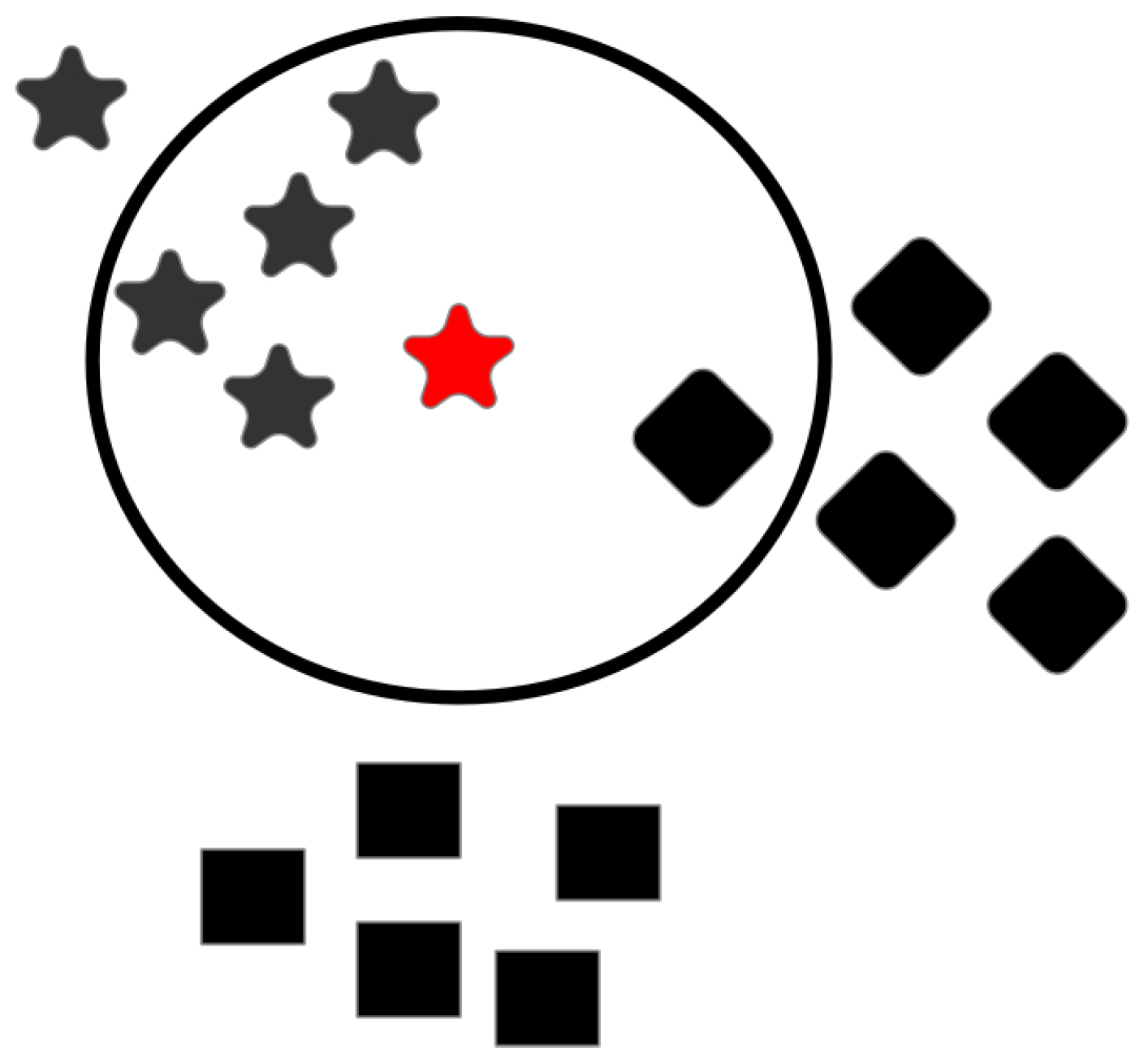
4.3. The Classifier Space
4.3.1. K-Nearest Neighbor
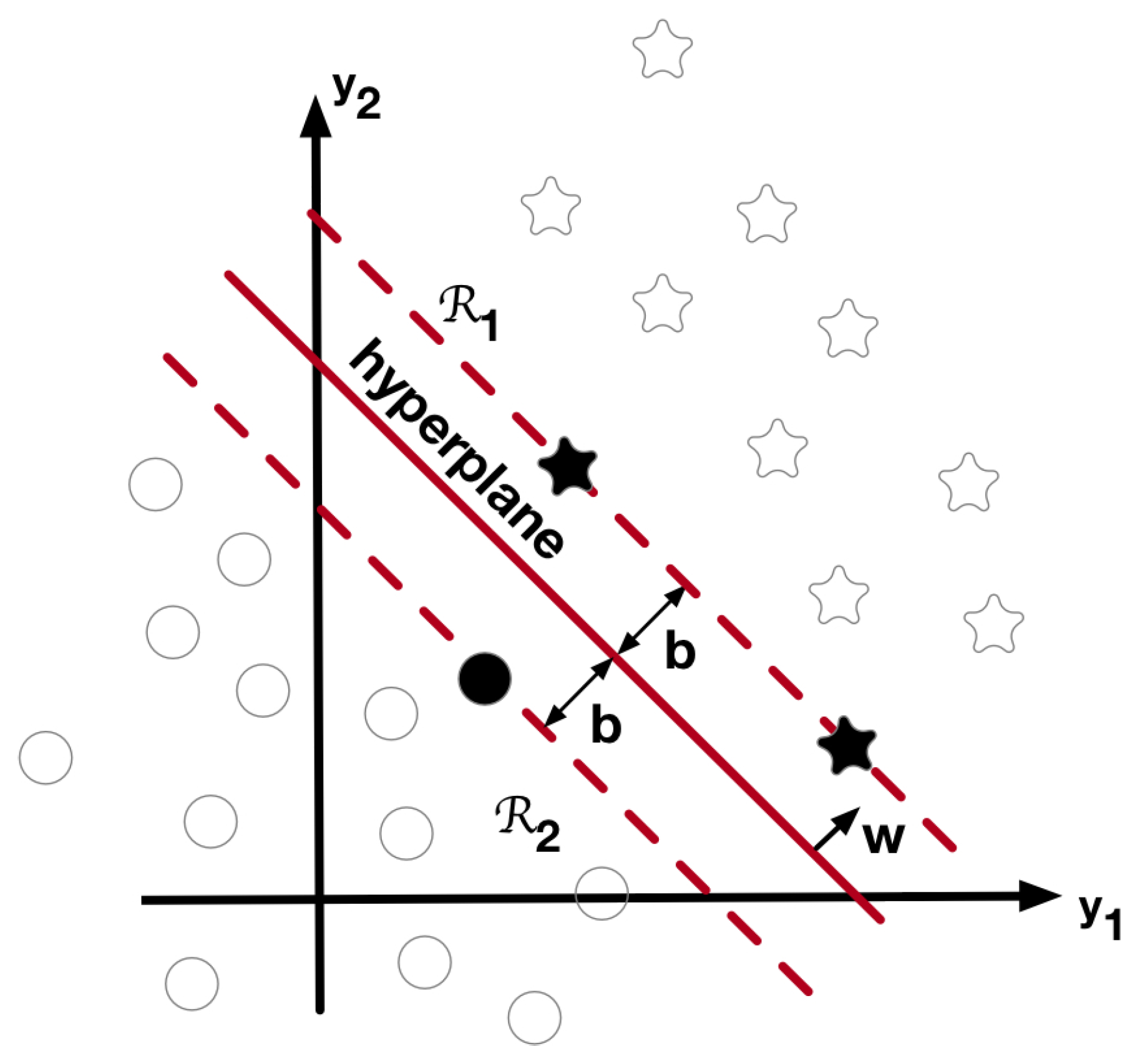
4.3.2. Support Vector Machines
4.4. The Ensemble Classifier
5. The Database
6. Evaluation Metrics
7. Results
8. Comparison with Other Methodologies
9. Discussion
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABCDE | criteria |
| A | asymmetry |
| B | border |
| C | color |
| D | differential structure |
| E | evolving |
| AK | actinic keratosis |
| BCC | basal cell carcinoma |
| IEC | intraepithelial carcinoma |
| MEL | malignant melanoma |
| SCC | squamous cell carcinoma |
| 1D fractal signature of the k-th channel | |
| 1D GLRL signature of the k-th channel | |
| energy of the 1D signature | |
| KNN | K-nearest neighbor |
| KNN-5 | 5-nearest neighbor |
| SVM | support vector machines |
| SVM-L | SVM with a linear kernel |
| SVM-G | SVM with a Gaussian kernel |
| CNN | convolutional neural network |
| TP | true positive |
| TN | true negative |
| FP | false positive |
| FN | false negative |
References
- Fitzpatrick, B.T. The validity and practicality of sun-reactive skin types I through VI. Arch. Dermatol. 1988, 124, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Department of Health. National Skin Cancer Prevention Plan 2019–2022. Available online: https://www.gov.ie/en/publication/4655d6-national-skin-cancer-prevention-plan-2019-2022/ (accessed on 27 May 2019).
- American Cancer Society. Skin Cancer. Available online: https://www.cancer.org/cancer/skin-cancer.html (accessed on 27 May 2019).
- Hay, R.J.; Johns, N.E.; Williams, H.C.; Bolliger, I.W.; Dellavalle, R.P.; Margolis, D.J.; Marks, R.; Naldi, L.; Weinstock, M.A.; Wulf, S.K.; et al. The Global burden of skin disease in 2010: An analysis of the prevalence and impact of skin conditions. J. Investig. Dermatol. 2014, 134, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.A.; Bonilla-Hernández, J.D.; Castillo, D.E.; Carrasquilla-Sotomayor, M.; Alvis-Zakzuk, N.J. Characterization of skin cancer in a dermatologic center in Colombia. J. Am. Acad. Dermatol. 2018, 79, AB70. [Google Scholar]
- Nachbar, F.; Stolz, W.; Merkle, T.; Cognetta, A.B.; Vogt, T.L.; Thaler, M.; Bilek, P.; Braun-Falco, O.; Plewig, G. The ABCD rule of dermatoscopy: High prospective value in the diagnosis of doubtful melanocytic skin lesions. J. Am. Acad. Dermatol. 1994, 30, 551–559. [Google Scholar] [CrossRef]
- Folland, G.B. Fourier Analysis and Its Applications; American Mathematical Society: Providence, RI, USA, 2000; pp. 314–318. [Google Scholar]
- Al-masni, M.A.; Al-antari, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Meth. Prog. Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Haralick, R.M. Statistical and Structural Approaches to Texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Sun, C.; Wee, W.G. Neighboring gray level dependence matrix for texture classification. Comput. Vis. Graph. Image Process. 1983, 23, 341–352. [Google Scholar] [CrossRef]
- Amadasun, M.; King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Chu, A.; Sehgal, C.; Greenleaf, J. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit. Lett. 1990, 11, 415–419. [Google Scholar] [CrossRef]
- Dasarathy, B.V.; Holder, E.B. Image characterizations based on joint gray level-run length distributions. Pattern Recognit. Lett. 1991, 12, 497–502. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikäinen, M.; Harwood, D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Tang, X. Texture Information in run-length matrices. IEEE Trans. Image Process. 1998, 7, 1602–1609. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, X.; Zhou, J.; You, J. Robust texture image representation by scale selective local binary patterns. IEEE Trans. Image Process. 2016, 25, 687–699. [Google Scholar] [CrossRef]
- Liu, L.; Lao, S.; Fieguth, P.W.; Guo, Y.; Wang, X.; Pietikäinen, M. Median Robust Extended Local Binary Pattern for Texture Classification. IEEE Trans. Image Process. 2016, 25, 1368–1381. [Google Scholar] [CrossRef]
- Dash, S.; Senapati, M.R. Gray level run length matrix based on various illumination normalization techniques for texture classification. Evol. Intell. 2018, 1–10. [Google Scholar] [CrossRef]
- Al-antari, M.A.; Al-Masni, M.A.; Park, S.; Park, J.; Metwally, M.K.; Kadah, Y.M.; Han, S.M.; Kim, T.S. An Automatic Computer-Aided Diagnosis System for Breast Cancer in Digital Mammograms via Deep Belief Network. J. Med. Biol. Eng. 2018, 38, 443–456. [Google Scholar] [CrossRef]
- Al-antari, M.A.; Han, S.M.; Kim, T.S. Evaluation of deep learning detection and classification towards computer-aided diagnosis of breast lesions in digital X-ray mammograms. Comput. Meth. Prog. Biomed. 2020, 196, 105584. [Google Scholar] [CrossRef]
- Arivazhagan, S.; Ganesan, L.; Priyal, S.P. Texture classification using Gabor wavelets based rotation invariant features. Pattern Recognit. Lett. 2006, 27, 1976–1982. [Google Scholar] [CrossRef]
- Regniers, O.; Bombrun, L.; Lafon, V.; Germain, C. Supervised Classification of Very High Resolution Optical Images Using Wavelet-Based Textural Features. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3722–3735. [Google Scholar] [CrossRef]
- Guerra-Rosas, E.; Álvarez-Borrego, J. Methodology for diagnosis of skin cancer on images of dermatologic spots by spectral analysis. Biomed. Opt. Express 2015, 10, 3876–3891. [Google Scholar] [CrossRef] [PubMed]
- Depeursinge, A.; Püspöki, Z.; Ward, J.P.; Unser, M. Steerable wavelet machines (SWM): Learning moving frames for texture classification. IEEE Trans. Image Process. 2017, 26, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Rosas, E.; Álvarez-Borrego, J.; Angulo-Molina, A. Identification of melanoma cells: A method based in mean variance of signatures via spectral densities. Biomed. Opt. Express 2017, 4, 2185–2194. [Google Scholar] [CrossRef]
- Pardo, A.; Gutiérrez-Gutiérrez, J.A.; Lihacova, I.; López-Higuera, J.M.; Conde, O.M. On the spectral signature of melanoma: A non-parametric classification framework for cancer detection in hyperspectral imaging of melanocytic lesions. Biomed. Opt. Express 2018, 12, 6283–6301. [Google Scholar] [CrossRef]
- Emerson, C.W.; Lam, N.S.N.; Quattrochi, D.A. Multi-scale fractal analysis of image texture and pattern. Photogramm. Eng. Remote Sens. 1999, 65, 51–61. [Google Scholar]
- Lam, N.S.N.; Qiu, H.L.; Quattrochi, D.A.; Emerson, C.W. An Evaluation of Fractal Methods for Characterizing Image Complexity. Cartogr. Geogr. Inf. Sci. 2002, 29, 25–35. [Google Scholar] [CrossRef]
- Chen, X.W.; Zeng, X.; van Alphen, D. Multi-class feature selection for texture classification. Pattern Recogn. Lett. 2006, 27, 1685–1691. [Google Scholar] [CrossRef]
- Ashour, M.W.; Khalid, F.; Halin, A.B.; Darwish, S.H. Multi-class support vector machines for texture classification using gray-level histogram and edge detection features. IJAECS 2016, 3, 1–5. [Google Scholar]
- Garnavi, R.; Aldeen, M.; Bailey, J. Computer-aided diganosis of Melanoma using border- and wavelet-based texture analysis. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 1239–1252. [Google Scholar] [CrossRef]
- Barata, C.; Ruela, M.; Francisco, M.; Mendoza, T.; Marques, J.S. Two systems for the detection of melanomas in dermoscopy images using texture and color features. IEEE Syst. J. 2014, 8, 965–979. [Google Scholar] [CrossRef]
- Amelard, R.; Glaister, J.; Wong, A.; Clausi, D. High-level intuitive Features (HLIFs) for intuitive skin lesion description. IEEE Trans. Biomed. Eng. 2015, 62, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Spyridonos, P.; Gaitanis, G.; Likas, A.; Bassukas, I. Automatic discrimination of actinic keratoses from clinical photographs. Comput. Biol. Med. 2017, 88, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Q.; Hussain, A.; Rehman, S.U.; Khan, U.; Maqsood, M.; Mehmood, K.; Khan, M.A. Classification of melanoma and nevus in digital images for diagnosis of skin cancer. IEEE Access 2019, 7, 90132–90144. [Google Scholar] [CrossRef]
- Albahar, M.A. Skin lesion classification using convolutional neural network with novel regularizer. IEEE Access 2019, 7, 38306–38313. [Google Scholar] [CrossRef]
- Marka, A.; Carter, J.B.; Toto, E.; Hassanpour, S. Automated detection of nonmelanoma skin cancer using digital images: A systematic review. BMC Med. Imaging 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Shimizu, K.; Iyatomi, H.; Celebi, M.E.; Norton, K.A.; Tanaka, M. Four-class classification of skin lesions with task decomposition strategy. IEEE Trans. Biomed. Eng. 2015, 62, 274–283. [Google Scholar] [CrossRef]
- Wahba, M.A.; Ashour, A.S.; Guo, Y.; Napoleon, S.A.; Abd-Elnaby, M.M. A novel cumulative level difference mean based GLDM and modified ABCD features ranked using eigenvector centrality approach for four skin lesion types classification. Comput. Meth. Prog. Biomed. 2018, 165, 163–174. [Google Scholar] [CrossRef]
- Wu, Z.; Zhao, S.; Peng, Y.; He, X.; Zhao, X.; Huang, K.; Wu, X.; Fan, W.; Li, F.; Chen, M.; et al. Studies on different CNN algorithms for face skin disease classification based on clinical images. IEEE Access 2019, 7, 66505–66511. [Google Scholar] [CrossRef]
- Garza-Flores, E.; Guerra-Rosas, E.; Álvarez-Borrego, J. Spectral indexes obtained by implementation of the fractional Fourier and Hermite transform for the diagnosis of malignant melanoma. Biomed. Opt. Express 2019, 10, 6043–6056. [Google Scholar] [CrossRef]
- Gessert, N.; Nielsen, M.; Shaikh, M.; Werner, R.; Schlaefer, A. Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data. MethodsX 2020, 1, 1008641-8. [Google Scholar] [CrossRef] [PubMed]
- Bajwa, M.N.; Muta, K.; Malik, M.I.; Siddiqui, S.A.; Braun, S.A.; Homey, B.; Dengel, A.; Ahmed, S. Computer-aided diagnosis of skin diseases using deep neural networks. Appl. Sci. 2020, 10, 2488. [Google Scholar] [CrossRef]
- Barnsley, M.F.; Devaney, R.L.; Mandelbrot, B.B.; Peitgen, H.O.; Saupe, D.; Voss, R.F. The Science of Fractal Images; Springer: New York, NY, USA, 1998; pp. 22–70. [Google Scholar]
- Edgar, G. Measure, Topology, and Fractal Geometry, 2nd ed.; Springer: New York, NY, USA, 2000; pp. 165–224. [Google Scholar]
- Backes, A.R.; Casanova, D.; Bruno, M.O. Plant leaf identification based on volumetric fractal dimension. Int. J. Pattern Recog. 2009, 23, 1145–1160. [Google Scholar] [CrossRef]
- Florindo, J.B.; Bruno, O.M.; Landini, G. Morphological classification of odontogenic keratocyst using Bouligand-Minkowski fractal descriptors. Comput. Biol. Med. 2017, 81, 1–10. [Google Scholar] [CrossRef]
- Florindo, J.B.; Bruno, O.M. Fractal Descriptors of Texture Images Based on the Triangular Prism Dimension. J. Math. Imaging Vis. 2019, 61, 140–159. [Google Scholar]
- Silveira, M.; Nascimento, J.C.; Marques, J.S.; Marçal, A.R.S.; Mendonça, T.; Yamauchi, S.; Maeda, J.; Rozeira, J. Comparison of segmentation methods for melanoma diagnosis in dermoscopy images. IEEE J. Sel. Top. Signal Process. 2009, 3, 35–45. [Google Scholar] [CrossRef]
- Celebi, M.E.; Wen, Q.; Iyatomi, H.; Shimizu, K.; Zhou, H.; Schaefer, G. Dermoscopy Image Analysis; Chapter A State-of-the-Art Survey on Lesion Border Detection in Dermoscopy Images; Celebi, M.E., Mendonca, T., Marques, J.S., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 97–129. [Google Scholar]
- Deep, R. Probability and Statistics; Elsevier Academic Press: Cambridge, MA, USA, 2006; pp. 19–22, 104, 287. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Gonzalez, R.C.; Wood, R.E.; Eddins, S.L. Digital Image Processing with MATLAB; Tata McGraw-Hill: New Delhi, India, 2010; pp. 237–240, 253–259. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006; pp. 67–357. [Google Scholar]
- Rogers, S.; Girolami, M. A First Course in Machine Learning; CRC Press: Boca Raton, FL, USA, 2011; pp. 169–205. [Google Scholar]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification, 2nd ed.; Wiley-Interscience: New York, NY, USA, 2001; pp. 20–513. [Google Scholar]
- Galar, M.; Fernández, A.; Barrenechea, E.; Bustince, H.; Herrera, F. A review on ensembles for the class imbalance problem: Bagging-, boosting-, and hybrid-based approaches. IEEE Trans. Syst. Man Cybern. C 2012, 42, 463–484. [Google Scholar] [CrossRef]
- Salunkhe, U.R.; Mali, S.N. Classifier ensemble design for imbalance data classification: A hybrid approach. Procedia Comput. Sci. 2016, 85, 725–732. [Google Scholar] [CrossRef]
- Li, Y.; Guo, H.; Liu, X.; Li, Y.; Li, J. Adapted ensemble classification algorithm based on multiple classifier system and feature selection for classifying multi-class imbalanced data. Knowl.-Based Syst. 2016, 94, 88–104. [Google Scholar] [CrossRef]
- Wang, Q.; Luo, Z.; Huang, J.; Feng, Y.; Liu, Z. A Novel Ensemble Method for Imbalanced Data Learning: Bagging of Extrapolation-SMOTE SVM. Comput. Intel. Neurosc. 2017, 2017, 1827016. [Google Scholar] [CrossRef]
- Leon, F.; Floria, S.; Badica, C. Evaluating the effect of voting methods on ensemble-based classification. In Proceedings of the 2017 IEEE International Conference on INnovations in Intelligent SysTems and Applications (INISTA), Gdynia, Poland, 3–5 July 2017; pp. 1–6. [Google Scholar]
- Wang, H.; Shen, Y.; Wang, S.; Xiao, T.; Deng, L.; Wang, X.; Zhao, X. Ensemble of 3D densely connected convolutional network for diagnosis of mild cognitive imparient and Alzheimer’s disease. Neurocomputing 2019, 333, 145–156. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Meth. Prog. Biomed. 2020, 193, 105475. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Kittler, H. Data Descriptor: The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.; Dusza, S.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), hosted by the International Skin Imaging Collaboration (ISIC). In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- MacKenzie-Wood, A.R.; Milton, G.W.; Launey, J.W. Melanoma: Accuracy of clinical diagnosis. Australas. J. Dermat. 1998, 39, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Ackerman, A.B. How accurate are dermatologists in the diagnosis of melanoma? Degree of accuracy and implications. Arch. Dermatol. 1992, 128, 559–560. [Google Scholar] [CrossRef]
- Lindelöf, B.; Hedblad, M.A. Accuracy in the clinical diagnosis and pattern of malignant melanoma at a dermatological clinic. J. Dermatol. 1994, 21, 461–464. [Google Scholar] [CrossRef]
- Grin, C.M.; Kopf, A.W.; Welkovich, B.; Bart, R.S.; Levenstein, M.J. Accuracy in the clinical diagnosis of malignant melanoma. Arch. Dermatol. 1990, 126, 763–766. [Google Scholar] [CrossRef]
- Anderson, A.M.; Matsumoto, M.; Saul, M.I.; Secrest, A.M.; Ferris, L.K. Accuracy of skin cancer diagnosis by physician assistants compared with dermatologists in a large health care system. JAMA Dermatol. 2018, 154, 569–573. [Google Scholar] [CrossRef]
| Class Name | Class Abbrevation | Number of Images |
|---|---|---|
| actinic keratosis | AK | 867 |
| basal cell carcinoma | BCC | 3323 |
| squamous cell carcinoma | SCC | 628 |
| benign keratosis | BKL | 2624 |
| melanoma | MEL | 4522 |
| melanocytic nevus | NEV | |
| dermatofibroma | DER | 239 |
| vascular lesion | VASC | 253 |
| Total | 25,331 |
| Training Dataset | ||||||||
|---|---|---|---|---|---|---|---|---|
| AK | BCC | SCC | BKL | MEL | NEV | DER | VASC | |
| 867 | 3323 | 628 | 2624 | 4522 | 12,875 | 239 | 253 | |
| AK | 662 | 91 | 3 | 16 | 5 | 90 | 0 | 0 |
| BCC | 22 | 3203 | 0 | 13 | 14 | 70 | 1 | 0 |
| SCC | 34 | 224 | 276 | 11 | 2 | 81 | 0 | 0 |
| BKL | 27 | 144 | 4 | 2105 | 39 | 305 | 0 | 0 |
| MEL | 37 | 111 | 5 | 85 | 3597 | 687 | 0 | 0 |
| NEV | 14 | 96 | 1 | 40 | 65 | 12,659 | 0 | 0 |
| DER | 3 | 54 | 3 | 10 | 14 | 108 | 47 | 0 |
| VASC | 3 | 33 | 3 | 5 | 10 | 105 | 0 | 94 |
| Exp-1 response | 802 | 3956 | 295 | 2285 | 3746 | 14,105 | 48 | 94 |
| Class | Training Dataset | Exp-1 Response | TP | FP | FN | TN |
|---|---|---|---|---|---|---|
| AK | 867 | 802 | 662 | 140 | 205 | 24,324 |
| BCC | 3323 | 3956 | 3203 | 753 | 120 | 21,255 |
| SCC | 628 | 295 | 276 | 19 | 352 | 24,684 |
| BKL | 2624 | 2285 | 2105 | 180 | 519 | 22,527 |
| MEL | 4522 | 3746 | 3597 | 149 | 925 | 20,660 |
| NEV | 12,875 | 14,105 | 12,659 | 1446 | 216 | 11,010 |
| DER | 239 | 48 | 47 | 1 | 192 | 25,091 |
| VASC | 253 | 94 | 94 | 0 | 159 | 25,078 |
| Class | (%) | (%) | (%) | (%) |
|---|---|---|---|---|
| AK | 98.64 | 82.54 | 76.36 | 99.43 |
| BCC | 96.55 | 80.97 | 96.39 | 96.58 |
| SCC | 98.54 | 93.56 | 43.95 | 99.92 |
| BKL | 97.24 | 92.12 | 80.22 | 99.21 |
| MEL | 95.76 | 96.02 | 79.54 | 99.28 |
| NEV | 93.44 | 89.75 | 98.32 | 88.39 |
| DER | 99.24 | 97.92 | 19.67 | 100.00 |
| VASC | 99.37 | 100.00 | 37.15 | 100.0 |
| Mean ± SD | 97.35 ± 2.04 | 91.61 ± 6.89 | 66.45 ± 29.09 | 97.85 ± 3.98 |
| Class | (%) | (%) | (%) | (%) |
|---|---|---|---|---|
| AK | 98.62 | 82.68 | 76.47 | 99.42 |
| BCC | 96.78 | 82.63 | 96.21 | 96.88 |
| SCC | 98.55 | 94.98 | 45.22 | 99.94 |
| BKL | 97.22 | 92.78 | 79.88 | 99.27 |
| MEL | 95.73 | 96.53 | 79.39 | 99.27 |
| NEV | 94.16 | 91.12 | 98.31 | 89.69 |
| Mean ± SD | 96.85 ± 1.71 | 90.12 ± 6.07 | 79.25 ± 19.06 | 97.42 ± 3.93 |
| Class | (%) | (%) | (%) | (%) |
|---|---|---|---|---|
| AK | 98.87 ± 0.07 | 85.78 ± 0.81 | 80.21 ± 1.80 | 99.54 ± 0.02 |
| BCC | 96.56 ± 0.13 | 81.00 ± 0.44 | 96.62 ± 0.29 | 96.55 ± 0.12 |
| SCC | 98.45 ± 0.05 | 92.94 ± 1.35 | 40.43 ± 2.05 | 99.92 ± 0.02 |
| BKL | 97.36 ± 0.07 | 92.92 ± 0.49 | 80.72 ± 0.65 | 99.29 ± 0.05 |
| MEL | 95.58 ± 0.10 | 95.54 ± 0.13 | 78.92 ± 0.60 | 99.20 ± 0.02 |
| NEV | 93.14 ± 0.08 | 89.34 ± 0.09 | 98.21 ± 0.12 | 87.91 ± 0.07 |
| DER | 99.16 ± 0.03 | 98.00 ± 2.75 | 10.87 ± 1.67 | 100.00 ± 0.00 |
| VASC | 99.33 ± 0.01 | 99.69 ± 0.70 | 29.16 ± 385 | 100.00 ± 0.00 |
| Mean ± SD | 97.31 ± 2.14 | 91.90 ± 6.28 | 64.39 ± 32.93 | 97.80 ± 4.15 |
| Class | (%) | (%) | (%) | (%) |
|---|---|---|---|---|
| AK | 95.61 ± 0.26 | 38.81 ± 2.15 | 47.56 ± 2.79 | 97.33 ± 0.13 |
| BCC | 89.48 ± 0.28 | 57.72 ± 1.29 | 69.07 ± 0.63 | 92.51 ± 0.28 |
| SCC | 97.19 ± 0.13 | 34.48 ± 4.10 | 13.84 ± 1.19 | 99.32 ± 0.11 |
| BKL | 90.30 ± 0.21 | 55.55 ± 3.59 | 28.97 ± 1.90 | 97.34 ± 0.29 |
| MEL | 86.85 ± 0.18 | 81.83 ± 2.11 | 33.84 ± 0.59 | 98.37 ± 0.19 |
| NEV | 80.06 ± 0.55 | 73.36 ± 0.65 | 95.58 ± 0.39 | 63.95 ± 0.86 |
| DER | 99.07 ± 0.07 | 90.00 ± 22.36 | 3.08 ± 1.95 | 99.99 ± 0.01 |
| VASC | 99.06 ± 0.07 | 98.57 ± 3.14 | 15.49 ± 1.70 | 99.99 ± 0.01 |
| Mean ± SD | 92.20 ± 6.74 | 66.30 ± 23.46 | 38.43 ± 31.12 | 93.60 ± 12.22 |
| Class | (%) | (%) | (%) | (%) |
|---|---|---|---|---|
| AK | 98.87 ± 0.08 | 85.88 ± 1.56 | 80.52 ± 1.21 | 99.52 ± 0.07 |
| BCC | 96.83 ± 0.04 | 82.93 ± 0.14 | 96.29 ± 0.23 | 96.91 ± 0.05 |
| SCC | 98.48 ± 0.05 | 94.68 ± 1.86 | 41.87 ± 1.05 | 99.93 ± 0.02 |
| BKL | 97.28 ± 0.04 | 92.88 ± 0.28 | 80.55 ± 0.53 | 99.27 ± 0.03 |
| MEL | 95.55 ± 0.07 | 96.21 ± 0.30 | 78.69 ± 0.67 | 99.31 ± 0.06 |
| NEV | 93.92 ± 0.19 | 90.83 ± 0.29 | 98.14 ± 0.09 | 89.39 ± 0.37 |
| Mean ± SD | 96.82 ± 1.85 | 90.57 ± 5.19 | 79.35 ± 20.24 | 97.39 ± 4.06 |
| Class | (%) | (%) | (%) | (%) |
|---|---|---|---|---|
| AK | 95.38 ± 0.30 | 38.25 ± 3.57 | 45.15 ± 1.91 | 97.26 ± 0.40 |
| BCC | 89.66 ± 0.21 | 59.15 ± 0.30 | 68.38 ± 0.83 | 92.87 ± 0.14 |
| SCC | 97.23 ± 0.23 | 40.12 ± 6.49 | 16.26 ± 3.14 | 99.36 ± 0.11 |
| BKL | 90.39 ± 0.16 | 58.13 ± 1.94 | 29.27 ± 0.80 | 97.53 ± 0.20 |
| MEL | 86.43 ± 0.28 | 81.66 ± 1.43 | 32.62 ± 1.15 | 98.37 ± 0.14 |
| NEV | 80.59 ± 0.45 | 74.40 ± 0.40 | 95.68 ± 0.28 | 64.17 ± 0.83 |
| Mean ± SD | 89.95 ± 6.05 | 58.62 ± 17.54 | 47.89 ± 29.30 | 91.60 ± 13.62 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina-Molina, E.O.; Solorza-Calderón, S.; Álvarez-Borrego, J. Classification of Dermoscopy Skin Lesion Color-Images Using Fractal-Deep Learning Features. Appl. Sci. 2020, 10, 5954. https://doi.org/10.3390/app10175954
Molina-Molina EO, Solorza-Calderón S, Álvarez-Borrego J. Classification of Dermoscopy Skin Lesion Color-Images Using Fractal-Deep Learning Features. Applied Sciences. 2020; 10(17):5954. https://doi.org/10.3390/app10175954
Chicago/Turabian StyleMolina-Molina, Edgar Omar, Selene Solorza-Calderón, and Josué Álvarez-Borrego. 2020. "Classification of Dermoscopy Skin Lesion Color-Images Using Fractal-Deep Learning Features" Applied Sciences 10, no. 17: 5954. https://doi.org/10.3390/app10175954
APA StyleMolina-Molina, E. O., Solorza-Calderón, S., & Álvarez-Borrego, J. (2020). Classification of Dermoscopy Skin Lesion Color-Images Using Fractal-Deep Learning Features. Applied Sciences, 10(17), 5954. https://doi.org/10.3390/app10175954





